

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147537.13 + phase: 0
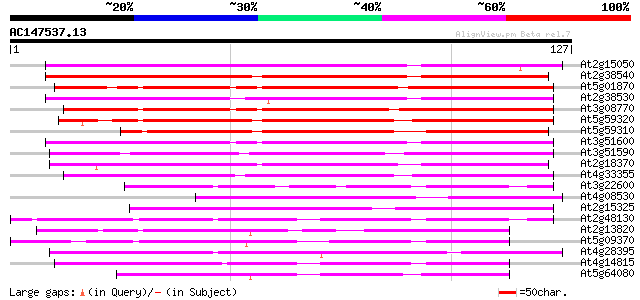
(127 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g15050 putative lipid transfer protein 119 4e-28
At2g38540 putative nonspecific lipid-transfer protein 112 7e-26
At5g01870 lipid-transfer protein-like 100 2e-22
At2g38530 putative nonspecific lipid-transfer protein 100 3e-22
At3g08770 putative nonspecific lipid-transfer protein 99 4e-22
At5g59320 nonspecific lipid-transfer protein precursor - like 98 1e-21
At5g59310 nonspecific lipid-transfer protein precursor - like 91 1e-19
At3g51600 non-specific lipid transfer protein 91 1e-19
At3g51590 lipid transfer protein-like protein 84 1e-17
At2g18370 putative lipid transfer protein 77 2e-15
At4g33355 non-specific lipid transfer protein 69 5e-13
At3g22600 predicted GPI-anchored protein 54 3e-08
At4g08530 ipid transfer protein like 51 2e-07
At2g15325 putative protein 51 2e-07
At2g48130 predicted GPI-anchored protein 50 3e-07
At2g13820 predicted GPI-anchored protein 50 4e-07
At5g09370 putative lipid transfer protein, predicted GPI-anchore... 48 1e-06
At4g28395 ATA7 47 3e-06
At4g14815 Expressed protein 44 3e-05
At5g64080 predicted GPI-anchored protein 42 1e-04
>At2g15050 putative lipid transfer protein
Length = 123
Score = 119 bits (298), Expect = 4e-28
Identities = 60/119 (50%), Positives = 71/119 (59%), Gaps = 5/119 (4%)
Query: 9 RLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTM 68
+L CL V ++ GP A SCG V ++L PC Y+ NGG T P QCCNG+R LN M
Sbjct: 6 KLGCLVFVFVIAAGPITAKAALSCGEVNSNLKPCTGYLTNGGITSPGPQCCNGVRKLNGM 65
Query: 69 AQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPY--QISPNTDCNRY 125
TT DRR C CIKNA G LN + AAG+PR+CG+ IPY QI NT CN Y
Sbjct: 66 VLTTLDRRQACRCIKNAARNVG---PGLNADRAAGIPRRCGIKIPYSTQIRFNTKCNTY 121
>At2g38540 putative nonspecific lipid-transfer protein
Length = 118
Score = 112 bits (279), Expect = 7e-26
Identities = 51/114 (44%), Positives = 74/114 (64%), Gaps = 5/114 (4%)
Query: 9 RLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTM 68
+L CL + C++ GP A SCG V ++L C+ Y++ GG PA CC+G++NLN++
Sbjct: 6 KLACLLLACMIVAGPITSNAALSCGSVNSNLAACIGYVLQGGVIPPA--CCSGVKNLNSI 63
Query: 69 AQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
A+TT DR+ C CI+ A G + LN AAG+P+ CGVNIPY+IS +T+C
Sbjct: 64 AKTTPDRQQACNCIQGAARALG---SGLNAGRAAGIPKACGVNIPYKISTSTNC 114
>At5g01870 lipid-transfer protein-like
Length = 116
Score = 100 bits (249), Expect = 2e-22
Identities = 51/113 (45%), Positives = 76/113 (67%), Gaps = 7/113 (6%)
Query: 11 VCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQ 70
+CL + + +G +EAA+ SC V +LYPCV Y++ GG +P + CCNGIR L+ A
Sbjct: 8 LCLLLASIFAWG--SEAAI-SCNAVQANLYPCVVYVVQGG-AIPYS-CCNGIRMLSKQAT 62
Query: 71 TTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
+ +D++ VC CIK+ V + SY+++ L AA LP KCGV +PY+I P+T+CN
Sbjct: 63 SASDKQGVCRCIKSVVGR--VSYSSIYLKKAAALPGKCGVKLPYKIDPSTNCN 113
>At2g38530 putative nonspecific lipid-transfer protein
Length = 118
Score = 100 bits (248), Expect = 3e-22
Identities = 50/116 (43%), Positives = 68/116 (58%), Gaps = 7/116 (6%)
Query: 9 RLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQ-CCNGIRNLNT 67
