

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147517.7 - phase: 0
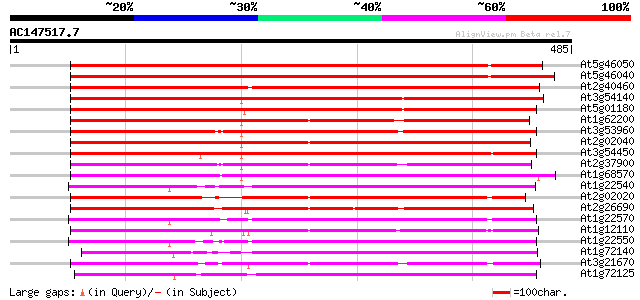
(485 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g46050 peptide transporter 499 e-141
At5g46040 peptide transporter 474 e-134
At2g40460 putative PTR2 family peptide transporter 399 e-111
At3g54140 peptide transport - like protein 358 3e-99
At5g01180 oligopeptide transporter - like protein 353 2e-97
At1g62200 unknown protein (At1g62200) 332 2e-91
At3g53960 transporter like protein 331 5e-91
At2g02040 histidine transport protein (PTR2-B) 323 1e-88
At3g54450 oligopeptide transporter -like protein 314 8e-86
At2g37900 putative peptide/amino acid transporter 308 6e-84
At1g68570 peptide transporter like 303 1e-82
At1g22540 Similar to peptide transporter 301 5e-82
At2g02020 putative peptide/amino acid transporter 295 3e-80
At2g26690 putative nitrate transporter 290 9e-79
At1g22570 Similar to LeOPT1 [Lycopersicon esculentum 285 3e-77
At1g12110 nitrate/chlorate transporter CHL1 285 3e-77
At1g22550 Similar to LeOPT1 [Lycopersicon esculentum 277 1e-74
At1g72140 peptide transporter like protein (PTR2-B) 272 3e-73
At3g21670 nitrate transporter 271 5e-73
At1g72125 oligopeptide like transporter 271 5e-73
>At5g46050 peptide transporter
Length = 582
Score = 499 bits (1284), Expect = e-141
Identities = 238/409 (58%), Positives = 315/409 (76%), Gaps = 2/409 (0%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G P ST+GADQFD F+ KE++QKLSF+NWW+F I GT+ A T+LVY+QDNVG+ LG
Sbjct: 161 GGTKPNISTIGADQFDVFDPKEKTQKLSFFNWWMFSIFFGTLFANTVLVYVQDNVGWTLG 220
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
YG+PT+ L +SI +F+LGTP YRH+LP+GSP T+M +V VA+ RK + D HE+
Sbjct: 221 YGLPTLGLAISITIFLLGTPFYRHKLPTGSPFTKMARVIVASFRKANAPMTHDITSFHEL 280
Query: 173 SIEEYTSKGRYRINHSSSLRFLDKAAVKTGQTSPWMLCTVTQIEETKQMTKMLPVLITTC 232
EY KG + I+ + SLRFLD+A++KTG W LCT T++EETKQM +MLPVL T
Sbjct: 281 PSLEYERKGAFPIHPTPSLRFLDRASLKTGTNHKWNLCTTTEVEETKQMLRMLPVLFITF 340
Query: 233 IPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRVFVPVIRR 292
+PS +++Q TLF++QGTTLDR + F IPPA L FV + MLIS+V+YDRVFV + R+
Sbjct: 341 VPSMMLAQINTLFVKQGTTLDRKVTGSFSIPPASLSGFVTLSMLISIVLYDRVFVKITRK 400
Query: 293 YTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDT-IPLTIFILV 351
+T NPRGIT+LQR+GIGL+ H+++MIVA + ER RL VA ++ L+ +PLTIF L+
Sbjct: 401 FTGNPRGITLLQRMGIGLIFHILIMIVASVTERYRLKVAADHGLIHQTGVKLPLTIFALL 460
Query: 352 PQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSIVADLTS 411
PQF LMG+AD+F+++AKLEFFYDQAPESMKSLGTSY+TT+L+IGNF+S+FLLS V+++T
Sbjct: 461 PQFVLMGMADSFLEVAKLEFFYDQAPESMKSLGTSYSTTSLAIGNFMSSFLLSTVSEITK 520
Query: 412 KNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVYNDDVT 460
K G +GWILNNLN SR+DYYY F A+L+ VNF+ F V+ KF+VY +VT
Sbjct: 521 KRG-RGWILNNLNESRLDYYYLFFAVLNLVNFVLFLVVVKFYVYRAEVT 568
>At5g46040 peptide transporter
Length = 586
