

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147712.15 + phase: 0 /pseudo/partial
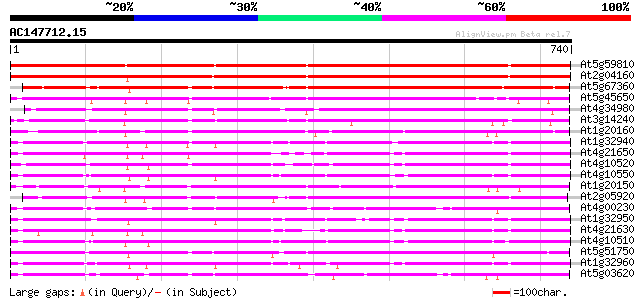
(740 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g59810 subtilisin-like protease - like protein 805 0.0
At2g04160 subtilisin-like serine protease AIR3 796 0.0
At5g67360 cucumisin-like serine protease (gb|AAC18851.1) 564 e-161
At5g45650 subtilisin-like protease 550 e-156
At4g34980 subtilisin proteinase - like 531 e-151
At3g14240 unknown protein 528 e-150
At1g20160 unknown protein 500 e-141
At1g32940 unknown protein 499 e-141
At4g21650 subtilisin proteinase like protein 498 e-141
At4g10520 putative subtilisin-like protease 494 e-140
At4g10550 subtilisin-like protease -like protein 492 e-139
At1g20150 unknown protein 487 e-138
At2g05920 serine protease like protein 485 e-137
At4g00230 subtilisin-type serine endopeptidase XSP1 484 e-137
At1g32950 putative serine proteinase (At1g32950) 484 e-137
At4g21630 serine protease - like protein 483 e-136
At4g10510 putative subtilisin-like protease 483 e-136
At5g51750 serine protease-like protein 482 e-136
At1g32960 unknown protein 481 e-136
At5g03620 cucumisin precursor -like protein 479 e-135
>At5g59810 subtilisin-like protease - like protein
Length = 778
Score = 805 bits (2080), Expect = 0.0
Identities = 409/741 (55%), Positives = 519/741 (69%), Gaps = 8/741 (1%)
Query: 1 SYIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVL 60
SYIVY+GSH+H P S++ L SH L S +GSHE AKEAIFYSY +HINGFAA+L
Sbjct: 41 SYIVYLGSHAHLPQISSAHLDGVAHSHRTFLASFVGSHENAKEAIFYSYKRHINGFAAIL 100
Query: 61 EVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTII 120
+ EAA+IAKHP+VVSVF NKG +L TT SW F+ L N GVV K S+W K YGE TII
Sbjct: 101 DENEAAEIAKHPDVVSVFPNKGRKLHTTHSWNFMLLAKN-GVVHKSSLWNKAGYGEDTII 159
Query: 121 ANIDSGVSPESKSFSDDGMGPVPSRWRGICQLDNFHCNRKLIGARFYSQGYESKFGRLNQ 180
AN+D+GV PESKSFSD+G G VP+RW+G C D CNRKLIGAR++++GY + G +
Sbjct: 160 ANLDTGVWPESKSFSDEGYGAVPARWKGRCHKD-VPCNRKLIGARYFNKGYLAYTGLPSN 218
Query: 181 SLYNA-RDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI*I 239
+ Y RD GHG+ TLS A GNFV GANVFG+ NGTA GGSP++ VAAYKVCW
Sbjct: 219 ASYETCRDHDGHGSHTLSTAAGNFVPGANVFGIGNGTASGGSPKARVAAYKVCWPPVDGA 278
Query: 240 ECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNS 299
EC DADI+ A E AI DGVD++S S+G + ++ DGI+IG+FHA++NGV VV GNS
Sbjct: 279 ECFDADILAAIEAAIEDGVDVLSASVGGDAG-DYMSDGIAIGSFHAVKNGVTVVCSAGNS 337
Query: 300 GPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEKFYSLVSSVD 359
GPK GTV+NVAPW+ +V AS++DR F ++++L + GTSLS LP EK YSL+S+ D
Sbjct: 338 GPKSGTVSNVAPWVITVGASSMDREFQAFVELKNGQSFKGTSLSKPLPEEKMYSLISAAD 397
Query: 360 AKVGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLGNDK 419
A V N + DA +CK GSLDP KVKGKIL CL R + V +A + G+ G+VL NDK
Sbjct: 398 ANVANGNVTDALLCKKGSLDPKKVKGKILVCL-RGDNARVDKGMQAAAAGAAGMVLCNDK 456
Query: 420 QRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSS 479
GN+I++ AH+LP S I+Y DGE + SY+ +TK P Y+ + KPAP +AS SS
Sbjct: 457 ASGNEIISDAHVLPASQIDYKDGETLFSYLSSTKDPKGYIKAPTATLNTKPAPFMASFSS 516
Query: 480 RGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAI 539
RGPN I P ILKPDITAPGV+I+ A+ A PT L SDN+ P+N SGTS+SCPH+S +
Sbjct: 517 RGPNTITPGILKPDITAPGVNIIAAFTEATGPTDLDSDNRRTPFNTESGTSMSCPHISGV 576
Query: 540 VALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKEDATPFGYGAGHIQPELAMDP 599
V LLKT++P+WSPAA +SAIMTT+ + N +P+ D+S + A PF YG+GH+QP A P
Sbjct: 577 VGLLKTLHPHWSPAAIRSAIMTTSRTRNNRRKPMVDESFKKANPFSYGSGHVQPNKAAHP 636
Query: 600 GLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKP-YICPKSYNMLDFNYPSITVPNLGKHFV 658
GLVYDL DYL+FLCA GYN T +++F+ P Y C + N+LDFNYPSITVPNL
Sbjct: 637 GLVYDLTTGDYLDFLCAVGYNNTVVQLFAEDPQYTCRQGANLLDFNYPSITVPNLTGSIT 696
Query: 659 QEVTRTVTNVGSPGTYRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTKPTSSGY 718
VTR + NVG P TY + EP G+ V ++P+ LTFN+ GE K F++ + T SGY
Sbjct: 697 --VTRKLKNVGPPATYNARFREPLGVRVSVEPKQLTFNKTGEVKIFQMTLRPLPVTPSGY 754
Query: 719 VFGHLLWSDGRHKVMSPLVVK 739
VFG L W+D H V SP+VV+
Sbjct: 755 VFGELTWTDSHHYVRSPIVVQ 775
>At2g04160 subtilisin-like serine protease AIR3
Length = 772
Score = 796 bits (2055), Expect = 0.0
Identities = 406/745 (54%), Positives = 524/745 (69%), Gaps = 10/745 (1%)
Query: 1 SYIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVL 60
SY+VY G+HSH + + ++HY+ LGS GS E+A +AIFYSY KHINGFAA L
Sbjct: 31 SYVVYFGAHSHVGEITEDAMDRVKETHYDFLGSFTGSRERATDAIFYSYTKHINGFAAHL 90
Query: 61 EVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTII 120
+ + A +I+KHP VVSVF NK +L TTRSW+FLGLE+N VP SIW K R+GE TII
Sbjct: 91 DHDLAYEISKHPEVVSVFPNKALKLHTTRSWDFLGLEHN-SYVPSSSIWRKARFGEDTII 149
Query: 121 ANIDSGVSPESKSFSDDGMGPVPSRWRGICQLDN---FHCNRKLIGARFYSQGYESKFGR 177
AN+D+GV PESKSF D+G+GP+PSRW+GICQ FHCNRKLIGAR++++GY + G
Sbjct: 150 ANLDTGVWPESKSFRDEGLGPIPSRWKGICQNQKDATFHCNRKLIGARYFNKGYAAAVGH 209
Query: 178 LNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI 237
LN S + RD+ GHG+ TLS A G+FV G ++FG NGTAKGGSPR+ VAAYKVCW
Sbjct: 210 LNSSFDSPRDLDGHGSHTLSTAAGDFVPGVSIFGQGNGTAKGGSPRARVAAYKVCWPPVK 269
Query: 238 *IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGG 297
EC DAD++ AF+ AI DG D+IS SLG P FF D ++IG+FHA + ++VV G
Sbjct: 270 GNECYDADVLAAFDAAIHDGADVISVSLGG-EPTSFFNDSVAIGSFHAAKKRIVVVCSAG 328
Query: 298 NSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLS-TGLPNEKFYSLVS 356
NSGP TV+NVAPW +V AST+DR F S L LG+ G SLS T LP+ KFY +++
Sbjct: 329 NSGPADSTVSNVAPWQITVGASTMDREFASNLVLGNGKHYKGQSLSSTALPHAKFYPIMA 388
Query: 357 SVDAKVGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLG 416
SV+AK NA+ DA++CK+GSLDP K KGKIL CL R +G V GG IG+VL
Sbjct: 389 SVNAKAKNASALDAQLCKLGSLDPIKTKGKILVCL-RGQNGRVEKGRAVALGGGIGMVLE 447
Query: 417 NDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIAS 476
N GND++A H+LP + + D V YI TK P+A++T ++T++G+KPAPV+AS
Sbjct: 448 NTYVTGNDLLADPHVLPATQLTSKDSFAVSRYISQTKKPIAHITPSRTDLGLKPAPVMAS 507
Query: 477 LSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHV 536
SS+GP+ + P ILKPDITAPGV ++ AY GA+SPT D + + +N SGTS+SCPH+
Sbjct: 508 FSSKGPSIVAPQILKPDITAPGVSVIAAYTGAVSPTNEQFDPRRLLFNAISGTSMSCPHI 567
Query: 537 SAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKEDATPFGYGAGHIQPELA 596
S I LLKT YP+WSPAA +SAIMTT TI + PI++ + ATPF +GAGH+QP LA
Sbjct: 568 SGIAGLLKTRYPSWSPAAIRSAIMTTATIMDDIPGPIQNATNMKATPFSFGAGHVQPNLA 627
Query: 597 MDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYICPK-SYNMLDFNYPSITVPNLGK 655
++PGLVYDL I DYLNFLC+ GYN +Q+ +FS + C ++++ NYPSITVPNL
Sbjct: 628 VNPGLVYDLGIKDYLNFLCSLGYNASQISVFSGNNFTCSSPKISLVNLNYPSITVPNLTS 687
Query: 656 HFVQEVTRTVTNVGSPGTYRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTK-PT 714
V V+RTV NVG P Y V+VN P G++V +KP SL F +VGE+KTFK+I +K
Sbjct: 688 SKV-TVSRTVKNVGRPSMYTVKVNNPQGVYVAVKPTSLNFTKVGEQKTFKVILVKSKGNV 746
Query: 715 SSGYVFGHLLWSDGRHKVMSPLVVK 739
+ GYVFG L+WSD +H+V SP+VVK
Sbjct: 747 AKGYVFGELVWSDKKHRVRSPIVVK 771
>At5g67360 cucumisin-like serine protease (gb|AAC18851.1)
Length = 757
Score = 564 bits (1453), Expect = e-161
Identities = 320/735 (43%), Positives = 457/735 (61%), Gaps = 33/735 (4%)
Query: 18 SDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLEVEEAAKIAKHPNVVSV 77
S + S+ D H N S L S + E + Y+Y I+GF+ L EEA + P V+SV
Sbjct: 39 SQMPSSFDLHSNWYDSSLRSISDSAE-LLYTYENAIHGFSTRLTQEEADSLMTQPGVISV 97
Query: 78 FENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIANIDSGVSPESKSFSDD 137
+EL TTR+ FLGL+ + D E G Y + ++ +D+GV PESKS+SD+
Sbjct: 98 LPEHRYELHTTRTPLFLGLDEHTA----DLFPEAGSYSD-VVVGVLDTGVWPESKSYSDE 152
Query: 138 GMGPVPSRWRGICQLD-NFH---CNRKLIGARFYSQGYESKFGRLNQSLYNA--RDVLGH 191
G GP+PS W+G C+ NF CNRKLIGARF+++GYES G +++S + RD GH
Sbjct: 153 GFGPIPSSWKGGCEAGTNFTASLCNRKLIGARFFARGYESTMGPIDESKESRSPRDDDGH 212
Query: 192 GTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI*IECTDADIMQAFE 251
GT T S A G+ V GA++ G A+GTA+G +PR+ VA YKVCWLG C +DI+ A +
Sbjct: 213 GTHTSSTAAGSVVEGASLLGYASGTARGMAPRARVAVYKVCWLGG----CFSSDILAAID 268
Query: 252 DAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNSGPKFGTVTNVAP 311
AI+D V+++S SLG +++ DG++IGAF A+E G++V GN+GP +++NVAP
Sbjct: 269 KAIADNVNVLSMSLGG-GMSDYYRDGVAIGAFAAMERGILVSCSAGNAGPSSSSLSNVAP 327
Query: 312 WLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTG--LPNEKFYSLVSSVDAKVGNATIED 369
W+ +V A T+DR+F + LG+ G SL G LP +K + + +A NAT +
Sbjct: 328 WITTVGAGTLDRDFPALAILGNGKNFTGVSLFKGEALP-DKLLPFIYAGNAS--NAT--N 382
Query: 370 AKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLGNDKQRGNDIMAYA 429
+C G+L P KVKGKI+ C R ++ V + + G +G++L N G +++A A
Sbjct: 383 GNLCMTGTLIPEKVKGKIVMCD-RGINARVQKGDVVKAAGGVGMILANTAANGEELVADA 441
Query: 430 HLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPII 489
HLLP + + G+ + Y+ P A ++ T VGVKP+PV+A+ SSRGPN I P I
Sbjct: 442 HLLPATTVGEKAGDIIRHYVTTDPNPTASISILGTVVGVKPSPVVAAFSSRGPNSITPNI 501
Query: 490 LKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPN 549
LKPD+ APGV+IL A+ GA PTGLASD++ + +NI SGTS+SCPHVS + ALLK+++P
Sbjct: 502 LKPDLIAPGVNILAAWTGAAGPTGLASDSRRVEFNIISGTSMSCPHVSGLAALLKSVHPE 561
Query: 550 WSPAAFKSAIMTTTTIQGNNHRPIKD-QSKEDATPFGYGAGHIQPELAMDPGLVYDLNIV 608
WSPAA +SA+MTT + +P+ D + + +TPF +GAGH+ P A +PGL+YDL
Sbjct: 562 WSPAAIRSALMTTAYKTYKDGKPLLDIATGKPSTPFDHGAGHVSPTTATNPGLIYDLTTE 621
Query: 609 DYLNFLCAHGYNQTQMKMFSRKPYIC--PKSYNMLDFNYPSITVPNLGKHFVQEVTRTVT 666
DYL FLCA Y Q++ SR+ Y C KSY++ D NYPS V N+ + TRTVT
Sbjct: 622 DYLGFLCALNYTSPQIRSVSRRNYTCDPSKSYSVADLNYPSFAV-NVDGVGAYKYTRTVT 680
Query: 667 NVGSPGTYRVQV-NEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKV--TKPTSSGYVFGHL 723
+VG GTY V+V +E G+ + ++P L F E EKK++ + F V +KP+ S FG +
Sbjct: 681 SVGGAGTYSVKVTSETTGVKISVEPAVLNFKEANEKKSYTVTFTVDSSKPSGSN-SFGSI 739
Query: 724 LWSDGRHKVMSPLVV 738
WSDG+H V SP+ +
Sbjct: 740 EWSDGKHVVGSPVAI 754
>At5g45650 subtilisin-like protease
Length = 791
Score = 550 bits (1416), Expect = e-156
Identities = 326/779 (41%), Positives = 443/779 (56%), Gaps = 59/779 (7%)
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
YIVY G H + H++ L S S E A+ ++ YSY INGFAA L
Sbjct: 27 YIVYFGEHK-----GDKAFHEIEEHHHSYLQSVKESEEDARASLLYSYKHSINGFAAELT 81
Query: 62 VEEAAKIAKHPNVVSVFEN--KGHELQTTRSWEFLGLENNY--GVVPKD----------- 106
++A+K+ K VVSVF++ + +E TTRSWEF+GLE VP+
Sbjct: 82 PDQASKLEKLAEVVSVFKSHPRKYEAHTTRSWEFVGLEEEETDSDVPRRKNDADDRFRVG 141
Query: 107 -SIWEKGRYGEGTIIANIDSGVSPESKSFSDDGMGPVPSRWRGICQ----LDNFHCNRKL 161
+ +K ++G+G I+ +DSGV PESKSF+D GMGPVP W+GICQ ++ HCNRK+
Sbjct: 142 RNFLKKAKHGDGIIVGVLDSGVWPESKSFNDKGMGPVPKSWKGICQTGVAFNSSHCNRKI 201
Query: 162 IGARFYSQGYESKFGRLN----QSLYNARDVLGHGTPTLSVAGGNFVSGANVFG-LANGT 216
IGAR+Y +GYE +G N + + RD GHG+ T S A G V GA+ G A G+
Sbjct: 202 IGARYYVKGYERYYGAFNATANKDFLSPRDPDGHGSHTASTAVGRRVLGASALGGFAKGS 261
Query: 217 AKGGSPRSHVAAYKVCWL--------GTI*IECTDADIMQAFEDAISDGVDIISCSLGQT 268
A GG+P + +A YK CW G I C + D++ A +DAI+DGV +IS S+G T
Sbjct: 262 ASGGAPLARLAIYKACWAKPNAEKVEGNI---CLEEDMLAAIDDAIADGVHVISISIGTT 318
Query: 269 SPKEFFEDGISIGAFHAIENGVIVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSY 328
P F +DGI++GA HA++ ++V A GNSGPK GT++N+APW+ +V AST+DR FV
Sbjct: 319 EPFPFTQDGIAMGALHAVKRNIVVAASAGNSGPKPGTLSNLAPWIITVGASTLDRAFVGG 378
Query: 329 LQLGDKHIIMGTSLSTGLPNEKFYSLVSSVDAKVGNATIEDAKICKVGSLDPNKVKGKIL 388
L LG+ + I S+ T +KF LV + + V + + C SL P V GK++
Sbjct: 379 LVLGNGYTIKTDSI-TAFKMDKFAPLVYASNVVVPGIALNETSQCLPNSLKPELVSGKVV 437
Query: 389 FCLLRELDGLVYAEEEAISGGSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSY 448
CL R + E G G++LGN GN++ + +H +PT+ + T + + Y
Sbjct: 438 LCL-RGAGSRIGKGMEVKRAGGAGMILGNIAANGNEVPSDSHFVPTAGVTPTVVDKILEY 496
Query: 449 IKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGA 508
IK K P A++ KT + AP + SSRGPN + P ILKPDITAPG+ IL A+ GA
Sbjct: 497 IKTDKNPKAFIKPGKTVYKYQAAPSMTGFSSRGPNVVDPNILKPDITAPGLYILAAWSGA 556
Query: 509 ISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGN 568
SP+ ++ D + YNI SGTS+SCPHV+ +ALLK I+P WS AA +SA+MTT + +
Sbjct: 557 DSPSKMSVDQRVAGYNIYSGTSMSCPHVAGAIALLKAIHPKWSSAAIRSALMTTAWMTND 616
Query: 569 NHRPIKDQSKEDATPFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFS 628
+PI+D + A PF G+GH +P A DPGLVYD + YL + C+ N T +
Sbjct: 617 KKKPIQDTTGLPANPFALGSGHFRPTKAADPGLVYDASYRAYLLYGCS--VNITNIDPTF 674
Query: 629 RKPYICPKSYNMLDFNYPSITVPNLGKHFVQEVTRTVTNVG---SPGTYRVQVNEPHGIF 685
+ P P YN NYPSI VPNL K V RTVTNVG S TY V P GI
Sbjct: 675 KCPSKIPPGYN---HNYPSIAVPNLKK--TVTVKRTVTNVGTGNSTSTYLFSVKPPSGIS 729
Query: 686 VLIKPRSLTFNEVGEKKTFKIIFK-----VTKPTSSG-YVFGHLLWSDGRHKVMSPLVV 738
V P L+FN +G+K+ FKI+ K V T G Y FG W+D H V SP+ V
Sbjct: 730 VKAIPNILSFNRIGQKQRFKIVIKPLKNQVMNATEKGQYQFGWFSWTDKVHVVRSPIAV 788
>At4g34980 subtilisin proteinase - like
Length = 764
Score = 531 bits (1367), Expect = e-151
Identities = 305/748 (40%), Positives = 448/748 (59%), Gaps = 55/748 (7%)
Query: 20 LQSATDSHYNLLGSHLGSHEKAKEA-IFYSYNKHINGFAAVLEVEEAAKIAKHPNVVSVF 78
+ S +HY H S E A+E+ I + Y+ +GF+AV+ +EA + HP V++VF
Sbjct: 37 MPSIFPTHY-----HWYSTEFAEESRIVHVYHTVFHGFSAVVTPDEADNLRNHPAVLAVF 91
Query: 79 ENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIANIDSGVSPESKSFSDDG 138
E++ EL TTRS +FLGL+N G +W + YG II D+G+ PE +SFSD
Sbjct: 92 EDRRRELHTTRSPQFLGLQNQKG------LWSESDYGSDVIIGVFDTGIWPERRSFSDLN 145
Query: 139 MGPVPSRWRGICQ----LDNFHCNRKLIGARFYSQGYESK-FGRLNQSL--YNARDVLGH 191
+GP+P RWRG+C+ +CNRK+IGARF+++G ++ G +N+++ + RD GH
Sbjct: 146 LGPIPKRWRGVCESGARFSPRNCNRKIIGARFFAKGQQAAVIGGINKTVEFLSPRDADGH 205
Query: 192 GTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI*IECTDADIMQAFE 251
GT T S A G A++ G A+G AKG +P++ +AAYKVCW + C D+DI+ AF+
Sbjct: 206 GTHTSSTAAGRHAFKASMSGYASGVAKGVAPKARIAAYKVCWKDS---GCLDSDILAAFD 262
Query: 252 DAISDGVDIISCSLGQ----TSPKEFFEDGISIGAFHAIENGVIVVAGGGNSGPKFGTVT 307
A+ DGVD+IS S+G TSP ++ D I+IG++ A G+ V + GN GP +VT
Sbjct: 263 AAVRDGVDVISISIGGGDGITSP--YYLDPIAIGSYGAASKGIFVSSSAGNEGPNGMSVT 320
Query: 308 NVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLP-NEKFYSLVSSVDAKVGNAT 366
N+APW+ +V ASTIDRNF + LGD H + G SL G+P N + + +V G +
Sbjct: 321 NLAPWVTTVGASTIDRNFPADAILGDGHRLRGVSLYAGVPLNGRMFPVVYP-----GKSG 375
Query: 367 IEDAKICKVGSLDPNKVKGKILFC----LLRELDGLVYAEEEAISGGSIGLVLGNDKQRG 422
+ A +C +LDP +V+GKI+ C R GLV + G +G++L N G
Sbjct: 376 MSSASLCMENTLDPKQVRGKIVICDRGSSPRVAKGLVVKK-----AGGVGMILANGASNG 430
Query: 423 NDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGP 482
++ AHL+P + +G+ + +Y + P+A + T VG+KPAPVIAS S RGP
Sbjct: 431 EGLVGDAHLIPACAVGSNEGDRIKAYASSHPNPIASIDFRGTIVGIKPAPVIASFSGRGP 490