+L C+ + C++ GP A+ SCG V +L C++Y+ G P Q CCNG+ NL
Sbjct: 6 KLACMVLACMIVAGPITANALMSCGTVNGNLAGCIAYLTRGA---PLTQGCCNGVTNLKN 62
Query: 68 MAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
MA TT DR+ C C+++A G LN AAGLP C VNIPY+IS +T+CN
Sbjct: 63 MASTTPDRQQACRCLQSAAKAVG---PGLNTARAAGLPSACKVNIPYKISASTNCN 115
>At3g08770 putative nonspecific lipid-transfer protein
Length = 113
Score = 99.4 bits (246), Expect = 4e-22
Identities = 53/111 (47%), Positives = 70/111 (62%), Gaps = 5/111 (4%)
Query: 13 LAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQTT 72
L VCLV EAAV SC V+ LYPC+SY+ GG VP CCNG+ L + AQT+
Sbjct: 5 LLAVCLVLALHCGEAAV-SCNTVIADLYPCLSYVTQGG-PVPTL-CCNGLTTLKSQAQTS 61
Query: 73 NDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
DR+ VC CIK+A+ G + + + A LP KCGV++PY+ SP+TDC+
Sbjct: 62 VDRQGVCRCIKSAI--GGLTLSPRTIQNALELPSKCGVDLPYKFSPSTDCD 110
>At5g59320 nonspecific lipid-transfer protein precursor - like
Length = 115
Score = 98.2 bits (243), Expect = 1e-21
Identities = 53/113 (46%), Positives = 73/113 (63%), Gaps = 11/113 (9%)
Query: 12 CLAI-VCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQ 70
CL + VC+V +AA+ SCG V SL PC +Y+ GG P+ CC G++ LN+MA+
Sbjct: 10 CLVLTVCIVA---SVDAAI-SCGTVAGSLAPCATYLSKGGLVPPS--CCAGVKTLNSMAK 63
Query: 71 TTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
TT DR+ C CI++ S + LN +LA+GLP KCGV+IPY IS +T+CN
Sbjct: 64 TTPDRQQACRCIQSTAK----SISGLNPSLASGLPGKCGVSIPYPISMSTNCN 112
>At5g59310 nonspecific lipid-transfer protein precursor - like
Length = 112
Score = 91.3 bits (225), Expect = 1e-19
Identities = 46/97 (47%), Positives = 63/97 (64%), Gaps = 10/97 (10%)
Query: 26 EAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQTTNDRRAVCTCIKNA 85
+AA+T CG V +SL PC+ Y+ GG P CC G++ LN MAQTT DR+ C C+++A
Sbjct: 22 DAAIT-CGTVASSLSPCLGYLSKGGVVPPP--CCAGVKKLNGMAQTTPDRQQACRCLQSA 78
Query: 86 VSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
+N +LA+GLP KCGV+IPY IS +T+C
Sbjct: 79 AK-------GVNPSLASGLPGKCGVSIPYPISTSTNC 108
>At3g51600 non-specific lipid transfer protein
Length = 118
Score = 91.3 bits (225), Expect = 1e-19
Identities = 49/115 (42%), Positives = 65/115 (55%), Gaps = 5/115 (4%)
Query: 9 RLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTM 68
+L L IVC++ P A A SCG V SL C +Y+ GG +P CC+G++ LN++
Sbjct: 6 KLSTLVIVCMLVTAPMASEAAISCGAVTGSLGQCYNYLTRGG-FIPRG-CCSGVQRLNSL 63
Query: 69 AQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
A+TT DR+ C CI+ A G + LN AA LP C V I Y IS T+CN
Sbjct: 64 ARTTRDRQQACRCIQGAARALG---SRLNAGRAARLPGACRVRISYPISARTNCN 115
>At3g51590 lipid transfer protein-like protein
Length = 119
Score = 84.3 bits (207), Expect = 1e-17
Identities = 44/114 (38%), Positives = 66/114 (57%), Gaps = 8/114 (7%)
Query: 10 LVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMA 69
+ CL ++ + P + CG V ++L C++Y+ N G +QCC G+++L +A
Sbjct: 8 ITCLIVLTIYMASPTE--STIQCGTVTSTLAQCLTYLTNSGPL--PSQCCVGVKSLYQLA 63
Query: 70 QTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
QTT DR+ VC C+K A G LN +L A LP CGV+IPY IS +T+C+
Sbjct: 64 QTTPDRKQVCECLKLA----GKEIKGLNTDLVAALPTTCGVSIPYPISFSTNCD 113
>At2g18370 putative lipid transfer protein
Length = 116
Score = 77.0 bits (188), Expect = 2e-15