Score = 474 bits (1220), Expect = e-134
Identities = 230/420 (54%), Positives = 312/420 (73%), Gaps = 2/420 (0%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G P ST+GADQFDEF+ K++ K SF+NWW+F I GT A T+LVY+QDNVG+A+G
Sbjct: 161 GGTKPNISTIGADQFDEFDPKDKIHKHSFFNWWMFSIFFGTFFATTVLVYVQDNVGWAIG 220
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
YG+ T+ L SI +F+LGT LYRH+LP GSP T+M +V VA++RK + + DS +E+
Sbjct: 221 YGLSTLGLAFSIFIFLLGTRLYRHKLPMGSPFTKMARVIVASLRKAREPMSSDSTRFYEL 280
Query: 173 SIEEYTSKGRYRINHSSSLRFLDKAAVKTGQTSPWMLCTVTQIEETKQMTKMLPVLITTC 232
EY SK + I+ +SSLRFL++A++KTG T W LCT+T++EETKQM KMLPVL T
Sbjct: 281 PPMEYASKRAFPIHSTSSLRFLNRASLKTGSTHKWRLCTITEVEETKQMLKMLPVLFVTF 340
Query: 233 IPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRVFVPVIRR 292
+PS +++Q TLFI+QGTTLDR + +F IPPA L+ F ML+S+VIYDRVFV +R+
Sbjct: 341 VPSMMLAQIMTLFIKQGTTLDRRLTNNFSIPPASLLGFTTFSMLVSIVIYDRVFVKFMRK 400
Query: 293 YTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDT-IPLTIFILV 351
T NPRGIT+LQR+GIG+++H+++MI+A + ER RL VA E+ L IPL+IF L+
Sbjct: 401 LTGNPRGITLLQRMGIGMILHILIMIIASITERYRLKVAAEHGLTHQTAVPIPLSIFTLL 460
Query: 352 PQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSIVADLTS 411
PQ+ LMG+AD F++IAKLEFFYDQAPESMKSLGTSY +T++++G F+S+ LLS V+ +T
Sbjct: 461 PQYVLMGLADAFIEIAKLEFFYDQAPESMKSLGTSYTSTSMAVGYFMSSILLSSVSQITK 520
Query: 412 KNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVYNDDVTRTKMDLEMNPN 471
K G +GWI NNLN SR+D YY F A+L+ +NFI F V+ +F+ Y DVT++ + PN
Sbjct: 521 KQG-RGWIQNNLNESRLDNYYMFFAVLNLLNFILFLVVIRFYEYRADVTQSANVEQKEPN 579
>At2g40460 putative PTR2 family peptide transporter
Length = 583
Score = 399 bits (1026), Expect = e-111
Identities = 193/407 (47%), Positives = 284/407 (69%), Gaps = 4/407 (0%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G P ST GADQFD + +E+ QK+SF+NWW+F +G + A LVYIQ+N+G+ LG
Sbjct: 154 GGTKPNISTFGADQFDSYSIEEKKQKVSFFNWWMFSSFLGALFATLGLVYIQENLGWGLG 213
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTR-MVQVFVAAMRKWKLNVPIDSKELHE 171
YGIPT+ L+VS++VF +GTP YRH++ L + +VQV +AA + KL P D EL+E
Sbjct: 214 YGIPTVGLLVSLVVFYIGTPFYRHKVIKTDNLAKDLVQVPIAAFKNRKLQCPDDHLELYE 273
Query: 172 VSIEEYTSKGRYRINHSSSLRFLDKAAVKTGQTSPWMLCTVTQIEETKQMTKMLPVLITT 231
+ Y S G+++++H+ RFLDKAA+KT P CTVT++E K++ ++ + + T
Sbjct: 274 LDSHYYKSNGKHQVHHTPVFRFLDKAAIKTSSRVP---CTVTKVEVAKRVLGLIFIWLVT 330
Query: 232 CIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRVFVPVIR 291
IPST+ +Q TLF++QGTTLDR +G++F+IP A L +FV + ML+SV +YD+ FVP +R
Sbjct: 331 LIPSTLWAQVNTLFVKQGTTLDRKIGSNFQIPAASLGSFVTLSMLLSVPMYDQSFVPFMR 390
Query: 292 RYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDTIPLTIFILV 351
+ T NPRGIT+LQRLG+G + ++ + +A +E KR+ V +E ++ P +P++IF L+
Sbjct: 391 KKTGNPRGITLLQRLGVGFAIQIVAIAIASAVEVKRMRVIKEFHITSPTQVVPMSIFWLL 450
Query: 352 PQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSIVADLTS 411
PQ++L+GI D F I LEFFYDQ+PE M+SLGT++ T+ + +GNFL++FL++++ +TS