Query: 483 NPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVAL 542
N + P ILKPD+ APGV+IL A+ A+ PTGL SD + +NI SGTS++CPHVS AL
Sbjct: 491 NGLSPEILKPDLIAPGVNILAAWTDAVGPTGLPSDPRKTEFNILSGTSMACPHVSGAAAL 550
Query: 543 LKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSK-EDATPFGYGAGHIQPELAMDPGL 601
LK+ +P+WSPA +SA+MTTT + N++R + D+S + ATP+ YG+GH+ AM+PGL
Sbjct: 551 LKSAHPDWSPAVIRSAMMTTTNLVDNSNRSLIDESTGKSATPYDYGSGHLNLGRAMNPGL 610
Query: 602 VYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYICP--KSYNMLDFNYPSITV--PNLGKHF 657
VYD+ DY+ FLC+ GY +++ +R P CP + + + NYPSIT P +
Sbjct: 611 VYDITNDDYITFLCSIGYGPKTIQVITRTPVRCPTTRKPSPGNLNYPSITAVFPTNRRGL 670
Query: 658 VQE-VTRTVTNVG-SPGTYRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTKPT- 714
V + V RT TNVG + YR ++ P G+ V +KP L F +++++ + V
Sbjct: 671 VSKTVIRTATNVGQAEAVYRARIESPRGVTVTVKPPRLVFTSAVKRRSYAVTVTVNTRNV 730
Query: 715 ---SSGYVFGHLLWSD-GRHKVMSPLVV 738
+G VFG + W D G+H V SP+VV
Sbjct: 731 VLGETGAVFGSVTWFDGGKHVVRSPIVV 758
>At3g14240 unknown protein
Length = 775
Score = 528 bits (1361), Expect = e-150
Identities = 302/768 (39%), Positives = 450/768 (58%), Gaps = 55/768 (7%)
Query: 1 SYIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVL 60
+YIV++ H PS +H++ S L S + +I ++Y+ +GF+A L
Sbjct: 27 TYIVHV---DHEAKPSIFP------THFHWYTSSLASLTSSPPSIIHTYDTVFHGFSARL 77
Query: 61 EVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTII 120
++A+++ HP+V+SV + L TTRS EFLGL + K + E+ +G +I
Sbjct: 78 TSQDASQLLDHPHVISVIPEQVRHLHTTRSPEFLGLRST----DKAGLLEESDFGSDLVI 133
Query: 121 ANIDSGVSPESKSFSDDGMGPVPSRWRGIC----QLDNFHCNRKLIGARFYSQGYESKFG 176
ID+GV PE SF D G+GPVP +W+G C CNRKL+GARF+ GYE+ G
Sbjct: 134 GVIDTGVWPERPSFDDRGLGPVPIKWKGQCIASQDFPESACNRKLVGARFFCGGYEATNG 193
Query: 177 RLNQS--LYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWL 234
++N++ + RD GHGT T S++ G +V A+ G A+G A G +P++ +AAYKVCW
Sbjct: 194 KMNETTEFRSPRDSDGHGTHTASISAGRYVFPASTLGYAHGVAAGMAPKARLAAYKVCWN 253
Query: 235 GTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVA 294
C D+DI+ AF+ A++DGVD+IS S+G ++ D I+IGAF AI+ G+ V A
Sbjct: 254 SG----CYDSDILAAFDTAVADGVDVISLSVGGVVVP-YYLDAIAIGAFGAIDRGIFVSA 308
Query: 295 GGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSL--STGLPNEKFY 352
GN GP TVTNVAPW+ +V A TIDR+F + ++LG+ +I G S+ GL + Y
Sbjct: 309 SAGNGGPGALTVTNVAPWMTTVGAGTIDRDFPANVKLGNGKMISGVSVYGGPGLDPGRMY 368
Query: 353 SLVSSVDAKVGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIG 412
LV G+ + +C GSLDPN VKGKI+ C R ++ E G +G
Sbjct: 369 PLVYGGSLLGGDGY--SSSLCLEGSLDPNLVKGKIVLCD-RGINSRATKGEIVRKNGGLG 425
Query: 413 LVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYI------KATKTPMAYMTKAKTEV 466
+++ N G ++A H+LP + + + G+ + YI +++K P A + T +
Sbjct: 426 MIIANGVFDGEGLVADCHVLPATSVGASGGDEIRRYISESSKSRSSKHPTATIVFKGTRL 485
Query: 467 GVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIG 526
G++PAPV+AS S+RGPNP P ILKPD+ APG++IL A+ I P+G+ SDN+ +NI
Sbjct: 486 GIRPAPVVASFSARGPNPETPEILKPDVIAPGLNILAAWPDRIGPSGVTSDNRRTEFNIL 545
Query: 527 SGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSK-EDATPFG 585
SGTS++CPHVS + ALLK +P+WSPAA +SA++TT N+ P+ D+S ++
Sbjct: 546 SGTSMACPHVSGLAALLKAAHPDWSPAAIRSALITTAYTVDNSGEPMMDESTGNTSSVMD 605
Query: 586 YGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYICP---KSYNMLD 642
YG+GH+ P AMDPGLVYD+ DY+NFLC Y +T + +R+ C ++ ++ +
Sbjct: 606 YGSGHVHPTKAMDPGLVYDITSYDYINFLCNSNYTRTNIVTITRRQADCDGARRAGHVGN 665
Query: 643 FNYPSITV-------PNLGKHFVQEVTRTVTNVG-SPGTYRVQVNEPHGIFVLIKPRSLT 694
NYPS +V + HF+ RTVTNVG S Y +++ P G V ++P L+
Sbjct: 666 LNYPSFSVVFQQYGESKMSTHFI----RTVTNVGDSDSVYEIKIRPPRGTTVTVEPEKLS 721
Query: 695 FNEVGEKKTFKIIFKVTK----PTSSGYVFGHLLWSDGRHKVMSPLVV 738
F VG+K +F + K T+ P ++ GH++WSDG+ V SPLVV
Sbjct: 722 FRRVGQKLSFVVRVKTTEVKLSPGATNVETGHIVWSDGKRNVTSPLVV 769
>At1g20160 unknown protein
Length = 769
Score = 500 bits (1287), Expect = e-141
Identities = 300/756 (39%), Positives = 421/756 (55%), Gaps = 49/756 (6%)
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
YIVY+GS S N + + + T ++ + ++Y +GFAA L
Sbjct: 36 YIVYMGSASSAANANRAQILINT------------MFKRRANDLLHTYKHGFSGFAARLT 83
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVP-KDSIWEKGRYGEGTII 120
EEA IAK P VVSVF + +L TT SW+FL + + V S G Y +I+
Sbjct: 84 AEEAKVIAKKPGVVSVFPDPHFQLHTTHSWDFLKYQTSVKVDSGPPSSASDGSYD--SIV 141
Query: 121 ANIDSGVSPESKSFSDDGMGPVPSRWRGICQ----LDNFHCNRKLIGARFYSQGYESKFG 176
+D+G+ PES+SF+D MGP+PSRW+G C + +CNRK+IGAR+Y +
Sbjct: 142 GILDTGIWPESESFNDKDMGPIPSRWKGTCMEAKDFKSSNCNRKIIGARYYKNPDD---- 197
Query: 177 RLNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGT 236
+ Y RDV+GHG+ S G+ V A+ +G+A+GTAKGGS + +A YKVC G
Sbjct: 198 --DSEYYTTRDVIGHGSHVSSTIAGSAVENASYYGVASGTAKGGSQNARIAMYKVCNPGG 255
Query: 237 I*IECTDADIMQAFEDAISDGVDIISCSLGQTSPK--EFFEDGISIGAFHAIENGVIVVA 294
CT + I+ AF+DAI+DGVD++S SLG + + D I+IGAFHA+E G++V+
Sbjct: 256 ----CTGSSILAAFDDAIADGVDVLSLSLGAPAYARIDLNTDPIAIGAFHAVEQGILVIC 311
Query: 295 GGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLS-TGLPNEKFYS 353
GN GP GTVTN APW+ +VAA+TIDR+F S + LG +I G + + + Y
Sbjct: 312 SAGNDGPDGGTVTNTAPWIMTVAANTIDRDFESDVVLGGNKVIKGEGIHFSNVSKSPVYP 371
Query: 354 LVSSVDAKVGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAE---EEAISGGS 410
L+ AK +A+ A+ C SLD KVKGKI+ C + G YA +E S G
Sbjct: 372 LIHGKSAKSADASEGSARACDSDSLDQEKVKGKIVLC--ENVGGSYYASSARDEVKSKGG 429
Query: 411 IGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKP 470
G V +D+ R + + PT+ I+ + + SY+ +TK P+A + T P
Sbjct: 430 TGCVFVDDRTRA--VASAYGSFPTTVIDSKEAAEIFSYLNSTKDPVATILPTATVEKFTP 487
Query: 471 APVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTS 530
AP +A SSRGP+ + ILKPDITAPGV IL A+ G S L YN+ SGTS
Sbjct: 488 APAVAYFSSRGPSSLTRSILKPDITAPGVSILAAWTGNDSSISLEGKPA-SQYNVISGTS 546
Query: 531 ISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKEDATPFGYGAGH 590
++ PHVSA+ +L+K+ +P W P+A +SAIMTT T N+ I ++ ATP+ GAG
Sbjct: 547 MAAPHVSAVASLIKSQHPTWGPSAIRSAIMTTATQTNNDKGLITTETGATATPYDSGAGE 606
Query: 591 IQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSR---KPYICPKSYNM---LDFN 644
+ +M PGLVY+ DYLNFLC +GYN T +K S+ + + CP N+ N
Sbjct: 607 LSSTASMQPGLVYETTETDYLNFLCYYGYNVTTIKAMSKAFPENFTCPADSNLDLISTIN 666
Query: 645 YPSITVPNLGKHFVQEVTRTVTNVGSPG--TYRVQVNEPHGIFVLIKPRSLTFNEVGEKK 702
YPSI + + + VTRTVTNVG G Y V V P G + + P L F + GEK
Sbjct: 667 YPSIGISGFKGNGSKTVTRTVTNVGEDGEAVYTVSVETPPGFNIQVTPEKLQFTKDGEKL 726
Query: 703 TFKIIFKVTKPTSSGYVFGHLLWSDGRHKVMSPLVV 738
T+++I T VFG L WS+ ++KV SP+V+
Sbjct: 727 TYQVIVSATASLKQD-VFGALTWSNAKYKVRSPIVI 761
>At1g32940 unknown protein
Length = 774
Score = 499 bits (1285), Expect = e-141
Identities = 295/761 (38%), Positives = 446/761 (57%), Gaps = 50/761 (6%)
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
+IVY+G H D + ++SH+ +L S LGS A E++ YSY +GFAA L
Sbjct: 30 HIVYLGEKQH------DDPEFVSESHHQMLSSLLGSKVDAHESMVYSYRHGFSGFAAKLT 83
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIA 121
+A K+A P VV V + +EL TTR+W++LGL V +++ G+ II
Sbjct: 84 ESQAKKLADSPEVVHVMADSFYELATTRTWDYLGLS----VANPNNLLNDTNMGDQVIIG 139
Query: 122 NIDSGVSPESKSFSDDGMGPVPSRWRGICQLD----NFHCNRKLIGARFYSQGYESKFGR 177
ID+GV PES+SF+D+G+GP+PS W+G C+ + +CNRKLIGA+++ G+ ++