Identities = 39/115 (33%), Positives = 59/115 (50%), Gaps = 8/115 (6%)
Query: 10 LVCLAIVCL--VTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNT 67
L CLAI+ + + F P+ + SC V+ L PCVSY+ +G P CC+G+++L
Sbjct: 4 LKCLAIISVLGIFFIPRYSESAISCSVVLQDLQPCVSYLTSGSGNPPET-CCDGVKSLAA 62
Query: 68 MAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
T+ D++A C CIK+ + + LA L CG ++P SP DC
Sbjct: 63 ATTTSADKKAACQCIKSVANS-----VTVKPELAQALASNCGASLPVDASPTVDC 112
>At4g33355 non-specific lipid transfer protein
Length = 109
Score = 69.3 bits (168), Expect = 5e-13
Identities = 37/111 (33%), Positives = 52/111 (46%), Gaps = 6/111 (5%)
Query: 13 LAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQTT 72
L +V + G +C V L C+ Y+ GGN P+ CCNG+ +L A
Sbjct: 2 LLLVITILLGIAYHGEAIACPQVNMYLAQCLPYLKAGGN--PSPMCCNGLNSLKAAAPEK 59
Query: 73 NDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
DR+ C C+K+ + + +N + A LP KCGVNI S DCN
Sbjct: 60 ADRQVACNCLKSVAN----TIPGINDDFAKQLPAKCGVNIGVPFSKTVDCN 106
>At3g22600 predicted GPI-anchored protein
Length = 170
Score = 53.5 bits (127), Expect = 3e-08
Identities = 35/97 (36%), Positives = 50/97 (51%), Gaps = 13/97 (13%)
Query: 27 AAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQTTNDRRAVCTCIKNAV 86
+A +SC + S+ PC++YI G +T P QCCN L+ + Q++ D +C +
Sbjct: 23 SAQSSCTNALISMSPCLNYI-TGNSTSPNQQCCN---QLSRVVQSSPD--CLCQVLNGGG 76
Query: 87 SQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
SQ G N+N A GLPR C V P P + CN
Sbjct: 77 SQLGI---NVNQTQALGLPRACNVQTP----PVSRCN 106
>At4g08530 ipid transfer protein like
Length = 103
Score = 50.8 bits (120), Expect = 2e-07
Identities = 24/83 (28%), Positives = 38/83 (44%), Gaps = 7/83 (8%)
Query: 43 VSYIMNGGNTVPAAQCCNGIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAA 102
+S + N +P A CC I ++N +T R A+C C + + S F+
Sbjct: 19 ISACLQQANGLPHADCCYAINDVNRYVETIYGRLALCKCFQEILKDSRFT-------KLI 71
Query: 103 GLPRKCGVNIPYQISPNTDCNRY 125
G+P KC + P TDC+R+
Sbjct: 72 GMPEKCAIPNAVPFDPKTDCDRF 94
>At2g15325 putative protein
Length = 118
Score = 50.8 bits (120), Expect = 2e-07
Identities = 25/96 (26%), Positives = 45/96 (46%), Gaps = 5/96 (5%)
Query: 28 AVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQTTNDRRAVCTCIKNAVS 87
++T C L PC+ Y+ P+ +CC+G+ +N +T +DR +C C+
Sbjct: 27 SLTPCEEATNLLTPCLRYLWAPPEAKPSPECCSGLDKVNKGVKTYDDRHDMCICL----- 81
Query: 88 QSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
S + T+ + LP+ C V + + P DC+
Sbjct: 82 SSEAAITSADQYKFDNLPKLCNVALFAPVGPKFDCS 117
>At2g48130 predicted GPI-anchored protein
Length = 183
Score = 50.1 bits (118), Expect = 3e-07
Identities = 34/123 (27%), Positives = 60/123 (48%), Gaps = 15/123 (12%)
Query: 1 MGYSTMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCN 60
MGY + + +A+V + KA+ + +SC +T+L PC+SYI G +T P+ CC+
Sbjct: 1 MGYRR-SYAITFVALVAALWSVTKAQPS-SSCVSTLTTLSPCLSYI-TGNSTTPSQPCCS 57
Query: 61 GIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNT 120
+ ++ + + +C+ + + + G N+N A LP C + P P T
Sbjct: 58 RLDSV-----IKSSPQCICSAVNSPIPNIGL---NINRTQALQLPNACNIQTP----PLT 105
Query: 121 DCN 123
CN
Sbjct: 106 QCN 108
>At2g13820 predicted GPI-anchored protein
Length = 169
Score = 49.7 bits (117), Expect = 4e-07
Identities = 34/108 (31%), Positives = 57/108 (52%), Gaps = 12/108 (11%)