Sbjct: 451 PQYSLLGIGDVFNAIGLLEFFYDQSPEEMQSLGTTFFTSGIGLGNFLNSFLVTMIDKITS 510
Query: 412 KNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVYNDD 458
K G K WI NNLN SR+DYYY FL ++S VN F A +VY D
Sbjct: 511 KGGGKSWIGNNLNDSRLDYYYGFLVVISIVNMGLFVWAASKYVYKSD 557
>At3g54140 peptide transport - like protein
Length = 570
Score = 358 bits (920), Expect = 3e-99
Identities = 180/416 (43%), Positives = 271/416 (64%), Gaps = 8/416 (1%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G + P S+ GADQFDE + E+ +K SF+NW+ F I +G + A T+LV+IQ NVG+ G
Sbjct: 154 GGIKPCVSSFGADQFDENDENEKIKKSSFFNWFYFSINVGALIAATVLVWIQMNVGWGWG 213
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
+G+PT+ +V+++ F G+ YR + P GSPLTR+ QV VAA RK + VP D L E
Sbjct: 214 FGVPTVAMVIAVCFFFFGSRFYRLQRPGGSPLTRIFQVIVAAFRKISVKVPEDKSLLFET 273
Query: 173 SIEEYTSKGRYRINHSSSLRFLDKAAV-------KTGQTSPWMLCTVTQIEETKQMTKML 225
+ +E KG ++ H+ +L+F DKAAV K G+ +PW LC+VTQ+EE K + +L
Sbjct: 274 ADDESNIKGSRKLVHTDNLKFFDKAAVESQSDSIKDGEVNPWRLCSVTQVEELKSIITLL 333
Query: 226 PVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRV 285
PV T + +T+ SQ +T+F+ QG T+D+ MG +FEIP A L F + +L +YD+
Sbjct: 334 PVWATGIVFATVYSQMSTMFVLQGNTMDQHMGKNFEIPSASLSLFDTVSVLFWTPVYDQF 393
Query: 286 FVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDTIPL 345
+P+ R++T+N RG T LQR+GIGLV+ + MI A ++E RL + +N I +
Sbjct: 394 IIPLARKFTRNERGFTQLQRMGIGLVVSIFAMITAGVLEVVRLDYVKTHNAYDQ-KQIHM 452
Query: 346 TIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSI 405
+IF +PQ+ L+G A+ F I +LEFFYDQAP++M+SL ++ + TT+++GN+LST L+++
Sbjct: 453 SIFWQIPQYLLIGCAEVFTFIGQLEFFYDQAPDAMRSLCSALSLTTVALGNYLSTVLVTV 512
Query: 406 VADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVYNDDVTR 461
V +T KNG GWI +NLN +DY++ LA LS +NF+ + I+K + Y V R
Sbjct: 513 VMKITKKNGKPGWIPDNLNRGHLDYFFYLLATLSFLNFLVYLWISKRYKYKKAVGR 568
>At5g01180 oligopeptide transporter - like protein
Length = 570
Score = 353 bits (905), Expect = 2e-97
Identities = 177/410 (43%), Positives = 268/410 (65%), Gaps = 8/410 (1%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G + P S+ GADQFD+ + KE+ K SF+NW+ F I +G + A ++LV+IQ NVG+ G
Sbjct: 154 GGIKPCVSSFGADQFDDTDEKEKESKSSFFNWFYFVINVGAMIASSVLVWIQMNVGWGWG 213
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
G+PT+ + ++++ F G+ YR + P GSPLTRM+QV VA+ RK K+ +P D L+E
Sbjct: 214 LGVPTVAMAIAVVFFFAGSNFYRLQKPGGSPLTRMLQVIVASCRKSKVKIPEDESLLYEN 273
Query: 173 SIEEYTSKGRYRINHSSSLRFLDKAAVKT-------GQTSPWMLCTVTQIEETKQMTKML 225
E + G ++ H+ L F DKAAV+T ++S W LCTVTQ+EE K + ++L
Sbjct: 274 QDAESSIIGSRKLEHTKILTFFDKAAVETESDNKGAAKSSSWKLCTVTQVEELKALIRLL 333
Query: 226 PVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRV 285
P+ T + +++ SQ T+F+ QG TLD+ MG +F+IP A L F + +L +YD++
Sbjct: 334 PIWATGIVFASVYSQMGTVFVLQGNTLDQHMGPNFKIPSASLSLFDTLSVLFWAPVYDKL 393
Query: 286 FVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDTIPL 345
