

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147712.11 + phase: 0
(476 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
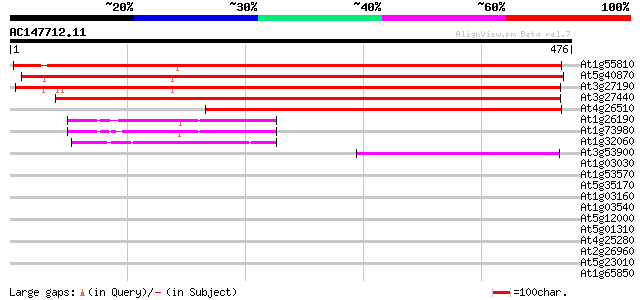
Score E
Sequences producing significant alignments: (bits) Value
At1g55810 uracil phosphoribosyltransferase 1, putative 777 0.0
At5g40870 uridine kinase-like protein 721 0.0
At3g27190 uracil phosphoribosyltransferase, putative 721 0.0
At3g27440 uridine kinase/uracil phosphoribosyl transferas, putative 655 0.0
At4g26510 uracil phosphoribosyl transferase like protein 569 e-162
At1g26190 unknown protein (At1g26190) 84 2e-16
At1g73980 unknown protein 78 1e-14
At1g32060 phosphoribulokinase (EC 2.7.1.19) precursor like protein 72 9e-13
At3g53900 uracil phosphoribosyltransferase-like protein 69 4e-12
At1g03030 unknown protein 39 0.005
At1g53570 MEK kinase MAP3Ka, putative 35 0.12
At5g35170 adenylate kinase -like protein 30 2.2
At1g03160 hypothetical protein 30 2.2
At1g03540 hypothetical protein 29 5.0
At5g12000 putative receptor - like kinase 29 6.5
At5g01310 putative protein 29 6.5
At4g25280 UMP/CMP kinase like protein 29 6.5
At2g26960 putative transcription factor (MYB81) 29 6.5
At5g23010 2-isopropylmalate synthase-like; homocitrate synthase-... 28 8.5
At1g65850 28 8.5
>At1g55810 uracil phosphoribosyltransferase 1, putative
Length = 466
Score = 777 bits (2007), Expect = 0.0
Identities = 385/468 (82%), Positives = 424/468 (90%), Gaps = 7/468 (1%)
Query: 4 AKSAVDLMGSSSEVHFSGFH-MDGFEKREASIEQPTTSATDVYNQPFVIGVAGGAASGKK 62
+KS V+++ +SS+VHFSGFH MDG AS + + + QPFVIGVAGGAASGK
Sbjct: 3 SKSDVNIIETSSKVHFSGFHQMDGL----ASNRPEQMAEEEEHGQPFVIGVAGGAASGKT 58
Query: 63 TVCDMIVQQLHDQRVVLVNQDSFYNNLTEEERARVQDYNFDHPDAFDTEELLRVMDKLKH 122
TVCDMI+QQLHDQR V+VNQDSFY+N+ E E RV DYNFDHPDAFDTE+LL M+KL+
Sbjct: 59 TVCDMIMQQLHDQRAVVVNQDSFYHNVNEVELVRVHDYNFDHPDAFDTEQLLSSMEKLRK 118
Query: 123 GEAVDIPKYDFKSYKSDAL--RRVNPSDVIILEGILVFHDPRVRELMNMKIFVDTDADVR 180
G+AVDIP YDFKSYK++ RRVNPSDVIILEGIL+FHDPRVR+LMNMKIFVD DADVR
Sbjct: 119 GQAVDIPNYDFKSYKNNVFPPRRVNPSDVIILEGILIFHDPRVRDLMNMKIFVDADADVR 178
Query: 181 LARRIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPTKKYADIIIPRGGDNHVAVDLIV 240
LARRI+RDT EK RDI +LDQYSKFVKPAF+DFILPTKKYADIIIPRGGDNHVA+DLIV
Sbjct: 179 LARRIKRDTVEKGRDIATVLDQYSKFVKPAFEDFILPTKKYADIIIPRGGDNHVAIDLIV 238
Query: 241 QHIRTKLGQHDLCKIYPNLYVIHSTFQIRGMHTLIRDAQITKHDFVFYSDRLIRLVVEHG 300