Sbjct: 140 FIDTGVWPESESFNDNGVGPIPSHWKGGCESGEKFISTNCNRKLIGAKYFINGFLAENEG 199
Query: 178 LN----QSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCW 233
N + +ARD +GHGT T S+AGG+FV + GLA G +GG+PR+ +A YK CW
Sbjct: 200 FNTTESRDYISARDFIGHGTHTASIAGGSFVPNISYKGLAGGNLRGGAPRARIAIYKACW 259
Query: 234 ----LGTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSP---KEFFEDGISIGAFHAI 286
LG + C+ +DI++A ++++ DGVD++S SLG P + D I+ GAFHA+
Sbjct: 260 YVDQLGAV--ACSSSDILKAMDESMHDGVDVLSLSLGAQIPLYPETDLRDRIATGAFHAV 317
Query: 287 ENGVIVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGL 346
G+IVV GGNSGP TV N APW+ +VAA+T+DR+F + + LG++ +I+G +L TG
Sbjct: 318 AKGIIVVCAGGNSGPAAQTVLNTAPWIITVAATTLDRSFPTPITLGNRKVILGQALYTG- 376
Query: 347 PNEKFYSLVSSVDAKVGNATIEDAKICKVGSLDPNK-VKGKILFCLLRE--LDGLVYAEE 403
F SLV +A N T + +C+ +L+PN+ + GK++ C + A
Sbjct: 377 QELGFTSLVYPENAGFTNETF--SGVCERLNLNPNRTMAGKVVLCFTTNTLFTAVSRAAS 434
Query: 404 EAISGGSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAK 463
+ G +G+++ + G ++ P I+Y G V YI++T++P+ + ++
Sbjct: 435 YVKAAGGLGVIIA--RNPGYNLTPCRDDFPCVAIDYELGTDVLLYIRSTRSPVVKIQPSR 492
Query: 464 TEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPY 523
T VG +A+ SSRGPN I P ILKPDI APGV IL A T S++ +
Sbjct: 493 TLVGQPVGTKVATFSSRGPNSISPAILKPDIGAPGVSILAA-------TSPDSNSSVGGF 545
Query: 524 NIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTT--TIQGNNHRPIKDQSKEDA 581
+I +GTS++ P V+ +VALLK ++PNWSPAAF+SAI+TT T + S++ A
Sbjct: 546 DILAGTSMAAPVVAGVVALLKALHPNWSPAAFRSAIVTTAWRTDPFGEQIFAEGSSRKVA 605
Query: 582 TPFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYIC--PKSYN 639
PF YG G + PE A DPGL+YD+ DY+ +LC+ GYN + + +C PK+ +
Sbjct: 606 DPFDYGGGIVNPEKAADPGLIYDMGPRDYILYLCSAGYNDSSITQLVGNVTVCSTPKT-S 664
Query: 640 MLDFNYPSITVPNLGKHFVQEVTRTVTNVGS-PGTYRVQVNEPHGIFVLIKPRSLTFNEV 698
+LD N PSIT+P+L +TRTVTNVG+ Y+V V P GI V++ P +L FN
Sbjct: 665 VLDVNLPSITIPDLKDEVT--LTRTVTNVGTVDSVYKVVVEPPLGIQVVVAPETLVFNSK 722
Query: 699 GEKKTFKIIFKVTKPTSSGYVFGHLLWSDGRHKVMSPLVVK 739
+ +F + T ++G+ FG+L+W+D H V P+ V+
Sbjct: 723 TKNVSFTVRVSTTHKINTGFYFGNLIWTDSMHNVTIPVSVR 763
>At4g21650 subtilisin proteinase like protein
Length = 766
Score = 498 bits (1283), Expect = e-141
Identities = 302/764 (39%), Positives = 422/764 (54%), Gaps = 70/764 (9%)
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
YIVY+G H D + T SH+ +L S L S E A+ ++ YSY +GFAA+L
Sbjct: 42 YIVYLGEREH------DDPELVTASHHQMLESLLQSKEDAQNSLIYSYQHGFSGFAALLT 95
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLE----NNYGVVPKDSIWEKGRYGEG 117
+A KI++HP V+ V N+ +L+TTR+W+ LGL + + + G
Sbjct: 96 SSQAKKISEHPEVIHVIPNRIRKLKTTRAWDHLGLSPIPTSFSSLSSVKGLLHDTNLGSE 155
Query: 118 TIIANIDSGVSPESKSFSDDGMGPVPSRWRGICQLDN-----FHCNRKLIGARFYSQGYE 172
II IDSG+ PESK+ +D G+GP+P RWRG C+ HCN KLIGAR+Y G
Sbjct: 156 AIIGVIDSGIWPESKAVNDQGLGPIPKRWRGKCEPGEQFNATIHCNNKLIGARYYLNGVV 215
Query: 173 S----KFGR-LNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVA 227
+ KF R + Q + RD GHGT T ++AGG+FV + FGLA G +GG+PR+ +A
Sbjct: 216 AAIGGKFNRTIIQDFQSTRDANGHGTHTATIAGGSFVPNVSYFGLAQGLVRGGAPRARIA 275
Query: 228 AYKVCWL------GTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGIS-I 280
+YK CW G CT AD+ +AF+DAI DGVD++S S+G P++ D + I
Sbjct: 276 SYKACWNVMRDEGGGTDGRCTSADMWKAFDDAIHDGVDVLSVSIGGGIPEDSEVDKLDYI 335
Query: 281 GAFHAIENGVIVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGT 340
AFHA+ G+ VVA GN GP TV NVAPWL +VAA+T+DR+F + + LG+ +
Sbjct: 336 AAFHAVAKGITVVAAAGNEGPGAHTVDNVAPWLLTVAATTLDRSFPTKITLGNNQTLFAE 395
Query: 341 SLSTGLPNEKFYSLVSSVDAKVGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVY 400
SL TG + G A ++ S D VKGK + LV+
Sbjct: 396 SLFTG------------PEISTGLAFLDS------DSDDTVDVKGKTV---------LVF 428
Query: 401 AEEEAISGGSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMT 460
I+G + V+ Q+ +D+++ + +P +Y G + YI+ T++P +T
Sbjct: 429 DSATPIAGKGVAAVI--LAQKPDDLLSRCNGVPCIFPDYEFGTEILKYIRTTRSPTVRIT 486
Query: 461 KAKTEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQW 520
A T G +A+ S RGPN + P ILKPDI APGV IL AISP N
Sbjct: 487 AATTLTGQPATTKVAAFSCRGPNSVSPAILKPDIAAPGVSIL----AAISPLNPEEQN-- 540
Query: 521 IPYNIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPI--KDQSK 578
+ + SGTS+S P VS I+ALLK+++P WSPAA +SA++TT + PI + +K
Sbjct: 541 -GFGLLSGTSMSTPVVSGIIALLKSLHPKWSPAAVRSALVTTAWRTSPSGEPIFAEGSNK 599
Query: 579 EDATPFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYICP-KS 637
+ A PF YG G + PE A PGLVYD+ IVDY+ ++C+ GYN + + K CP
Sbjct: 600 KLADPFDYGGGLVNPEKAAKPGLVYDMGIVDYIKYMCSAGYNDSSISRVLGKKTNCPIPK 659
Query: 638 YNMLDFNYPSITVPNLGKHFVQEVTRTVTNVGS-PGTYRVQVNEPHGIFVLIKPRSLTFN 696
+MLD N PSIT+PNL K +TRTVTNVG YR + P GI + + P +L F
Sbjct: 660 PSMLDINLPSITIPNLEKEVT--LTRTVTNVGPIKSVYRAVIESPLGITLTVNPTTLVFK 717
Query: 697 EVGEK-KTFKIIFKVTKPTSSGYVFGHLLWSDGRHKVMSPLVVK 739
++ TF + K + ++GY FG L WSDG H V+ P+ VK
Sbjct: 718 SAAKRVLTFSVKAKTSHKVNTGYFFGSLTWSDGVHDVIIPVSVK 761
>At4g10520 putative subtilisin-like protease
Length = 756
Score = 494 bits (1271), Expect = e-140
Identities = 288/752 (38%), Positives = 427/752 (56%), Gaps = 50/752 (6%)
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
Y+VY+G H NP +S T+SH+ +L S LGS E ++I YSY +GFAA L
Sbjct: 30 YVVYLGEKEHD-NP-----ESVTESHHQMLWSLLGSKEAVLDSIVYSYRHGFSGFAAKLT 83
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIA 121
+A +I++ P VV V N +E+ TTR+W++LG+ DS+ +K G I+
Sbjct: 84 ESQAQQISELPEVVQVIPNTLYEMTTTRTWDYLGVSPGNS----DSLLQKANMGYNVIVG 139
Query: 122 NIDSGVSPESKSFSDDGMGPVPSRWRGICQLD-----NFHCNRKLIGARFYSQGYESKFG 176
IDSGV PES+ F+D G GP+PSRW+G C+ + HCNRKLIGA+++ G ++FG
Sbjct: 140 VIDSGVWPESEMFNDKGFGPIPSRWKGGCESGELFNASIHCNRKLIGAKYFVDGLVAEFG 199
Query: 177 RLNQS----LYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVC 232
+N++ + RD GHGT S GG+F+ + GL GTA+GG+P H+A YK C
Sbjct: 200 VVNRTQNPEYLSPRDFAGHGTHVASTIGGSFLPNVSYVGLGRGTARGGAPGVHIAVYKAC 259
Query: 233 WLGTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIV 292
W G C+ AD+++A ++AI DGVDI+S SLG + P + S+GAFHA+ G+ V
Sbjct: 260 WSGY----CSGADVLKAMDEAIHDGVDILSLSLGPSVPLFPETEHTSVGAFHAVAKGIPV 315
Query: 293 VAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEKFY 352
V GN+GP T++NVAPW+ +VAA+T DR+F + + LG+ I+G ++ G P F
Sbjct: 316 VIAAGNAGPTAQTISNVAPWVLTVAATTQDRSFPTAITLGNNITILGQAIYGG-PELGFV 374
Query: 353 SLVSSVDAKVGNATIEDAKICKVGSLDPNK-VKGKILFCLLRELDGLVYAEEEAISGGSI 411
L G+ C+ S +PN ++GK++ C A I+ G +
Sbjct: 375 GLTYPESPLSGD--------CEKLSANPNSTMEGKVVLCFAASTPSNA-AIAAVINAGGL 425
Query: 412 GLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPA 471
GL++ + + P I++ G + YI++T++P+ + +KT G +
Sbjct: 426 GLIMAKNPTHS---LTPTRKFPWVSIDFELGTDILFYIRSTRSPIVKIQASKTLFGQSVS 482
Query: 472 PVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSI 531
+A+ SSRGPN + P ILKPDI APGV+IL AISP +D + + SGTS+
Sbjct: 483 TKVATFSSRGPNSVSPAILKPDIAAPGVNIL----AAISPNSSINDG---GFAMMSGTSM 535
Query: 532 SCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPI--KDQSKEDATPFGYGAG 589
+ P VS +V LLK+++P+WSP+A KSAI+TT + PI S++ A PF YG G
Sbjct: 536 ATPVVSGVVVLLKSLHPDWSPSAIKSAIVTTAWRTDPSGEPIFADGSSRKLADPFDYGGG 595
Query: 590 HIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYICPK-SYNMLDFNYPSI 648
I PE A+ PGL+YD+ DY+ ++C+ Y+ + K +CP ++LD N PSI