Query: 7 ATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTV-PAAQCCNGIRNL 65
AT L+ ++V L++ G +A AAV C ++ ++ C+S++ +G V P CC+G++
Sbjct: 4 ATILMIFSVVALMS-GERAHAAV-DCSSLILNMADCLSFVTSGSTVVKPEGTCCSGLK-- 59
Query: 66 NTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIP 113
T+ +T + C+ A SG L+L+ AA LP C V P
Sbjct: 60 -TVVRTGPE------CLCEAFKNSGSLGLTLDLSKAASLPSVCKVAAP 100
>At5g09370 putative lipid transfer protein, predicted GPI-anchored
protein
Length = 129
Score = 48.1 bits (113), Expect = 1e-06
Identities = 34/114 (29%), Positives = 52/114 (44%), Gaps = 15/114 (13%)
Query: 1 MGYSTMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNT-VPAAQCC 59
M Y + AT L+ L L P A + C +V +L+PC+ +I GG P A CC
Sbjct: 1 MAYFSTATSLLLLV---LSVSSPYVHGA-SDCDTLVITLFPCLPFISIGGTADTPTASCC 56
Query: 60 NGIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIP 113
+ ++N+ D + +C C + G LN+ +A LP C +N P
Sbjct: 57 SSLKNI-------LDTKPICLCEGLKKAPLGI---KLNVTKSATLPVACKLNAP 100
>At4g28395 ATA7
Length = 180
Score = 46.6 bits (109), Expect = 3e-06
Identities = 29/126 (23%), Positives = 55/126 (43%), Gaps = 14/126 (11%)
Query: 10 LVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMA 69
LV +V + T + + C V S PC+ ++ G P+ CC G+ +LN +
Sbjct: 47 LVVAFLVLMKTAVSQDNNPLEHCRDVFVSFMPCMGFV-EGIFQQPSPDCCRGVTHLNNVV 105
Query: 70 ----------QTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPN 119
Q + + VC CI+ + + + +N LP +C + + + IS +
Sbjct: 106 KFTSPGSRNRQDSGETERVCLCIEIMGNANHLPFLPAAIN---NLPLRCSLTLSFPISVD 162
Query: 120 TDCNRY 125
DC+++
Sbjct: 163 MDCSQF 168
>At4g14815 Expressed protein
Length = 156
Score = 43.5 bits (101), Expect = 3e-05
Identities = 28/103 (27%), Positives = 47/103 (45%), Gaps = 9/103 (8%)
Query: 11 VCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQ 70
+CL + + +A +SC V+ S+ PC+S+I ++P+ QCCN + ++
Sbjct: 5 MCLILFIALMRVMSIVSAQSSCTNVLISMAPCLSFITQ-NTSLPSQQCCNQLAHV----- 58
Query: 71 TTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIP 113
+C + SQ G N+N A LP+ C V P
Sbjct: 59 VRYSSECLCQVLDGGGSQLGI---NVNETQALALPKACHVETP 98
>At5g64080 predicted GPI-anchored protein
Length = 173
Score = 41.6 bits (96), Expect = 1e-04
Identities = 22/90 (24%), Positives = 42/90 (46%), Gaps = 10/90 (11%)
Query: 25 AEAAVTSCGPVVTSLYPCVSYIMNGGNTV-PAAQCCNGIRNLNTMAQTTNDRRAVCTCIK 83
A A C ++ ++ C+S++ +GG P CC+G++ + D + +C K
Sbjct: 36 APAPSVDCSTLILNMADCLSFVSSGGTVAKPEGTCCSGLKTV-----LKADSQCLCEAFK 90
Query: 84 NAVSQSGFSYTNLNLNLAAGLPRKCGVNIP 113
++ S LN+ A+ LP C ++ P
Sbjct: 91 SSASLG----VTLNITKASTLPAACKLHAP 116
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.134 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,772,704
Number of Sequences: 26719
Number of extensions: 93312
Number of successful extensions: 239
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 188
Number of HSP's gapped (non-prelim): 44
length of query: 127
length of database: 11,318,596
effective HSP length: 87
effective length of query: 40
effective length of database: 8,994,043
effective search space: 359761720
effective search space used: 359761720
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 54 (25.4 bits)
Medicago: description of AC147537.13