QHI TKLGQHDLCKIYPNLYVI STFQIRGMHTLIRD++ TKHDF+FYSDRLIRLVVEHG
Sbjct: 239 QHIHTKLGQHDLCKIYPNLYVIQSTFQIRGMHTLIRDSKTTKHDFIFYSDRLIRLVVEHG 298
Query: 301 LGHLPFTEKQVITPTGSVYTGVDFCKRLCGVSVIRSGESMENALRACCKGIKIGKILIHR 360
LGHLPFTEKQV+TPTGSVY+GVDFCK+LCGVSVIRSGESMENALRACCKGIKIGKILIHR
Sbjct: 299 LGHLPFTEKQVVTPTGSVYSGVDFCKKLCGVSVIRSGESMENALRACCKGIKIGKILIHR 358
Query: 361 EGDNGQQLIYEKLPNDISERHVLLLDPILGTGNSAVQAISLLIQKGVPESNIIFLNLISA 420
EGDNGQQLIYEKLP+DISERHVLLLDPILGTGNSAVQAI LLI KGVPESNIIFLNLISA
Sbjct: 359 EGDNGQQLIYEKLPSDISERHVLLLDPILGTGNSAVQAIRLLISKGVPESNIIFLNLISA 418
Query: 421 PKGVHVVCKCFPRIKIVTSEIEIGLNEDFRVIPGMGEFGDRYFGTDDD 468
P+GV+VVCK FPRIKIVTSEIE+GLN++FRV+PGMGEFGDRYFGTDD+
Sbjct: 419 PEGVNVVCKKFPRIKIVTSEIELGLNDEFRVVPGMGEFGDRYFGTDDE 466
>At5g40870 uridine kinase-like protein
Length = 486
Score = 721 bits (1860), Expect = 0.0
Identities = 353/475 (74%), Positives = 402/475 (84%), Gaps = 15/475 (3%)
Query: 11 MGSSSEVHFSGFHMDGFE-------------KREASIEQPTTSATDVYNQPFVIGVAGGA 57
M +S HFSG DG + S P++S + QPF+IGV+GG
Sbjct: 12 MEKASGPHFSGLRFDGLLSSSPPNSSVVSSLRSAVSSSSPSSSDPEAPKQPFIIGVSGGT 71
Query: 58 ASGKKTVCDMIVQQLHDQRVVLVNQDSFYNNLTEEERARVQDYNFDHPDAFDTEELLRVM 117
ASGK TVCDMI+QQLHD RVVLVNQDSFY LT EE RVQ+YNFDHPDAFDTE+LL
Sbjct: 72 ASGKTTVCDMIIQQLHDHRVVLVNQDSFYRGLTSEELQRVQEYNFDHPDAFDTEQLLHCA 131
Query: 118 DKLKHGEAVDIPKYDFKSY--KSDALRRVNPSDVIILEGILVFHDPRVRELMNMKIFVDT 175
+ LK G+ +P YDFK++ +SD R+VN SDVIILEGILVFHD RVR LMNMKIFVDT
Sbjct: 132 ETLKSGQPYQVPIYDFKTHQRRSDTFRQVNASDVIILEGILVFHDSRVRNLMNMKIFVDT 191
Query: 176 DADVRLARRIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPTKKYADIIIPRGGDNHVA 235
DADVRLARRIRRDT E+ RD+ ++L+QY+KFVKPAFDDF+LP+KKYAD+IIPRGGDNHVA
Sbjct: 192 DADVRLARRIRRDTVERGRDVNSVLEQYAKFVKPAFDDFVLPSKKYADVIIPRGGDNHVA 251
Query: 236 VDLIVQHIRTKLGQHDLCKIYPNLYVIHSTFQIRGMHTLIRDAQITKHDFVFYSDRLIRL 295
VDLI QHI TKLGQHDLCKIYPN+YVI STFQIRGMHTLIR+ I+KHDFVFYSDRLIRL
Sbjct: 252 VDLITQHIHTKLGQHDLCKIYPNVYVIQSTFQIRGMHTLIREKDISKHDFVFYSDRLIRL 311
Query: 296 VVEHGLGHLPFTEKQVITPTGSVYTGVDFCKRLCGVSVIRSGESMENALRACCKGIKIGK 355
VVEHGLGHLPFTEKQV+TPTG+VYTGVDFCK+LCGVS+IRSGESMENALRACCKGIKIGK
Sbjct: 312 VVEHGLGHLPFTEKQVVTPTGAVYTGVDFCKKLCGVSIIRSGESMENALRACCKGIKIGK 371
Query: 356 ILIHREGDNGQQLIYEKLPNDISERHVLLLDPILGTGNSAVQAISLLIQKGVPESNIIFL 415
ILIHR+GDNG+QLIYEKLP+DISERHVLLLDP+L TGNSA QAI LLIQKGVPE++IIFL
Sbjct: 372 ILIHRDGDNGKQLIYEKLPHDISERHVLLLDPVLATGNSANQAIELLIQKGVPEAHIIFL 431