Sbjct: 596 LINPEKAVKPGLIYDMTTDDYVMYMCSVDYSDISISRVLGKITVCPNPKPSVLDLNLPSI 655
Query: 649 TVPNLGKHFVQEVTRTVTNVGSPGT-YRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKII 707
T+PNL +TRTVTNVG + Y+V ++ P GI V + P L F+ K++F +
Sbjct: 656 TIPNLRGEVT--LTRTVTNVGPVNSVYKVVIDPPTGINVAVTPAELVFDYTTTKRSFTVR 713
Query: 708 FKVTKPTSSGYVFGHLLWSDGRHKVMSPLVVK 739
T ++GY FG L W+D H V P+ V+
Sbjct: 714 VSTTHKVNTGYYFGSLTWTDNMHNVAIPVSVR 745
>At4g10550 subtilisin-like protease -like protein
Length = 778
Score = 492 bits (1267), Expect = e-139
Identities = 300/759 (39%), Positives = 435/759 (56%), Gaps = 47/759 (6%)
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
+IVY+G H D + T+SH+ +L S LGS E A +++ YSY +GFAA L
Sbjct: 35 HIVYLGEKQH------DDPEFVTESHHRMLWSLLGSKEDANDSMVYSYRHGFSGFAAKLT 88
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIA 121
+A KIA P+VV V + ++L TTR+W++LGL PK S+ + GE II
Sbjct: 89 ESQAKKIADLPDVVHVIPDSFYKLATTRTWDYLGLS---AANPK-SLLHETNMGEQIIIG 144
Query: 122 NIDSGVSPESKSFSDDGMGPVPSRWRGICQL-DNFH---CNRKLIGARFYSQGYESKFGR 177
ID+GV PES+ F+D G GPVPS W+G C+ +NF+ CN+KLIGA+++ G+ ++
Sbjct: 145 VIDTGVWPESEVFNDSGFGPVPSHWKGGCETGENFNSSNCNKKLIGAKYFINGFLAENES 204
Query: 178 LNQS----LYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCW 233
N + + RD+ GHGT ++AGG+FV + GLA GT +GG+PR+H+A YK CW
Sbjct: 205 FNSTNSLDFISPRDLDGHGTHVSTIAGGSFVPNISYKGLAGGTVRGGAPRAHIAMYKACW 264
Query: 234 L--GTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSP---KEFFEDGISIGAFHAIEN 288
C+ ADI++A ++A+ DGVD++S SLG + P + DGI+ GAFHA+
Sbjct: 265 YLDDDDTTTCSSADILKAMDEAMHDGVDVLSISLGSSVPLYGETDIRDGITTGAFHAVLK 324
Query: 289 GVIVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPN 348
G+ VV GGNSGP TVTN APW+ +VAA+T+DR+F + L LG+ +I+G ++ TG P
Sbjct: 325 GITVVCSGGNSGPDSLTVTNTAPWIITVAATTLDRSFATPLTLGNNKVILGQAMYTG-PG 383
Query: 349 EKFYSLVSSVDAKVGNATIEDAKICKVGSLDPNK-VKGKILFCLLRELDG--LVYAEEEA 405
F SLV GN+ + C+ + N+ ++GK++ C G ++ A
Sbjct: 384 LGFTSLV--YPENPGNSNESFSGTCEELLFNSNRTMEGKVVLCFTTSPYGGAVLSAARYV 441
Query: 406 ISGGSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTE 465
G +G+++ + G I P +++ G + Y +++ +P+ + +KT
Sbjct: 442 KRAGGLGVIIA--RHPGYAIQPCLDDFPCVAVDWELGTDILLYTRSSGSPVVKIQPSKTL 499
Query: 466 VGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNI 525
VG +A+ SSRGPN I P ILKPDI APGV IL A + SD +I +
Sbjct: 500 VGQPVGTKVATFSSRGPNSIAPAILKPDIAAPGVSIL-----AATTNTTFSDQGFI---M 551
Query: 526 GSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTT--TIQGNNHRPIKDQSKEDATP 583
SGTS++ P +S + ALLK ++ +WSPAA +SAI+TT T + + A P
Sbjct: 552 LSGTSMAAPAISGVAALLKALHRDWSPAAIRSAIVTTAWKTDPFGEQIFAEGSPPKLADP 611
Query: 584 FGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYIC--PKSYNML 641
F YG G + PE + +PGLVYD+ + DY+ ++C+ GYN+T + K +C PK ++L
Sbjct: 612 FDYGGGLVNPEKSANPGLVYDMGLEDYVLYMCSVGYNETSISQLIGKTTVCSNPKP-SVL 670
Query: 642 DFNYPSITVPNLGKHFVQEVTRTVTNVGS-PGTYRVQVNEPHGIFVLIKPRSLTFNEVGE 700
DFN PSIT+PNL +TRTVTNVG YRV V P G V + P +L FN +
Sbjct: 671 DFNLPSITIPNLKDEVT--ITRTVTNVGPLNSVYRVTVEPPLGFQVTVTPETLVFNSTTK 728
Query: 701 KKTFKIIFKVTKPTSSGYVFGHLLWSDGRHKVMSPLVVK 739
K FK+ T T++GY FG L WSD H V PL V+
Sbjct: 729 KVYFKVKVSTTHKTNTGYYFGSLTWSDSLHNVTIPLSVR 767
>At1g20150 unknown protein
Length = 780
Score = 487 bits (1254), Expect = e-138
Identities = 303/768 (39%), Positives = 434/768 (56%), Gaps = 61/768 (7%)
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
YI+Y+G+ AS S + H LL S L +++ + + Y +GFAA L
Sbjct: 33 YIIYMGA--------ASSDGSTDNDHVELLSSLL---QRSGKTPMHRYKHGFSGFAAHLS 81
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGE----- 116
+EA IAK P V+SVF ++ +L TTRSW+FL E+ +D+ + + Y +
Sbjct: 82 EDEAHLIAKQPGVLSVFPDQMLQLHTTRSWDFLVQES----YQRDTYFTEMNYEQESEMH 137
Query: 117 --GTIIANIDSGVSPESKSFSDDGMGPVPSRWRGIC------QLDNFHCNRKLIGARFYS 168
TII +DSG+ PE++SF+D MGPVP +W+G C Q D+F CNRKLIGAR+Y+
Sbjct: 138 EGDTIIGFLDSGIWPEAQSFNDRHMGPVPEKWKGTCMRGKKTQPDSFRCNRKLIGARYYN 197
Query: 169 QGYESKFGRLNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAA 228
+ L+ RD LGHGT S+A G ++ A+ +GLA+G +GGSP S +A
Sbjct: 198 SSFF-----LDPDYETPRDFLGHGTHVASIAAGQIIANASYYGLASGIMRGGSPSSRIAM 252
Query: 229 YKVCWLGTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIEN 288
Y+ C L + C + I+ AF+DAI+DGVD+IS S+G P ED +SIG+FHA+E
Sbjct: 253 YRACSL----LGCRGSSILAAFDDAIADGVDVISISMGLW-PDNLLEDPLSIGSFHAVER 307
Query: 289 GVIVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLG--DKHIIMGTSLS-TG 345
G+ VV GNSGP +V N APW+ +VAASTIDR F S + LG + +I G ++
Sbjct: 308 GITVVCSVGNSGPSSQSVFNAAPWMITVAASTIDRGFESNILLGGDENRLIEGFGINIAN 367
Query: 346 LPNEKFYSLVSSVDAKVGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLV--YAEE 403
+ + Y L+ + AK +A E A+ C +LD VKGKI+ C +LD V + +
Sbjct: 368 IDKTQAYPLIHARSAKKIDANEEAARNCAPDTLDQTIVKGKIVVCD-SDLDNQVIQWKSD 426
Query: 404 EAISGGSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAK 463
E G IG+VL +D+ + + L+ I DG + SYI +T+ P+A + +
Sbjct: 427 EVKRLGGIGMVLVDDESMDLSFIDPSFLVTI--IKPEDGIQIMSYINSTREPIATIMPTR 484
Query: 464 TEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIP- 522
+ G AP I S SSRGP + ILKPDI APGV+IL +++ + A + + P
Sbjct: 485 SRTGHMLAPSIPSFSSRGPYLLTRSILKPDIAAPGVNILASWL--VGDRNAAPEGKPPPL 542
Query: 523 YNIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKEDAT 582
+NI SGTS+SCPHVS I A LK+ YP+WSPAA +SAIMTT N I ++ E AT
Sbjct: 543 FNIESGTSMSCPHVSGIAARLKSRYPSWSPAAIRSAIMTTAVQMTNTGSHITTETGEKAT 602
Query: 583 PFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRK---PYICPKSYN 639
P+ +GAG + PGL+Y+ N +DYLNFL +G+ Q+K S + + CP+ N
Sbjct: 603 PYDFGAGQVTIFGPSSPGLIYETNHMDYLNFLGYYGFTSDQIKKISNRIPQGFACPEQSN 662
Query: 640 MLD---FNYPSITVPNLGKHFVQEVTRTVTNVGS------PGTYRVQVNEPHGIFVLIKP 690
D NYPSI++ N + V+RTVTNV S Y V ++ P G+ V + P
Sbjct: 663 RGDISNINYPSISISNFNGKESRRVSRTVTNVASRLIGDEDTVYTVSIDAPEGLLVRVIP 722
Query: 691 RSLTFNEVGEKKTFKIIFKVTKPTSSGYVFGHLLWSDGRHKVMSPLVV 738
R L F ++G+K ++++IF T FG + WS+G + V SP VV
Sbjct: 723 RRLHFRKIGDKLSYQVIFSSTTTILKDDAFGSITWSNGMYNVRSPFVV 770
>At2g05920 serine protease like protein
Length = 754
Score = 485 bits (1249), Expect = e-137
Identities = 283/738 (38%), Positives = 425/738 (57%), Gaps = 46/738 (6%)
Query: 18 SDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLEVEEAAKIAKHPN-VVS 76
SD + +H++ S L S + ++ Y+Y +GF+A L+ EA + N ++
Sbjct: 37 SDKPESFLTHHDWYTSQLNS----ESSLLYTYTTSFHGFSAYLDSTEADSLLSSSNSILD 92
Query: 77 VFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIANIDSGVSPESKSFSD 136
+FE+ + L TTR+ EFLGL + +GV + G G II +D+GV PES+SF D
Sbjct: 93 IFEDPLYTLHTTRTPEFLGLNSEFGV------HDLGSSSNGVIIGVLDTGVWPESRSFDD 146
Query: 137 DGMGPVPSRWRGICQ----LDNFHCNRKLIGARFYSQGYESKFG---RLNQSLYNARDVL 189
M +PS+W+G C+ D+ CN+KLIGAR +S+G++ G + + RDV
Sbjct: 147 TDMPEIPSKWKGECESGSDFDSKLCNKKLIGARSFSKGFQMASGGGFSSKRESVSPRDVD 206
Query: 190 GHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI*IECTDADIMQA 249
GHGT T + A G+ V A+ G A GTA+G + R+ VA YKVCW C +DI+ A
Sbjct: 207 GHGTHTSTTAAGSAVRNASFLGYAAGTARGMATRARVATYKVCWS----TGCFGSDILAA 262
Query: 250 FEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNSGPKFGTVTNV 309
+ AI DGVD++S SLG S ++ D I+IGAF A+E GV V GNSGP +V NV
Sbjct: 263 MDRAILDGVDVLSLSLGGGSAP-YYRDTIAIGAFSAMERGVFVSCSAGNSGPTRASVANV 321
