

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147537.14 - phase: 0
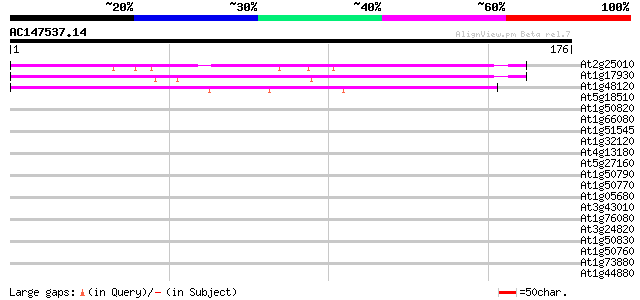
(176 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g25010 unknown protein 72 2e-13
At1g17930 unknown protein 59 1e-09
At1g48120 serine/threonine phosphatase PP7, putative 54 4e-08
At5g18510 putative protein 28 1.9
At1g50820 hypothetical protein 28 1.9
At1g66080 unknown protein 28 2.5
At1g51545 unknown protein 28 2.5
At1g32120 hypothetical protein 28 2.5
At4g13180 short-chain alcohol dehydrogenase like protein 28 3.3
At5g27160 putative protein 27 4.3
At1g50790 hypothetical protein 27 4.3
At1g50770 hypothetical protein 27 4.3
At1g05680 indole-3-acetate beta-glucosyltransferase like protein 27 4.3
At3g43010 putative protein 27 5.7
At1g76080 unknown protein 27 5.7
At3g24820 unknown protein 27 7.4
At1g50830 hypothetical protein 27 7.4
At1g50760 hypothetical protein 27 7.4
At1g73880 putative glucosyltransferase (At1g73880) 26 9.6
At1g44880 hypothetical protein 26 9.6
>At2g25010 unknown protein
Length = 509
Score = 72.0 bits (175), Expect = 2e-13
Identities = 59/182 (32%), Positives = 88/182 (47%), Gaps = 28/182 (15%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGI-----PVVGHRV--ELAEE-------QLF 46
+E W E + F + GE+ +L +VAL+LG+ P+VG +V E+A + +L
Sbjct: 80 VERWRRETNTFHLPLGEMTITLDEVALVLGLEIDGDPIVGSKVGDEVAMDMCGRLLGKLP 139
Query: 47 SELEEEYGASRAKRKVAMTSLEARLDSIGEDVSDDFV----RSFLLFTIG-TFLSSIDG- 100
S +E SR K + L+ ED S D V R++LL+ IG T ++ DG
Sbjct: 140 SAANKEVNCSRVK----LNWLKRTFSECPEDASFDVVKCHTRAYLLYLIGSTIFATTDGD 195
Query: 101 KVDSRYLSFLGNLDDVSGFAWGAAVIEDLCQWLDKRKDNNVQYVGGCLIFLQVSLICLSY 160
KV +YL + D +AWGAA + L + L + + GCL LQ C SY
Sbjct: 196 KVSVKYLPLFEDFDQAGRYAWGAAALACLYRALGNASLKSQSNICGCLTLLQ----CWSY 251
Query: 161 YH 162
+H
Sbjct: 252 FH 253
>At1g17930 unknown protein
Length = 478
Score = 59.3 bits (142), Expect = 1e-09
Identities = 45/175 (25%), Positives = 76/175 (42%), Gaps = 17/175 (9%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRVELAEEQ----------LFSELE 50
+E W E + F GE+ +L +V+LILG+ V G V +E+ L +L
Sbjct: 71 VERWRRETNTFHFPCGEMTITLDEVSLILGLAVDGKPVVGVKEKDEDPSQVCLRLLGKLP 130
Query: 51 E-EYGASRAKRKVAMTSLEARLDSIGEDVSDDFVRSFLLFTIGT--FLSSIDGKVDSRYL 107
+ E +R K S + R++L++ +G+ F ++ K+ YL
Sbjct: 131 KGELSGNRVTAKWLKESFAECPKGATMKEIEYHTRAYLIYIVGSTIFATTDPSKISVDYL 190
Query: 108 SFLGNLDDVSGFAWGAAVIEDLCQWLDKRKDNNVQYVGGCLIFLQVSLICLSYYH 162
+ + +AWGAA + L + + + +GGCL LQ C SY+H
Sbjct: 191 ILFEDFEKAGEYAWGAAALAFLYRQIGNASQRSQSIIGGCLTLLQ----CWSYFH 241
>At1g48120 serine/threonine phosphatase PP7, putative
Length = 1338