Query: 416 NLISAPKGVHVVCKCFPRIKIVTSEIEIGLNEDFRVIPGMGEFGDRYFGTDDDEQ 470
NLISAP+G+H VCK FP +KIVTSEI+ LN++FRVIPG+GEFGDRYFGTD+++Q
Sbjct: 432 NLISAPEGIHCVCKRFPALKIVTSEIDQCLNQEFRVIPGLGEFGDRYFGTDEEDQ 486
>At3g27190 uracil phosphoribosyltransferase, putative
Length = 483
Score = 721 bits (1860), Expect = 0.0
Identities = 360/478 (75%), Positives = 410/478 (85%), Gaps = 16/478 (3%)
Query: 6 SAVD-LMGSSSEVHFSGFHMDGF--EKREASIEQPTT--------SATD---VYNQPFVI 51
+A+D +M +S HFSG +DG ++S+ P+ SATD +QPFVI
Sbjct: 6 TAIDYVMEKASGPHFSGLRLDGLLSSPSKSSVSSPSHFRLSNSSFSATDDPAAPHQPFVI 65
Query: 52 GVAGGAASGKKTVCDMIVQQLHDQRVVLVNQDSFYNNLTEEERARVQDYNFDHPDAFDTE 111
GV GG ASGK TVCDMI+QQLHD R+VLVNQDSFY LT EE VQ+YNFDHPDAFDTE
Sbjct: 66 GVTGGTASGKTTVCDMIIQQLHDHRIVLVNQDSFYRGLTSEELEHVQEYNFDHPDAFDTE 125
Query: 112 ELLRVMDKLKHGEAVDIPKYDFKSY--KSDALRRVNPSDVIILEGILVFHDPRVRELMNM 169
+LL +D LK G+ IP YDFK++ K DA R+VN DVIILEGILVFHD RVR+LMNM
Sbjct: 126 QLLHCVDILKSGQPYQIPIYDFKTHQRKVDAFRQVNACDVIILEGILVFHDSRVRDLMNM 185
Query: 170 KIFVDTDADVRLARRIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPTKKYADIIIPRG 229
KIFVDTDADVRLARRIRRDT E+ RD+ ++L+QY+KFVKPAFDDF+LP+KKYAD+IIPRG
Sbjct: 186 KIFVDTDADVRLARRIRRDTVERGRDVDSVLEQYAKFVKPAFDDFVLPSKKYADVIIPRG 245
Query: 230 GDNHVAVDLIVQHIRTKLGQHDLCKIYPNLYVIHSTFQIRGMHTLIRDAQITKHDFVFYS 289
GDNHVAVDLIVQHI TKLGQHDLCKIYPN++VI +TFQIRGMHTLIR+ I+KHDFVFYS
Sbjct: 246 GDNHVAVDLIVQHIHTKLGQHDLCKIYPNVFVIETTFQIRGMHTLIREKDISKHDFVFYS 305
Query: 290 DRLIRLVVEHGLGHLPFTEKQVITPTGSVYTGVDFCKRLCGVSVIRSGESMENALRACCK 349
DRLIRLVVEHGLGHLPFTEKQV+TPTGSVY+GVDFCK+LCGVSVIRSGESMENALRACCK
Sbjct: 306 DRLIRLVVEHGLGHLPFTEKQVVTPTGSVYSGVDFCKKLCGVSVIRSGESMENALRACCK 365
Query: 350 GIKIGKILIHREGDNGQQLIYEKLPNDISERHVLLLDPILGTGNSAVQAISLLIQKGVPE 409
GIKIGKILIHR+GDNG QLIYEKLP+DISERHVLLLDP+LGTGNSA QAI LLIQKGVPE
Sbjct: 366 GIKIGKILIHRDGDNGMQLIYEKLPSDISERHVLLLDPVLGTGNSANQAIELLIQKGVPE 425
Query: 410 SNIIFLNLISAPKGVHVVCKCFPRIKIVTSEIEIGLNEDFRVIPGMGEFGDRYFGTDD 467
++IIFLNLISAP+G+H VCK FP++KIVTSEI+ LNE+FRVIPG+GEFGDRYFGTD+
Sbjct: 426 AHIIFLNLISAPEGIHCVCKRFPKLKIVTSEIDQCLNEEFRVIPGLGEFGDRYFGTDE 483
>At3g27440 uridine kinase/uracil phosphoribosyl transferas, putative
Length = 465
Score = 655 bits (1690), Expect = 0.0
Identities = 320/429 (74%), Positives = 376/429 (87%), Gaps = 1/429 (0%)
Query: 40 SATDVYNQPFVIGVAGGAASGKKTVCDMIVQQLHDQRVVLVNQDSFYNNLTEEERARVQD 99