Query: 310 APWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGL-----PNEKFYSLVSSVDAKVGN 364
APW+ +V A T+DR+F ++ LG+ + G SL +G+ P E Y+ GN
Sbjct: 322 APWVMTVGAGTLDRDFPAFANLGNGKRLTGVSLYSGVGMGTKPLELVYNK--------GN 373
Query: 365 ATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLGNDKQRGND 424
++ + +C GSLD + V+GKI+ C R ++ V G +G+++ N G +
Sbjct: 374 SS--SSNLCLPGSLDSSIVRGKIVVCD-RGVNARVEKGAVVRDAGGLGMIMANTAASGEE 430
Query: 425 IMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNP 484
++A +HLLP + G+ + Y+K+ P A + T + VKP+PV+A+ SSRGPN
Sbjct: 431 LVADSHLLPAIAVGKKTGDLLREYVKSDSKPTALLVFKGTVLDVKPSPVVAAFSSRGPNT 490
Query: 485 IQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLK 544
+ P ILKPD+ PGV+IL + AI PTGL D++ +NI SGTS+SCPH+S + LLK
Sbjct: 491 VTPEILKPDVIGPGVNILAGWSDAIGPTGLDKDSRRTQFNIMSGTSMSCPHISGLAGLLK 550
Query: 545 TIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKED-ATPFGYGAGHIQPELAMDPGLVY 603
+P WSP+A KSA+MTT + N + P+ D + + P+ +G+GH+ P+ A+ PGLVY
Sbjct: 551 AAHPEWSPSAIKSALMTTAYVLDNTNAPLHDAADNSLSNPYAHGSGHVDPQKALSPGLVY 610
Query: 604 DLNIVDYLNFLCAHGYNQTQMKMFSRKPYI-CPKSY-NMLDFNYPSITVPNLGKHFVQEV 661
D++ +Y+ FLC+ Y + ++P + C K + + NYPS +V GK V+
Sbjct: 611 DISTEEYIRFLCSLDYTVDHIVAIVKRPSVNCSKKFSDPGQLNYPSFSVLFGGKRVVR-Y 669
Query: 662 TRTVTNVGSPGT-YRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTKPTS--SGY 718
TR VTNVG+ + Y+V VN + + +KP L+F VGEKK + + F K S +
Sbjct: 670 TREVTNVGAASSVYKVTVNGAPSVGISVKPSKLSFKSVGEKKRYTVTFVSKKGVSMTNKA 729
Query: 719 VFGHLLWSDGRHKVMSPL 736
FG + WS+ +H+V SP+
Sbjct: 730 EFGSITWSNPQHEVRSPV 747
>At4g00230 subtilisin-type serine endopeptidase XSP1
Length = 749
Score = 484 bits (1246), Expect = e-137
Identities = 289/746 (38%), Positives = 417/746 (55%), Gaps = 49/746 (6%)
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
YI+Y+G + + +H NLL S S E+AKE YSY K N FAA L
Sbjct: 38 YIIYLGDRP-------DNTEETIKTHINLLSSLNISQEEAKERKVYSYTKAFNAFAAKLS 90
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIA 121
EA K+ + VVSV N+ +L TT+SW+F+GL + K + + II
Sbjct: 91 PHEAKKMMEMEEVVSVSRNQYRKLHTTKSWDFVGLP----LTAKRHL----KAERDVIIG 142
Query: 122 NIDSGVSPESKSFSDDGMGPVPSRWRGIC-QLDNFH-CNRKLIGARFYSQGYESKFGRLN 179
+D+G++P+S+SF D G+GP P++W+G C NF CN K+IGA+++ G +
Sbjct: 143 VLDTGITPDSESFLDHGLGPPPAKWKGSCGPYKNFTGCNNKIIGAKYFKHDGNVPAGEVR 202
Query: 180 QSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI*I 239
+ D+ GHGT T S G V+ A+++G+ANGTA+G P + +A YKVCW +
Sbjct: 203 SPI----DIDGHGTHTSSTVAGVLVANASLYGIANGTARGAVPSARLAMYKVCWARS--- 255
Query: 240 ECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNS 299
C D DI+ FE AI DGV+IIS S+G ++ D IS+G+FHA+ G++ VA GN
Sbjct: 256 GCADMDILAGFEAAIHDGVEIISISIGGPIA-DYSSDSISVGSFHAMRKGILTVASAGND 314
Query: 300 GPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEKFYSLVSSVD 359
GP GTVTN PW+ +VAAS IDR F S + LG+ G +S P K Y LVS VD
Sbjct: 315 GPSSGTVTNHEPWILTVAASGIDRTFKSKIDLGNGKSFSGMGISMFSPKAKSYPLVSGVD 374
Query: 360 AKVGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLGNDK 419
A A+ C SLD KVKGK++ C + G E S G G ++ +D+
Sbjct: 375 AAKNTDDKYLARYCFSDSLDRKKVKGKVMVCRM----GGGGVESTIKSYGGAGAIIVSDQ 430
Query: 420 QRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSS 479
N A + P + +N + G+ ++ YI +T++ A + K + +V + PAP +AS SS
Sbjct: 431 YLDN---AQIFMAPATSVNSSVGDIIYRYINSTRSASAVIQKTR-QVTI-PAPFVASFSS 485
Query: 480 RGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAI 539
RGPNP +LKPDI APG+DIL A+ S TGL D Q+ + I SGTS++CPHV+ +
Sbjct: 486 RGPNPGSIRLLKPDIAAPGIDILAAFTLKRSLTGLDGDTQFSKFTILSGTSMACPHVAGV 545
Query: 540 VALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKEDATPFGYGAGHIQPELAMDP 599
A +K+ +P+W+PAA KSAI+T+ +PI + +DA F YG G I P A P
Sbjct: 546 AAYVKSFHPDWTPAAIKSAIITSA-------KPISRRVNKDA-EFAYGGGQINPRRAASP 597
Query: 600 GLVYDLNIVDYLNFLCAHGYNQTQM-KMFSRKPYICPKSYNML---DFNYPSI--TVPNL 653
GLVYD++ + Y+ FLC GYN T + + + C L NYP+I T+ +
Sbjct: 598 GLVYDMDDISYVQFLCGEGYNATTLAPLVGTRSVSCSSIVPGLGHDSLNYPTIQLTLRSA 657
Query: 654 GKHFVQEVTRTVTNVGSPGT-YRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTK 712
+ R VTNVG P + Y V P G+ + ++P+SL+F++ +K++FK++ K +
Sbjct: 658 KTSTLAVFRRRVTNVGPPSSVYTATVRAPKGVEITVEPQSLSFSKASQKRSFKVVVKAKQ 717
Query: 713 PTSSGYVFGHLLWSDGRHKVMSPLVV 738
T V G L+W RH V SP+V+
Sbjct: 718 MTPGKIVSGLLVWKSPRHSVRSPIVI 743
>At1g32950 putative serine proteinase (At1g32950)
Length = 763
Score = 484 bits (1245), Expect = e-137
Identities = 298/760 (39%), Positives = 440/760 (57%), Gaps = 59/760 (7%)
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
+IVY+G H D + T+SH+ +L S LGS + A E++ YSY +GFAA L
Sbjct: 30 HIVYLGEKQH------DDPKFVTESHHQMLSSLLGSKDDAHESMVYSYRHGFSGFAAKLT 83
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLG--LENNYGVVPKDSIWEKGRYGEGTI 119
+A KIA P V+ V + +EL TTR W++LG +N+ +V ++ G+ TI
Sbjct: 84 KSQAKKIADSPEVIHVIPDSYYELATTRIWDYLGPSADNSKNLVSDTNM------GDQTI 137
Query: 120 IANIDSGVSPESKSFSDDGMGPVPSRWRGICQL-DNF---HCNRKLIGARFYSQGY--ES 173
I ID+GV PES+SF+D G+GPVPS W+G C+ +NF +CNRKLIGA+++ G+ E+
Sbjct: 138 IGVIDTGVWPESESFNDYGVGPVPSHWKGGCEPGENFISTNCNRKLIGAKYFINGFLAEN 197
Query: 174 KFGRLNQSLY-NARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVC 232
+F Y +ARD GHGT S+AGG+FV + GL GT +GG+PR+ +A YK C
Sbjct: 198 QFNATESPDYISARDFDGHGTHVASIAGGSFVPNVSYKGLGRGTLRGGAPRARIAMYKAC 257
Query: 233 WL--GTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSP---KEFFEDGISIGAFHAIE 287
W + C+ +DIM+A ++AI DGVD++S SLG P + DGI+ GAFHA+
Sbjct: 258 WYINELDGVTCSFSDIMKAIDEAIHDGVDVLSISLGGRVPLNSETDLRDGIATGAFHAVA 317
Query: 288 NGVIVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLP 347
G++VV GGN+GP TV N APW+ +VAA+T+DR+F + + LG+ +I+G ++ G P
Sbjct: 318 KGIVVVCAGGNAGPSSQTVVNTAPWILTVAATTLDRSFATPIILGNNQVILGQAMYIG-P 376
Query: 348 NEKFYSLVSSVDAKVGNATIEDAKICKVGSLDPNK-VKGKILFCLLRELDGLVYAEEEAI 406
F SLV D GN+ + +C+ +L+ N+ + GK++ C D V + +I
Sbjct: 377 ELGFTSLVYPEDP--GNSIDTFSGVCESLNLNSNRTMAGKVVLCFTTARDFTVVSTAASI 434
Query: 407 --SGGSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKT 464
+ G +GL++ + G ++ + P I+ G + YI+ T T +
Sbjct: 435 VKAAGGLGLIIA--RNPGYNLAPCSDDFPCVAIDNELGTDILFYIRYTGTLVG------E 486
Query: 465 EVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYN 524
VG K +A+ SSRGPN I P ILKPDI APGV IL A SP + ++
Sbjct: 487 PVGTK----VATFSSRGPNSISPAILKPDIAAPGVSIL----AATSPNDTLNAGGFV--- 535
Query: 525 IGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTT--TIQGNNHRPIKDQSKEDAT 582
+ SGTS++ P +S ++ALLK+++P+WSPAAF+SAI+TT T + S +
Sbjct: 536 MRSGTSMAAPVISGVIALLKSLHPDWSPAAFRSAIVTTAWRTDPFGEQIAAESSSLKVPD 595
Query: 583 PFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYIC--PKSYNM 640
PF YG G + PE A +PGL+ D++ DY+ +LC+ GYN + + K +C PK ++
Sbjct: 596 PFDYGGGLVNPEKAAEPGLILDMDSQDYVLYLCSAGYNDSSISRLVGKVTVCSNPKP-SV 654
Query: 641 LDFNYPSITVPNLGKHFVQEVTRTVTNVGS-PGTYRVQVNEPHGIFVLIKPRSLTFNEVG 699
LD N PSIT+PNL +TRTVTNVG Y+V V P GI V++ P +L FN
Sbjct: 655 LDINLPSITIPNLKDEVT--LTRTVTNVGPVDSVYKVLVEPPLGIQVVVTPETLVFNSKT 712
Query: 700 EKKTFKIIFKVTKPTSSGYVFGHLLWSDGRHKVMSPLVVK 739
+ +F +I T ++G+ FG L W+D H V+ P+ V+
Sbjct: 713 KSVSFTVIVSTTHKINTGFYFGSLTWTDSIHNVVIPVSVR 752
>At4g21630 serine protease - like protein
Length = 772
Score = 483 bits (1244), Expect = e-136