Score = 53.9 bits (128), Expect = 4e-08
Identities = 44/165 (26%), Positives = 77/165 (46%), Gaps = 12/165 (7%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRVELAEEQLFSELEEEYGASRAKR 60
+E W E H F + +GEI +L DV ++LG+ V G V + + +++L E+ R
Sbjct: 89 VERWRPETHTFHLPAGEITVTLQDVNILLGLRVDGPAVTGSTKYNWADLCEDLLGHRPGP 148
Query: 61 K------VAMTSLEARLDSIGEDVSD----DFVRSFLLFTIGTFLSSIDGKVD--SRYLS 108
K V++ L ++ D + R+F+L + FL K D +L
Sbjct: 149 KDLHGSHVSLAWLRENFRNLPADPDEVTLKCHTRAFVLALMSGFLYGDKSKHDVALTFLP 208
Query: 109 FLGNLDDVSGFAWGAAVIEDLCQWLDKRKDNNVQYVGGCLIFLQV 153
L + D+V+ +WG+A + L + L + V + G L+ LQ+
Sbjct: 209 LLRDFDEVAKLSWGSATLALLYRELCRASKRTVSTICGPLVLLQL 253
>At5g18510 putative protein
Length = 702
Score = 28.5 bits (62), Expect = 1.9
Identities = 25/75 (33%), Positives = 33/75 (43%), Gaps = 12/75 (16%)
Query: 2 EAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRVELAEEQLFSELEEEYGASRAKRK 61
E W SE +F GE +L DV ++LG V+G V FS LE R
Sbjct: 103 EKWCSETKSFIFPWGEATITLEDVMVLLGFSVLGSPV-------FSPLE-----CSEMRD 150
Query: 62 VAMTSLEARLDSIGE 76
A + R DS+G+
Sbjct: 151 SAEKLEKVRRDSLGK 165
>At1g50820 hypothetical protein
Length = 528
Score = 28.5 bits (62), Expect = 1.9
Identities = 18/70 (25%), Positives = 35/70 (49%), Gaps = 4/70 (5%)
Query: 2 EAWNSERHAFEIGSGEIGFSLLDVALILGIPVVG----HRVELAEEQLFSELEEEYGASR 57
E W + F GE +L DV ++LG V+G V+ + +++ ++LE+E+ +
Sbjct: 103 EKWCPDTKTFIFPWGEATITLEDVMVLLGFSVLGLPVFATVDSSGKEIMAKLEKEWKKIK 162
Query: 58 AKRKVAMTSL 67
+ +T L
Sbjct: 163 NDKVCLVTKL 172
>At1g66080 unknown protein
Length = 190
Score = 28.1 bits (61), Expect = 2.5
Identities = 24/81 (29%), Positives = 35/81 (42%), Gaps = 7/81 (8%)
Query: 15 SGEIGFSLLDVALILGIPVVGHR-----VELAEEQLFSELEEEYGASRAKRKVAMTSLEA 69
S +IG S+ DVA + + VV R E LF+ ++ G +K V M L+
Sbjct: 110 SAKIGVSVEDVAALRSLDVVAERRIEKLAMKVGENLFNFMQSFCGVDGSKLVVPMDILDR 169
Query: 70 RLDSIGEDV--SDDFVRSFLL 88
E DF++SF L
Sbjct: 170 WFKKFQEKAKRDPDFLKSFAL 190
>At1g51545 unknown protein
Length = 629
Score = 28.1 bits (61), Expect = 2.5
Identities = 32/110 (29%), Positives = 50/110 (45%), Gaps = 19/110 (17%)
Query: 1 MEAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRVELAEEQLFSELEEEYGASRAKR 60
+E W E +F GE +L DV ++LG V G V F+ LE ++
Sbjct: 36 VEKWCPETKSFLFPWGEATITLEDVLVLLGFSVQGSPV-------FAPLES------SEM 82
Query: 61 KVAMTSLE-ARLDSIGEDVSDDFVRSFLLFTIGTFLSSIDGKVDSRYLSF 109
+ ++ LE ARL++ G+ D VR L + +FL D +L+F
Sbjct: 83 RDSVEKLEKARLENRGQ---DGLVRQNL--WVSSFLGRGDQMEHEAFLAF 127
>At1g32120 hypothetical protein
Length = 1206
Score = 28.1 bits (61), Expect = 2.5
Identities = 17/48 (35%), Positives = 24/48 (49%), Gaps = 2/48 (4%)