SA QPFVIGVAGG ASGK TVC+MI+ QLHDQRVVLVNQDSFY++LT+E+ +V +
Sbjct: 21 SAPAPLKQPFVIGVAGGTASGKTTVCNMIMSQLHDQRVVLVNQDSFYHSLTKEKLNKVHE 80
Query: 100 YNFDHPDAFDTEELLRVMDKLKHGEAVDIPKYDFKSYKS-DALRRVNPSDVIILEGILVF 158
YNFDHPDAF+TE LL M+KL+ G+ V+IP YDFK ++S ++ VNP DVIILEGILV
Sbjct: 81 YNFDHPDAFNTEVLLSCMEKLRSGQPVNIPSYDFKIHQSIESSSPVNPGDVIILEGILVL 140
Query: 159 HDPRVRELMNMKIFVDTDADVRLARRIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPT 218
+DPRVR+LMNMKIFVDTDADVRL+RRI+RDT E+ R+I +L+QY+KFVKP+FD++I P+
Sbjct: 141 NDPRVRDLMNMKIFVDTDADVRLSRRIQRDTVERGRNIQNVLEQYTKFVKPSFDEYIQPS 200
Query: 219 KKYADIIIPRGGDNHVAVDLIVQHIRTKLGQHDLCKIYPNLYVIHSTFQIRGMHTLIRDA 278
KYADIIIPRGGDN VA+DLIVQHIRTKL QH+LCKIY N+++I STFQI+GMHTLIRD
Sbjct: 201 MKYADIIIPRGGDNDVAIDLIVQHIRTKLCQHNLCKIYSNIFIISSTFQIKGMHTLIRDI 260
Query: 279 QITKHDFVFYSDRLIRLVVEHGLGHLPFTEKQVITPTGSVYTGVDFCKRLCGVSVIRSGE 338
TKHDFVFY+DRLIRLVVEHGLGHLPFTEKQ+ TPTGSVYTGVDFCKRLCGVSVIRSGE
Sbjct: 261 NTTKHDFVFYADRLIRLVVEHGLGHLPFTEKQITTPTGSVYTGVDFCKRLCGVSVIRSGE 320
Query: 339 SMENALRACCKGIKIGKILIHREGDNGQQLIYEKLPNDISERHVLLLDPILGTGNSAVQA 398
SMENALRACC GIKIGKILIHRE ++G+QLIYEKLP DIS RHV LLDP+L +G SAV+A
Sbjct: 321 SMENALRACCNGIKIGKILIHRENNDGRQLIYEKLPKDISSRHVFLLDPVLASGYSAVKA 380
Query: 399 ISLLIQKGVPESNIIFLNLISAPKGVHVVCKCFPRIKIVTSEIEIGLNEDFRVIPGMGEF 458
I+LL+ KGVPES+IIFLNLI+AP+G+H +CK FP +KIVTSEI+ LNED RVIPG+GEF
Sbjct: 381 ITLLLSKGVPESHIIFLNLIAAPQGIHALCKKFPMLKIVTSEIDSSLNEDSRVIPGLGEF 440
Query: 459 GDRYFGTDD 467
DRYFGT++
Sbjct: 441 ADRYFGTNN 449
>At4g26510 uracil phosphoribosyl transferase like protein
Length = 302
Score = 569 bits (1467), Expect = e-162
Identities = 279/302 (92%), Positives = 290/302 (95%)
Query: 167 MNMKIFVDTDADVRLARRIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPTKKYADIII 226
MNMKIFV TDADVRLARRI+RDT E RDIG +LDQYSKFVKPAFDDFILPTKKYADIII
Sbjct: 1 MNMKIFVCTDADVRLARRIKRDTVENGRDIGTVLDQYSKFVKPAFDDFILPTKKYADIII 60
Query: 227 PRGGDNHVAVDLIVQHIRTKLGQHDLCKIYPNLYVIHSTFQIRGMHTLIRDAQITKHDFV 286
PRGGDNHVA+DLIVQHI TKLGQHDLCKIYPNLYVIHSTFQIRGMHTLIRD+Q TKHDFV
Sbjct: 61 PRGGDNHVAIDLIVQHICTKLGQHDLCKIYPNLYVIHSTFQIRGMHTLIRDSQTTKHDFV 120
Query: 287 FYSDRLIRLVVEHGLGHLPFTEKQVITPTGSVYTGVDFCKRLCGVSVIRSGESMENALRA 346
FYSDRLIRLVVEHGLGHLPFTEKQVITPTG VY+GVDFCKRLCGVSVIRSGESMENALRA
Sbjct: 121 FYSDRLIRLVVEHGLGHLPFTEKQVITPTGCVYSGVDFCKRLCGVSVIRSGESMENALRA 180
Query: 347 CCKGIKIGKILIHREGDNGQQLIYEKLPNDISERHVLLLDPILGTGNSAVQAISLLIQKG 406