Identities = 294/774 (37%), Positives = 431/774 (54%), Gaps = 82/774 (10%)
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLG-------------SHEKAKEAIFYS 48
YIVY+G H D + T SH+ +L S L S + A ++ YS
Sbjct: 40 YIVYLGEREH------DDPELFTASHHQMLESLLQRSTSLTCVSNDIYSKDDAHNSLIYS 93
Query: 49 YNKHINGFAAVLEVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDS- 107
Y +GFAA+L +A KI++HP V+ V N+ +L+TTR+W+ LGL N S
Sbjct: 94 YQYGFSGFAALLTSSQAKKISEHPEVIHVIPNRILKLKTTRTWDHLGLSPNPTSFSSSSS 153
Query: 108 ---IWEKGRYGEGTIIANIDSGVSPESKSFSDDGMGPVPSRWRGICQLDN-----FHCNR 159
+ + G II +D+G+ PESK F+D G+GP+P RWRG C+ HCN
Sbjct: 154 AKGLLHETNMGSEAIIGVVDTGIWPESKVFNDHGLGPIPQRWRGKCESGEQFNAKIHCNN 213
Query: 160 KLIGARFYSQGYES----KFGR-LNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLAN 214
KLIGA++Y G + KF R + Q + RD +GHGT T ++AGG+FV + +GLA
Sbjct: 214 KLIGAKYYLSGLLAETGGKFNRTIIQDFKSNRDAIGHGTHTATIAGGSFVPNVSFYGLAR 273
Query: 215 GTAKGGSPRSHVAAYKVCW--LGTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSPKE 272
GT +GG+PR+ +A+YKVCW +G I CT AD+ +AF+DAI D VD++S S+G P+
Sbjct: 274 GTVRGGAPRARIASYKVCWNVVGYDGI-CTVADMWKAFDDAIHDQVDVLSVSIGAGIPEN 332
Query: 273 FFEDGIS-IGAFHAIENGVIVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQL 331
D + I AFHA+ G+ VVA GGN GP +TN APWL +VAA+T+DR+F + + L
Sbjct: 333 SEVDSVDFIAAFHAVAKGITVVAAGGNDGPGAQNITNAAPWLLTVAATTLDRSFPTKITL 392
Query: 332 GDKHIIMGTSLSTGLPNEKFYSLVSSVDAKVGNATIEDAKICKVGSLDPNKVKGKILFCL 391
G+ + SL TG + + ++ +D+ N ++ I + S P+ + G+
Sbjct: 393 GNNQTLFAESLFTG---PEISTSLAFLDSD-HNVDVKGKTILEFDSTHPSSIAGR----- 443
Query: 392 LRELDGLVYAEEEAISGGSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKA 451
G + ++L ++ +D++A + +P +Y G ++ YI+
Sbjct: 444 -----------------GVVAVILA---KKPDDLLARYNSIPYIFTDYEIGTHILQYIRT 483
Query: 452 TKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISP 511
T++P ++ A T G +A SSRGPN + P ILKPDI APGV IL A+SP
Sbjct: 484 TRSPTVRISAATTLNGQPAMTKVAEFSSRGPNSVSPAILKPDIAAPGVSIL----AAVSP 539
Query: 512 TGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHR 571
+ N + + SGTS+S P VS I+ALLK+++PNWSPAA +SA++TT +
Sbjct: 540 LDPDAFN---GFGLYSGTSMSTPVVSGIIALLKSLHPNWSPAAMRSALVTTAWRTSPSGE 596
Query: 572 PI--KDQSKEDATPFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSR 629
PI + +K+ A PF YG G + P+ A PGLVYD+ I DY+N++C+ GY + +
Sbjct: 597 PIFAQGSNKKLADPFDYGGGLVNPDKAAQPGLVYDMGIKDYINYMCSAGYIDSSISRVLG 656
Query: 630 KPYIC--PKSYNMLDFNYPSITVPNLGKHFVQEVTRTVTNVGS-PGTYRVQVNEPHGIFV 686
K C PK ++LD N PSIT+PNL K +TRTVTNVG Y+ + P GI +
Sbjct: 657 KKTKCTIPKP-SILDINLPSITIPNLEKEVT--LTRTVTNVGPIKSVYKAVIESPLGITL 713
Query: 687 LIKPRSLTFNEVGEK-KTFKIIFKVTKPTSSGYVFGHLLWSDGRHKVMSPLVVK 739
+ P +L FN ++ TF + K + +SGY FG L W+DG H V+ P+ VK
Sbjct: 714 TVNPTTLVFNSAAKRVLTFSVKAKTSHKVNSGYFFGSLTWTDGVHDVIIPVSVK 767
>At4g10510 putative subtilisin-like protease
Length = 765
Score = 483 bits (1242), Expect = e-136
Identities = 295/759 (38%), Positives = 431/759 (55%), Gaps = 48/759 (6%)
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
+IVY+G H D + T+SH+ +L S LGS E+A ++ +S+ +GFAA L
Sbjct: 23 HIVYLGEKQH------DDPEFVTESHHRMLWSLLGSKEEAHGSMVHSFRHGFSGFAAKLT 76
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIA 121
+A KIA P VV V ++ ++ TTR+W++LGL PK+ + + GE II
Sbjct: 77 ESQAKKIADLPEVVHVIPDRFYKPATTRTWDYLGLSPTN---PKNLL-NQTNMGEQMIIG 132
Query: 122 NIDSGVSPESKSFSDDGMGPVPSRWRGICQ----LDNFHCNRKLIGARFYSQGYESKFGR 177
IDSGV PES+ F+D+ +GPVPS W+G C+ ++ HCN+KLIGA+++ + +
Sbjct: 133 IIDSGVWPESEVFNDNEIGPVPSHWKGGCESGEDFNSSHCNKKLIGAKYFINAFLATHES 192
Query: 178 LNQS----LYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCW 233
N S + R GHGT ++AGG++V + GLA GT +GG+PR+ +A YK CW
Sbjct: 193 FNSSESLDFISPRGYNGHGTHVATIAGGSYVPNTSYKGLAGGTVRGGAPRARIAVYKTCW 252
Query: 234 LGTI*IE-CTDADIMQAFEDAISDGVDIISCSLG--QTSPKEFFEDGISIGAFHAIENGV 290
+ I C+ ADI++A ++AI DGVD++S SLG P+ DGI+ GAFHA+ G+
Sbjct: 253 YLDLDIAACSSADILKAMDEAIHDGVDVLSLSLGFEPLYPETDVRDGIATGAFHAVLKGI 312
Query: 291 IVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEK 350
VV GN+GP TV N APW+ +VAA+T+DR+FV+ + LG+ +I+G ++ TG
Sbjct: 313 TVVCAAGNAGPAAQTVGNTAPWILTVAATTLDRSFVTPMTLGNNKVILGQAIYTG-TEVG 371
Query: 351 FYSLVSSVDAKVGNATIEDAKICKVGSLDPNK-VKGKILFCLLRE--LDGLVYAEEEAIS 407
F SLV GN+ + C+ ++ N+ + GK++ C + A
Sbjct: 372 FTSLV--YPENPGNSNESFSGTCERLLINSNRTMAGKVVLCFTESPYSISVTRAAHYVKR 429
Query: 408 GGSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVG 467
G +G+++ Q GN + P ++Y G Y+ YI++ +P+ + ++T +G
Sbjct: 430 AGGLGVIIAG--QPGNVLRPCLDDFPCVAVDYELGTYILFYIRSNGSPVVKIQPSRTLIG 487
Query: 468 VKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGS 527
+AS SSRGPNPI ILKPDI APGV IL A + +D +I S
Sbjct: 488 QPVGTKVASFSSRGPNPISAAILKPDIAAPGVSIL----AATTTNTTFNDRGFI---FLS 540
Query: 528 GTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTT--TIQGNNHRPIKDQSKEDATPFG 585
GTS++ P +S IVALLK ++P+WSPAA +SAI+TT T + ++ A PF
Sbjct: 541 GTSMATPTISGIVALLKALHPDWSPAAIRSAIVTTAWRTDPFGEQIFAEGSPRKPADPFD 600
Query: 586 YGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYIC--PKSYNMLDF 643
YG G + PE A PGLVYDL + DY+ ++C+ GYN+T + K +C PK ++LDF
Sbjct: 601 YGGGLVNPEKATKPGLVYDLGLEDYVLYMCSVGYNETSISQLVGKGTVCSYPKP-SVLDF 659
Query: 644 NYPSITVPNLGKHFVQEVT--RTVTNVGS-PGTYRVQVNEPHGIFVLIKPRSLTFNEVGE 700
N PSIT+PNL +EVT RT+TNVG YRV V P G V + P +L FN +
Sbjct: 660 NLPSITIPNL----KEEVTLPRTLTNVGPLESVYRVAVEPPLGTQVTVTPETLVFNSTTK 715
Query: 701 KKTFKIIFKVTKPTSSGYVFGHLLWSDGRHKVMSPLVVK 739
+ +FK+ T ++GY FG L WSD H V PL V+
Sbjct: 716 RVSFKVSVSTTHKINTGYYFGSLTWSDSLHNVTIPLSVR 754
>At5g51750 serine protease-like protein
Length = 780
Score = 482 bits (1240), Expect = e-136
Identities = 288/757 (38%), Positives = 435/757 (57%), Gaps = 37/757 (4%)
Query: 1 SYIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKE-AIFYSYNKHINGFAAV 59
+Y++++ S P P + LQ + S N + H E+ I Y+Y +G AA
Sbjct: 36 TYVIHMDK-SAMPLPYTNHLQWYS-SKINSVTQHKSQEEEGNNNRILYTYQTAFHGLAAQ 93
Query: 60 LEVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTI 119
L EEA ++ + VV+V +EL TTRS FLGLE + +W + +
Sbjct: 94 LTQEEAERLEEEDGVVAVIPETRYELHTTRSPTFLGLERQ----ESERVWAERVTDHDVV 149
Query: 120 IANIDSGVSPESKSFSDDGMGPVPSRWRGICQLDNF----HCNRKLIGARFYSQGYESKF 175
+ +D+G+ PES+SF+D GM PVP+ WRG C+ +CNRK++GAR + +GYE+
Sbjct: 150 VGVLDTGIWPESESFNDTGMSPVPATWRGACETGKRFLKRNCNRKIVGARVFYRGYEAAT 209
Query: 176 GRLNQSLY--NARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCW 233
G++++ L + RD GHGT T + G+ V GAN+FG A GTA+G + ++ VAAYKVCW
Sbjct: 210 GKIDEELEYKSPRDRDGHGTHTAATVAGSPVKGANLFGFAYGTARGMAQKARVAAYKVCW 269
Query: 234 LGTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVV 293
+G C +DI+ A + A++DGV ++S SLG + D +SI F A+E GV V
Sbjct: 270 VGG----CFSSDILSAVDQAVADGVQVLSISLGG-GVSTYSRDSLSIATFGAMEMGVFVS 324
Query: 294 AGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTG---LPNEK 350
GN GP ++TNV+PW+ +V AST+DR+F + +++G G SL G LP K
Sbjct: 325 CSAGNGGPDPISLTNVSPWITTVGASTMDRDFPATVKIGTMRTFKGVSLYKGRTVLPKNK 384
Query: 351 FYSLVSSVDAKVG-NATIED-AKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISG 408
Y LV +G NA+ D C G+LD V GKI+ C R + V +
Sbjct: 385 QYPLVY-----LGRNASSPDPTSFCLDGALDRRHVAGKIVICD-RGVTPRVQKGQVVKRA 438
Query: 409 GSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGV 468
G IG+VL N G +++A +H+LP + +G+ + Y +K A + T +G+