Query: 116 VSGFAWGAAVIEDLCQWLDKRKDNNVQYVGGCLIFLQ-VSLICLSYYH 162
V+ F + ED C W+ R D N+ G CL F + V L C+ Y+
Sbjct: 354 VNNFRFPKFYFEDDC-WVRVRPDENIVAFGRCLRFAKLVGLDCIEPYY 400
>At4g13180 short-chain alcohol dehydrogenase like protein
Length = 263
Score = 27.7 bits (60), Expect = 3.3
Identities = 15/51 (29%), Positives = 26/51 (50%)
Query: 45 LFSELEEEYGASRAKRKVAMTSLEARLDSIGEDVSDDFVRSFLLFTIGTFL 95
LF + E+E+G+ L+ + S+ E +DF +F + T G+FL
Sbjct: 85 LFDQTEQEFGSKVHIVVNCAGVLDPKYPSLSETTLEDFDNTFTINTRGSFL 135
>At5g27160 putative protein
Length = 702
Score = 27.3 bits (59), Expect = 4.3
Identities = 24/120 (20%), Positives = 49/120 (40%), Gaps = 8/120 (6%)
Query: 11 FEIGSGEIGFSLLDVALILGIPVVGHRVELAEEQLFSELEEEYGASRAKRK--VAMTSLE 68
F + I +S+ + AL+ G + EL + + ++ K+ + + +E
Sbjct: 183 FVVNGVPIRYSIKEHALLCGFDCHDYPEELQPSMKIVDADLKFAKKIFKKVSGIKIVDVE 242
Query: 69 ARLDSI---GEDVSDDFVRSFLLFTIGTFL---SSIDGKVDSRYLSFLGNLDDVSGFAWG 122
+LD + GE D + +L + + S DG +D +L + ++ F WG
Sbjct: 243 KKLDELKKCGEKKKTDRKKLAILLFLCKVIAAKSKADGNIDKFFLKIVDDVRACETFPWG 302
>At1g50790 hypothetical protein
Length = 812
Score = 27.3 bits (59), Expect = 4.3
Identities = 17/60 (28%), Positives = 26/60 (43%)
Query: 2 EAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRVELAEEQLFSELEEEYGASRAKRK 61
E W + + F GE +L DV ++LG V+G V + E+ + G K K
Sbjct: 107 EKWCPDTNTFVFSWGEATITLEDVMVLLGFSVLGSPVFATLDSSGKEIMAKLGKEWLKIK 166
>At1g50770 hypothetical protein
Length = 632
Score = 27.3 bits (59), Expect = 4.3
Identities = 18/56 (32%), Positives = 28/56 (49%), Gaps = 4/56 (7%)
Query: 2 EAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRV----ELAEEQLFSELEEEY 53
E W + F GE +L DV L+LG V+G V + + E + +LE+E+
Sbjct: 106 EKWCPDTKTFVFPWGETTITLEDVMLLLGFSVLGSPVFVTLDSSGEIIREKLEKEW 161
>At1g05680 indole-3-acetate beta-glucosyltransferase like protein
Length = 453
Score = 27.3 bits (59), Expect = 4.3
Identities = 17/57 (29%), Positives = 30/57 (51%), Gaps = 1/57 (1%)
Query: 97 SIDGKVDSRYLSFLGNLDDVSGFAWGAAVIEDLCQWLDKRKDNNVQYVG-GCLIFLQ 152
+I V S YL + D GF+ A + + +WL+ ++ N+V Y+ G L+ L+
Sbjct: 228 NIGPTVPSMYLDKRLSEDKNYGFSLFNAKVAECMEWLNSKEPNSVVYLSFGSLVILK 284
>At3g43010 putative protein
Length = 264
Score = 26.9 bits (58), Expect = 5.7
Identities = 26/116 (22%), Positives = 49/116 (41%), Gaps = 7/116 (6%)
Query: 14 GSGEIGFSLLDVALILGIPVV--GHRVELAEEQLFSELEEEYGASRA--KRKVAMTSLEA 69
G + FSL + A + G+P + ++ +F + + Y + V + + A
Sbjct: 27 GGHPMRFSLTEFACVTGLPCAEFSEDYDPDDDSVFVDGMKSYWDELIGPDKTVTLRDVSA 86
Query: 70 RLDSIGEDVSDD--FVRSFLLFTIGTFLSSID-GKVDSRYLSFLGNLDDVSGFAWG 122
L + + +S D +FLL G ++S + +Y+ L +LD F WG
Sbjct: 87 MLTNKRKTLSSDHRLKLAFLLIVDGVLIASNQICRPTFKYVEMLADLDKFLSFPWG 142