CCKGIKIGKILIHREGDNGQQL+YEKLPNDISERHVLLLDPILGTGNSAV+AI+LLI KG
Sbjct: 181 CCKGIKIGKILIHREGDNGQQLVYEKLPNDISERHVLLLDPILGTGNSAVEAINLLISKG 240
Query: 407 VPESNIIFLNLISAPKGVHVVCKCFPRIKIVTSEIEIGLNEDFRVIPGMGEFGDRYFGTD 466
VPE NIIFLNLISAP+GVHVVCK FPRIKIVTSEI+ GLNE+FRVIPGMGEFGDRYFGTD
Sbjct: 241 VPEGNIIFLNLISAPQGVHVVCKKFPRIKIVTSEIDNGLNEEFRVIPGMGEFGDRYFGTD 300
Query: 467 DD 468
DD
Sbjct: 301 DD 302
>At1g26190 unknown protein (At1g26190)
Length = 674
Score = 84.0 bits (206), Expect = 2e-16
Identities = 55/179 (30%), Positives = 94/179 (51%), Gaps = 11/179 (6%)
Query: 50 VIGVAGGAASGKKTVCDMIVQQLHDQRVVLVNQDSFYNNLTEEERARVQDYNFDHPDAFD 109
++GVAG + +GK + I+ L V +++ D++ + +R+ D NFD P D
Sbjct: 67 LVGVAGPSGAGKTVFTEKILNFLPS--VAVISMDNY------NDSSRIVDGNFDDPRLTD 118
Query: 110 TEELLRVMDKLKHGEAVDIPKYDFKSYKSDALRR--VNPSDVIILEGILVFHDPRVRELM 167
+ LL+ ++ LK G+ V++P YDFKS R V PS ++I+EGI + ++R L+
Sbjct: 119 YDTLLKNLEDLKEGKQVEVPIYDFKSSSRVGYRTLDVPPSRIVIIEGIYALSE-KLRPLL 177
Query: 168 NMKIFVDTDADVRLARRIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPTKKYADIII 226
++++ V L +R+ RD + I+ Q S+ V P + FI P + A I I
Sbjct: 178 DLRVSVTGGVHFDLVKRVLRDIQRAGQQPEEIIHQISETVYPMYKAFIEPDLQTAQIKI 236
>At1g73980 unknown protein
Length = 643
Score = 77.8 bits (190), Expect = 1e-14
Identities = 57/179 (31%), Positives = 92/179 (50%), Gaps = 11/179 (6%)
Query: 50 VIGVAGGAASGKKTVCDMIVQQLHDQRVVLVNQDSFYNNLTEEERARVQDYNFDHPDAFD 109
++G+AG + +GK + I+ + + ++N D+ YN+ T RV D NFD P D
Sbjct: 67 LVGLAGPSGAGKTIFTEKILNFMPS--IAIINMDN-YNDGT-----RVIDGNFDDPRLTD 118
Query: 110 TEELLRVMDKLKHGEAVDIPKYDFKSYKSDALR--RVNPSDVIILEGILVFHDPRVRELM 167
+ LL + L+ G+ V +P YDFKS R V S ++ILEGI + ++R L+
Sbjct: 119 YDTLLDNIHGLRDGKPVQVPIYDFKSSSRIGYRTLEVPSSRIVILEGIYALSE-KLRPLL 177
Query: 168 NMKIFVDTDADVRLARRIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPTKKYADIII 226
++++ V L +R+ RD ++ I+ Q S+ V P + FI P K A I I
Sbjct: 178 DLRVSVTGGVHFDLVKRVLRDIQRAGQEPEEIIHQISETVYPMYKAFIEPDLKTAQIKI 236
>At1g32060 phosphoribulokinase (EC 2.7.1.19) precursor like protein
Length = 395
Score = 71.6 bits (174), Expect = 9e-13
Identities = 53/174 (30%), Positives = 87/174 (49%), Gaps = 4/174 (2%)
Query: 53 VAGGAASGKKTVCDMIVQQLHDQRVVLVNQDSFYNNLTEEERARVQDYNFDHPDAFDTEE 112
V GGAA K + D V+ D Y++L R + D P A D +
Sbjct: 73 VFGGAAKPPKGGNPDSNTLISDTTTVICLDD--YHSLDRYGRKEQKVTALD-PRANDFDL 129