Sbjct: 439 GGIGMVLTNTATNGEELVADSHMLPAVAVGEKEGKLIKQYAMTSKKATASLEILGTRIGI 498
Query: 469 KPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSG 528
KP+PV+A+ SSRGPN + ILKPD+ APGV+IL A+ G ++P+ L+SD + + +NI SG
Sbjct: 499 KPSPVVAAFSSRGPNFLSLEILKPDLLAPGVNILAAWTGDMAPSSLSSDPRRVKFNILSG 558
Query: 529 TSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQS-KEDATPFGYG 587
TS+SCPHVS + AL+K+ +P+WSPAA KSA+MTT + N +P+ D S ++P+ +G
Sbjct: 559 TSMSCPHVSGVAALIKSRHPDWSPAAIKSALMTTAYVHDNMFKPLTDASGAAPSSPYDHG 618
Query: 588 AGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYICPK---SYNMLDFN 644
AGHI P A DPGLVYD+ +Y FLC + +Q+K+F++ K + N + N
Sbjct: 619 AGHIDPLRATDPGLVYDIGPQEYFEFLCTQDLSPSQLKVFTKHSNRTCKHTLAKNPGNLN 678
Query: 645 YPSITVPNLGKHFVQEVT--RTVTNVGSP-GTYRVQVNEPHGIFVLIKPRSLTFNEVGEK 701
YP+I+ V+ +T RTVTNVG +Y+V V+ G V ++P++L F +K
Sbjct: 679 YPAISALFPENTHVKAMTLRRTVTNVGPHISSYKVSVSPFKGASVTVQPKTLNFTSKHQK 738
Query: 702 KTFKIIFKVTKPTSSGYVFGHLLWSDGRHKVMSPLVV 738
++ + F+ T+ FG L+W HKV SP+++
Sbjct: 739 LSYTVTFR-TRFRMKRPEFGGLVWKSTTHKVRSPVII 774
>At1g32960 unknown protein
Length = 777
Score = 481 bits (1237), Expect = e-136
Identities = 291/762 (38%), Positives = 435/762 (56%), Gaps = 52/762 (6%)
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
+IVY+G H D + T+SH+ +L S LGS + A +++ YSY +GFAA L
Sbjct: 33 HIVYLGEKKH------HDPEFVTESHHQMLASLLGSKKDADDSMVYSYRHGFSGFAAKLT 86
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIA 121
+A KIA P VV V + HEL TTR+WE+LGL + PK+ + + G+ II
Sbjct: 87 KSQAKKIADLPEVVHVIPDGFHELATTRTWEYLGLSS---ANPKNLLNDTNM-GDQVIIG 142
Query: 122 NIDSGVSPESKSFSDDGMGPVPSRWRGICQL-DNFH---CNRKLIGARFYSQGYESKFGR 177
ID+GV PES+SF+D+G+GP+P +W+G C+ +NF CNRKLIGA+++ G+ ++
Sbjct: 143 VIDTGVWPESESFNDNGVGPIPRKWKGGCESGENFRSTDCNRKLIGAKYFINGFLAENKG 202
Query: 178 LN----QSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCW 233
N + +ARD GHGT S+AGG+FV + GLA GT +GG+PR+ +A YK CW
Sbjct: 203 FNTTESRDYISARDFDGHGTHVASIAGGSFVPNVSYKGLAGGTLRGGAPRARIAMYKACW 262
Query: 234 LGTI*--IECTDADIMQAFEDAISDGVDIISCSLGQTSP---KEFFEDGISIGAFHAIEN 288
+ C+D+DIM+A ++AI DGVD++S SL P + D + G FHA+
Sbjct: 263 FHEELKGVTCSDSDIMKAIDEAIHDGVDVLSISLVGQIPLNSETDIRDEFATGLFHAVAK 322
Query: 289 GVIVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPN 348
G++VV GGN GP TV N+APW+ +VAA+T+DR+F + + LG+ +I+G + TG P
Sbjct: 323 GIVVVCAGGNDGPAAQTVVNIAPWILTVAATTLDRSFPTPITLGNNKVILGQATYTG-PE 381
Query: 349 EKFYSLVSSVDAKVGNATIEDAKICKVGSLDPN---KVKGKILFCLLRELDGLVYAEEEA 405
SLV +A+ N T + +C+ +L+PN +K + F R + A
Sbjct: 382 LGLTSLVYPENARNNNETF--SGVCESLNLNPNYTMAMKVVLCFTASRTNAAISRAASFV 439
Query: 406 ISGGSIGLVLGNDKQRGNDIMAYAHL---LPTSHINYTDGEYVHSYIKATKTPMAYMTKA 462
+ G +GL++ N + + P ++Y G + SYI++T++P+ + ++
Sbjct: 440 KAAGGLGLIISR-----NPVYTLSPCNDDFPCVAVDYELGTDILSYIRSTRSPVVKIQRS 494
Query: 463 KTEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIP 522
+T G + + SSRGPN + P ILKPDI APGV IL A SP +
Sbjct: 495 RTLSGQPVGTKVVNFSSRGPNSMSPAILKPDIAAPGVRIL----AATSPNDTLNVG---G 547
Query: 523 YNIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTT--TIQGNNHRPIKDQSKED 580
+ + SGTS++ P +S ++ALLK ++P WSPAAF+SAI+TT T + S++
Sbjct: 548 FAMLSGTSMATPVISGVIALLKALHPEWSPAAFRSAIVTTAWRTDPFGEQIFAEGSSRKV 607
Query: 581 ATPFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYIC--PKSY 638
+ PF YG G + PE A +PGL+YD+ DY+ +LC+ GYN + + + +C PK
Sbjct: 608 SDPFDYGGGIVNPEKAAEPGLIYDMGPQDYILYLCSAGYNDSSISQLVGQITVCSNPKP- 666
Query: 639 NMLDFNYPSITVPNLGKHFVQEVTRTVTNVG-SPGTYRVQVNEPHGIFVLIKPRSLTFNE 697
++LD N PSIT+PNL +TRTVTNVG Y+V V P G+ V++ P +L FN
Sbjct: 667 SVLDVNLPSITIPNLKDEVT--LTRTVTNVGLVDSVYKVSVEPPLGVRVVVTPETLVFNS 724
Query: 698 VGEKKTFKIIFKVTKPTSSGYVFGHLLWSDGRHKVMSPLVVK 739
+F + T ++GY FG L W+D H V+ PL V+
Sbjct: 725 KTISVSFTVRVSTTHKINTGYYFGSLTWTDSVHNVVIPLSVR 766
>At5g03620 cucumisin precursor -like protein
Length = 766
Score = 479 bits (1234), Expect = e-135
Identities = 307/767 (40%), Positives = 433/767 (56%), Gaps = 73/767 (9%)
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
YIVY+G + + L A ++H+NLL + +G KA+E YSY K+INGF A L
Sbjct: 35 YIVYMGEATE------NSLVEAAENHHNLLMTVIGDESKARELKIYSYGKNINGFVARLF 88
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGL-ENNYGVVPKDSIWEKGRYGEGTII 120
EA K+++ VVSVF+N +L TTRSW+FLGL E+ Y K S+ + I+
Sbjct: 89 PHEAEKLSREEGVVSVFKNTQRQLHTTRSWDFLGLVESKY----KRSVGIESNI----IV 140
Query: 121 ANIDSGVSPESKSFSDDGMGPVPSRWRGICQLDN--FHCNRKLIGARFY---SQGYESKF 175
+D+G+ ES SF+D G+GP P++W+G C N CN K+IGA+++ S+G
Sbjct: 141 GVLDTGIDVESPSFNDKGVGPPPAKWKGKCVTGNNFTRCNNKVIGAKYFHIQSEGLPDGE 200
Query: 176 GRLNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLG 235
G A D GHGT T S G VS A++FG+ANGTA+GG P + +AAYKVCW
Sbjct: 201 GD------TAADHDGHGTHTSSTIAGVSVSSASLFGIANGTARGGVPSARIAAYKVCWDS 254
Query: 236 TI*IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAG 295
CTD D++ AF++AISDGVDIIS S+G S FFED I+IGAFHA++ G++
Sbjct: 255 G----CTDMDMLAAFDEAISDGVDIISISIGGAS-LPFFEDPIAIGAFHAMKRGILTTCS 309
Query: 296 GGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEKFYSLV 355
GN+GP TV+N+APW+ +VAA+++DR F + ++LG+ G SL+ P +K Y L
Sbjct: 310 AGNNGPGLFTVSNLAPWVMTVAANSLDRKFETVVKLGNGLTASGISLNGFNPRKKMYPLT 369
Query: 356 S-SVDAKVGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLV 414
S S+ + + + C+ G+L +KV GK+++C EE G V
Sbjct: 370 SGSLASNLSAGGYGEPSTCEPGTLGEDKVMGKVVYCEAGR-------EEGGNGGQGQDHV 422
Query: 415 LGNDKQRGNDI-------MAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVG 467
+ + K G + MA + L+ S++ + DG + YI +TK P A + K KT
Sbjct: 423 VRSLKGAGVIVQLLEPTDMATSTLIAGSYVFFEDGTKITEYINSTKNPQAVIFKTKTTKM 482
Query: 468 VKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGS 527
+ AP I+S S+RGP I P ILKPDI+APG++IL AY S TG DN+ ++I S
Sbjct: 483 L--APSISSFSARGPQRISPNILKPDISAPGLNILAAYSKLASVTGYPDDNRRTLFSIMS 540
Query: 528 GTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKEDATPFGYG 587
GTS++CPH +A A +K+ +P+WSPAA KSA+MTT T P++ + E YG
Sbjct: 541 GTSMACPHAAAAAAYVKSFHPDWSPAAIKSALMTTAT-------PMRIKGNE--AELSYG 591
Query: 588 AGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMF--------SRKPYICPKSYN 639
+G I P A+ PGLVYD+ YL FLC GYN T + + ++K Y C
Sbjct: 592 SGQINPRRAIHPGLVYDITEDAYLRFLCKEGYNSTSIGLLTGDNSNNTTKKEYNCENIKR 651
Query: 640 ML---DFNYPSI-TVPNLGKHFVQEV-TRTVTNVG-SPGTYRVQVNEPHGIFVLIKPRSL 693
L NYPS+ N + V EV RTVTNVG P TY +V P G+ V + P+ +
Sbjct: 652 GLGSDGLNYPSLHKQVNSTEAKVSEVFYRTVTNVGYGPSTYVARVWAPKGLRVEVVPKVM 711
Query: 694 TFNEVGEKKTFKIIFK-VTKPTSSGYVFGHLLWSDGR-HKVMSPLVV 738
+F EK+ FK++ V T G V + W D R H V SP+++
Sbjct: 712 SFERPKEKRNFKVVIDGVWDETMKGIVSASVEWDDSRGHLVRSPILL 758
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,977,659
Number of Sequences: 26719
Number of extensions: 832353
Number of successful extensions: 2440
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 61
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 1814
Number of HSP's gapped (non-prelim): 87
length of query: 740
length of database: 11,318,596
effective HSP length: 107
effective length of query: 633
effective length of database: 8,459,663
effective search space: 5354966679
effective search space used: 5354966679
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC147712.15