>At1g76080 unknown protein
Length = 302
Score = 26.9 bits (58), Expect = 5.7
Identities = 13/51 (25%), Positives = 27/51 (52%)
Query: 57 RAKRKVAMTSLEARLDSIGEDVSDDFVRSFLLFTIGTFLSSIDGKVDSRYL 107
+ R ++ T + AR++ D +F++ + + TFL DG++ RY+
Sbjct: 231 KLSRSMSETVVFARMNGDENDSCMEFLKDMNVIEVPTFLFIRDGEIRGRYV 281
>At3g24820 unknown protein
Length = 186
Score = 26.6 bits (57), Expect = 7.4
Identities = 18/59 (30%), Positives = 33/59 (55%), Gaps = 3/59 (5%)
Query: 78 VSDDFVRSFLLFTIGTFLS-SIDGKVDSRYLSFLGNLDDVSGFAWGAAVIEDLCQWLDK 135
V+D+ + FTI TF + S++ + + ++ LG D+ +G + A V +DL W +K
Sbjct: 41 VTDELIDHVRSFTIDTFKNFSLEDEEEEVSVNPLG--DEDNGMSSSANVKKDLSDWQEK 97
>At1g50830 hypothetical protein
Length = 768
Score = 26.6 bits (57), Expect = 7.4
Identities = 14/37 (37%), Positives = 19/37 (50%)
Query: 2 EAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGHRV 38
E W E +F GE +L DV ++LG V+G V
Sbjct: 125 EKWCPETKSFVFPWGEATITLEDVMVLLGFSVLGSPV 161
>At1g50760 hypothetical protein
Length = 649
Score = 26.6 bits (57), Expect = 7.4
Identities = 20/98 (20%), Positives = 44/98 (44%), Gaps = 4/98 (4%)
Query: 2 EAWNSERHAFEIGSGEIGFSLLDVALILGIPVVGH----RVELAEEQLFSELEEEYGASR 57
E W+ + F GE +L DV ++ G V+G ++ + +++ L+ + +
Sbjct: 96 EKWSPDTKTFVFSWGEATITLEDVMVLSGFSVLGSPAFATLDSSGKKIMERLKNAWQTIK 155
Query: 58 AKRKVAMTSLEARLDSIGEDVSDDFVRSFLLFTIGTFL 95
+ KV + + A +D + +FL+ +G F+
Sbjct: 156 REGKVNLLTQVAWMDRFMNCGGELEHVAFLVLWLGYFV 193
>At1g73880 putative glucosyltransferase (At1g73880)
Length = 473
Score = 26.2 bits (56), Expect = 9.6
Identities = 9/19 (47%), Positives = 14/19 (73%)
Query: 126 IEDLCQWLDKRKDNNVQYV 144
++ + WLD R+DN+V YV
Sbjct: 267 VDHVMSWLDAREDNHVVYV 285
>At1g44880 hypothetical protein
Length = 1038
Score = 26.2 bits (56), Expect = 9.6
Identities = 13/39 (33%), Positives = 20/39 (50%), Gaps = 1/39 (2%)
Query: 85 SFLLFTIGTFLSSID-GKVDSRYLSFLGNLDDVSGFAWG 122
+FLL G ++S G+ +Y+ L +LD F WG
Sbjct: 178 AFLLIVDGVLIASNQIGRPTFKYVEMLADLDKFLSFPWG 216
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.142 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,959,189
Number of Sequences: 26719
Number of extensions: 161685
Number of successful extensions: 439
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 427
Number of HSP's gapped (non-prelim): 21
length of query: 176
length of database: 11,318,596
effective HSP length: 93
effective length of query: 83
effective length of database: 8,833,729
effective search space: 733199507
effective search space used: 733199507
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 56 (26.2 bits)
Medicago: description of AC147537.14