Query: 113 LLRVMDKLKHGEAVDIPKYDFKSYKSDALRRVNPSDVIILEGILVFHDPRVRELMNMKIF 172
+ + LK+G AV+ P Y+ + D + P ++++EG+ D RVR+L++ I+
Sbjct: 130 MYEQVKALKNGIAVEKPIYNHVTGLLDPPELIQPPKILVIEGLHPMFDERVRDLLDFSIY 189
Query: 173 VDTDADVRLARRIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPTKKYADIII 226
+D +V+ A +I+RD +E+ + +I + KP FD FI P K+YAD +I
Sbjct: 190 LDISNEVKFAWKIQRDMAERGHSLESIKASI-EARKPDFDAFIDPQKQYADAVI 242
>At3g53900 uracil phosphoribosyltransferase-like protein
Length = 296
Score = 69.3 bits (168), Expect = 4e-12
Identities = 47/175 (26%), Positives = 89/175 (50%), Gaps = 3/175 (1%)
Query: 295 LVVEHGLGHLPFTEKQVITPTGSVYTG-VDFCKRLCGVSVIRSGESMENALRACCKGIKI 353
L+ E LP ++++P G +D + + V ++R+G ++ + KI
Sbjct: 117 LMYEASREWLPTVVGEIMSPMGPASVEFIDPREPIAVVPILRAGLALAEHASSVLPANKI 176
Query: 354 GKILIHREGDNGQQLIY-EKLPNDISER-HVLLLDPILGTGNSAVQAISLLIQKGVPESN 411
+ + R+ +Y KLP++ + V L+DP+L TG + + A+ LL ++G+
Sbjct: 177 YHLGVSRDEKTLLPSVYLNKLPDEFPKNSRVFLVDPVLATGGTIMAAMDLLKERGLSVQQ 236
Query: 412 IIFLNLISAPKGVHVVCKCFPRIKIVTSEIEIGLNEDFRVIPGMGEFGDRYFGTD 466
I + I+AP + + + FP + + I+ +NE +IPG+G+ GDR FGT+
Sbjct: 237 IKVICAIAAPPALSKLNEKFPGLHVYAGIIDPEVNEKGYIIPGLGDAGDRSFGTE 291
>At1g03030 unknown protein
Length = 333
Score = 39.3 bits (90), Expect = 0.005
Identities = 53/232 (22%), Positives = 97/232 (40%), Gaps = 29/232 (12%)
Query: 21 GFHMDGFEKREASIEQPTTSATDVYNQPFVIGVAGGAASGKKTVCDMIVQQLH------- 73
G MD + A PT +A N ++G+AG +GK TV + +V++++
Sbjct: 100 GSCMDEIYDKLAERLVPTAAAMFSPNLKRLVGLAGPPGAGKSTVANEVVRRVNKLWPQKA 159
Query: 74 ---DQRV------VLVNQDSFY---NNLTEEERARVQDYNFDHPDAFDTEELLRVMDKLK 121
D V +++ D F+ + L E + P FD LL + KLK
Sbjct: 160 ASFDAEVNPPDVAIVLPMDGFHLYRSQLDAMEDPKEAHARRGAPWTFDPALLLNCLKKLK 219
Query: 122 HGEAVDIPKYDF---KSYKSDALRRVNPSDVIILEGILVFHDPRVRELMNM---KIFVDT 175
+ +V +P +D + D + VI+ ++ + +++ +M K F+D
Sbjct: 220 NEGSVYVPSFDHGVGDPVEDDIFVSLQHKVVIVEGNYILLEEGSWKDISDMFDEKWFIDV 279
Query: 176 DADVRLARRIRRDTSE-KDRDIGAILDQYSKFVKPAFDDFILPTKKYADIII 226
+ D + R R S K D+ Y+ +P + I+ +K AD++I
Sbjct: 280 NLDTAMQRVENRHISTGKPPDVAKWRVDYND--RPN-AELIIKSKTNADLLI 328
>At1g53570 MEK kinase MAP3Ka, putative
Length = 609
Score = 34.7 bits (78), Expect = 0.12
Identities = 19/58 (32%), Positives = 30/58 (50%), Gaps = 2/58 (3%)
Query: 2 GTAKSAVDLMGSSSEVHFSGFHMDGFEKREASIEQPTTSATDVYNQPFVIGVAGGAAS 59
GT + + + G+SS FSGF D EK+ + +P S +++Q V G G+ S
Sbjct: 44 GTPRCSREFAGASSA--FSGFDSDSTEKKGHPLPRPLLSPVSIHHQDHVSGSTSGSTS 99
>At5g35170 adenylate kinase -like protein
Length = 588
Score = 30.4 bits (67), Expect = 2.2
Identities = 43/190 (22%), Positives = 80/190 (41%), Gaps = 22/190 (11%)
Query: 46 NQPFVIGVAGGAASGKKTVCDMIVQQLHDQRVVLVNQDSFYNNLTEEERARVQDYNFDHP 105
N+P + ++G ASGK T C++IV + V + D ++ + F +
Sbjct: 77 NEPLKVMISGAPASGKGTQCELIVHKF--GLVHISTGDLLRAEVSSGTDIGKRAKEFMNS 134
Query: 106 DAFDTEELLRVM--------DKLKHGEAVDIPKYDFKSYKSDALRRVNPSDVIIL---EG 154
+ +E++ M D +HG +D F +S V P I+L +
Sbjct: 135 GSLVPDEIVIAMVAGRLSREDAKEHGWLLDGFPRSFAQAQSLDKLNVKPDIFILLDVPDE 194
Query: 155 ILVFH------DPRVRELMNMKIFVDTDADVRLARRIRR--DTSEKDRDIGAILDQYSKF 206
IL+ DP ++ ++K + +++ AR + R DT EK + I Q S+
Sbjct: 195 ILIDRCVGRRLDPVTGKIYHIKNYPPESDEIK-ARLVTRPDDTEEKVKARLQIYKQNSEA 253
Query: 207 VKPAFDDFIL 216
+ A+ D ++
Sbjct: 254 IISAYSDVMV 263
>At1g03160 hypothetical protein
Length = 642
Score = 30.4 bits (67), Expect = 2.2
Identities = 21/65 (32%), Positives = 36/65 (55%), Gaps = 8/65 (12%)
Query: 46 NQPFVIGVAGGAASGKKTVCDMIV--QQLHDQRVVLVNQDSF--YNNLTEEERARVQDYN 101
++PF++ + G SGK TV + ++ + L + V N+ +F Y++L EE+ R Q
Sbjct: 347 DEPFLMVIVGEFNSGKSTVINALLGKRYLKEGVVPTTNEITFLCYSDLESEEQQRCQ--- 403
Query: 102 FDHPD 106
HPD
Sbjct: 404 -THPD 407
>At1g03540 hypothetical protein
Length = 609
Score = 29.3 bits (64), Expect = 5.0
Identities = 22/103 (21%), Positives = 48/103 (46%), Gaps = 8/103 (7%)
Query: 326 KRLCGVSVIRSGESMENAL--RACCKGIKIGKILIHREGDNGQQLIYEKLPNDISERHVL 383
K G++ +R G+ + R C + + LI G +G ++ + +S R+++
Sbjct: 370 KACAGLAAVRLGKEIHGQYVRRGCFGNVIVESALIDLYGKSGCIDSASRVYSKMSIRNMI 429
Query: 384 LLDPIL------GTGNSAVQAISLLIQKGVPESNIIFLNLISA 420
+ +L G G AV + +++KG+ I F+ +++A
Sbjct: 430 TWNAMLSALAQNGRGEEAVSFFNDMVKKGIKPDYISFIAILTA 472
>At5g12000 putative receptor - like kinase
Length = 703
Score = 28.9 bits (63), Expect = 6.5
Identities = 11/40 (27%), Positives = 26/40 (64%)
Query: 184 RIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPTKKYAD 223
+ RR++ EKDR + A++ ++ K + D+ + T+++A+
Sbjct: 385 KARRESQEKDRALSALVQNDVRYRKYSIDEIEVATERFAN 424
>At5g01310 putative protein
Length = 912
Score = 28.9 bits (63), Expect = 6.5
Identities = 11/16 (68%), Positives = 12/16 (74%)
Query: 453 PGMGEFGDRYFGTDDD 468
PG GEFG + FG DDD
Sbjct: 119 PGAGEFGPKLFGYDDD 134
>At4g25280 UMP/CMP kinase like protein
Length = 227
Score = 28.9 bits (63), Expect = 6.5
Identities = 16/41 (39%), Positives = 21/41 (51%), Gaps = 11/41 (26%)
Query: 41 ATDVYNQ-----------PFVIGVAGGAASGKKTVCDMIVQ 70
ATD+ +Q PF+ V GG SGK T C+ IV+
Sbjct: 25 ATDIISQEERVSPPKEKAPFITFVLGGPGSGKGTQCEKIVE 65
>At2g26960 putative transcription factor (MYB81)
Length = 427
Score = 28.9 bits (63), Expect = 6.5
Identities = 21/83 (25%), Positives = 35/83 (41%), Gaps = 15/83 (18%)
Query: 101 NFDHPDAFDTEELLRVMDKLKHGEAVDIPKYDFK--------SYKSDALRRVNPSDVIIL 152
N D D ++E + + V+IP+ DF+ SY LR V P++V +
Sbjct: 149 NTDSLDGHHSQEYMEA-------DTVEIPEVDFEHLPLNRSSSYYQSMLRHVPPTNVFVR 201
Query: 153 EGILVFHDPRVRELMNMKIFVDT 175
+ F P V L+ ++ T
Sbjct: 202 QKPCFFQPPNVYNLIPPSPYMST 224
>At5g23010 2-isopropylmalate synthase-like; homocitrate
synthase-like
Length = 506
Score = 28.5 bits (62), Expect = 8.5
Identities = 24/93 (25%), Positives = 46/93 (48%), Gaps = 7/93 (7%)
Query: 10 LMGSSSEVHFSGFHMDGFEKREASIEQPTTSATDVYNQPFVIGVAGGAASGKKTVCDMIV 69
++G++ VH SG H DG K ++ E + + G+ G SG+ V D +
Sbjct: 379 IVGANCFVHESGIHQDGILKNRSTYEILSPEDIGIVKSQ-NSGLVLGKLSGRHAVKDRLK 437
Query: 70 Q---QLHDQRVVLVNQDSFYNNLTEEERARVQD 99
+ +L D+++ V S + +LT+ ++ R+ D
Sbjct: 438 ELGYELDDEKLNAV--FSLFRDLTKNKK-RITD 467
>At1g65850
Length = 926
Score = 28.5 bits (62), Expect = 8.5
Identities = 34/132 (25%), Positives = 57/132 (42%), Gaps = 19/132 (14%)
Query: 269 RGMHTL-------IRDAQITKHDFVFYSDRLIRLVVEHGLGHLPFTE---KQVITPTGSV 318
+G+H L I +I H+ + ++L + +V HGLGH P E +Q + T +
Sbjct: 378 QGLHVLTEKSLISIEGGRIKMHNLL---EQLGKEIVRHGLGHQPIREPGKRQFLVDTRDI 434
Query: 319 ---YTGVDFCKRLCGVSVIRSGESME-NALRACCKGIKIGKIL--IHREGDNGQQLIYEK 372
T K + G+ S S E N +G+ K L +R GD +L +
Sbjct: 435 CELLTNDTGSKSVIGIHFYSSELSSELNISERAFEGMPNLKFLRFYYRYGDESDKLYLPQ 494
Query: 373 LPNDISERHVLL 384
N +S++ +L
Sbjct: 495 GLNYLSQKLKIL 506
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.141 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,859,285
Number of Sequences: 26719
Number of extensions: 478833
Number of successful extensions: 1122
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1093
Number of HSP's gapped (non-prelim): 21
length of query: 476
length of database: 11,318,596
effective HSP length: 103
effective length of query: 373
effective length of database: 8,566,539
effective search space: 3195319047
effective search space used: 3195319047
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147712.11