VP R+YT + RG T LQR+GIGLV+ + M+ A ++E RL+ + +NL +TIP+
Sbjct: 394 IVPFARKYTGHERGFTQLQRIGIGLVISIFSMVSAGILEVARLNYVQTHNLYNE-ETIPM 452
Query: 346 TIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSI 405
TIF VPQ+ L+G A+ F I +LEFFYDQAP++M+SL ++ + T ++ GN+LSTFL+++
Sbjct: 453 TIFWQVPQYFLVGCAEVFTFIGQLEFFYDQAPDAMRSLCSALSLTAIAFGNYLSTFLVTL 512
Query: 406 VADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVY 455
V +T G GWI NLN +DY++ LA LS +NF+ + IAK++ Y
Sbjct: 513 VTKVTRSGGRPGWIAKNLNNGHLDYFFWLLAGLSFLNFLVYLWIAKWYTY 562
>At1g62200 unknown protein (At1g62200)
Length = 590
Score = 332 bits (852), Expect = 2e-91
Identities = 171/404 (42%), Positives = 265/404 (65%), Gaps = 16/404 (3%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G + P S+ GADQFD+ + +ER +K SF+NW+ F I IG+ + T+LV++Q+NVG+ LG
Sbjct: 186 GGIKPCVSSFGADQFDDTDPRERVRKASFFNWFYFSINIGSFISSTLLVWVQENVGWGLG 245
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
+ IPT+ + VSI F +GTPLYR + P GSP+TR+ QV VAA RK KLN+P D L+E
Sbjct: 246 FLIPTVFMGVSIASFFIGTPLYRFQKPGGSPITRVCQVLVAAYRKLKLNLPEDISFLYET 305
Query: 173 SIEEYTSKGRYRINHSSSLRFLDKAAV------KTGQ-TSPWMLCTVTQIEETKQMTKML 225
+ G +I H+ +FLDKAAV K+G ++PW LCTVTQ+EE K + +M
Sbjct: 306 REKNSMIAGSRKIQHTDGYKFLDKAAVISEYESKSGAFSNPWKLCTVTQVEEVKTLIRMF 365
Query: 226 PVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRV 285
P+ + + S + SQ +TLF++QG +++R + + FEIPPA F + +LIS+ IYDR
Sbjct: 366 PIWASGIVYSVLYSQISTLFVQQGRSMNRIIRS-FEIPPASFGVFDTLIVLISIPIYDRF 424
Query: 286 FVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDTIPL 345
VP +RR+T P+G+T LQR+GIGL + V+ + A ++E RL +A+ D + +
Sbjct: 425 LVPFVRRFTGIPKGLTDLQRMGIGLFLSVLSIAAAAIVETVRLQLAQ--------DFVAM 476
Query: 346 TIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSI 405
+IF +PQ+ LMGIA+ F I ++EFFYD++P++M+S+ ++ A ++G++LS+ +L++
Sbjct: 477 SIFWQIPQYILMGIAEVFFFIGRVEFFYDESPDAMRSVCSALALLNTAVGSYLSSLILTL 536
Query: 406 VADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVI 449
VA T+ G GW+ ++LN +DY++ L L VN + +I
Sbjct: 537 VAYFTALGGKDGWVPDDLNKGHLDYFFWLLVSLGLVNIPVYALI 580
>At3g53960 transporter like protein
Length = 620
Score = 331 bits (849), Expect = 5e-91
Identities = 171/412 (41%), Positives = 263/412 (63%), Gaps = 14/412 (3%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G P+ + GADQF++ +ER K+S++NWW + G ++A T++VYI+D +G+ +
Sbjct: 166 GGHKPSLESFGADQFEDGHPEERKMKMSYFNWWNAGLCAGILTAVTVIVYIEDRIGWGVA 225
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
I TI++ S +F +G P YR+R PSGSPLT M+QVFVAA+ K L P DS LHE+
Sbjct: 226 SIILTIVMATSFFIFRIGKPFYRYRAPSGSPLTPMLQVFVAAIAKRNLPCPSDSSLLHEL 285
Query: 173 SIEEYTSKGRYRINHSSSLRFLDKAAV--------KTGQTSPWMLCTVTQIEETKQMTKM 224
+ EEYT KGR ++ S +L+FLDKAAV K + SPW L TVT++EE K + M
Sbjct: 286 TNEEYT-KGRL-LSSSKNLKFLDKAAVIEDRNENTKAEKQSPWRLATVTKVEEVKLLINM 343
Query: 225 LPVLITTCIPSTIISQTTTLFIRQGTTLDRGM-GAHFEIPPACLIAFVNIFMLISVVIYD 283
+P+ T +Q++TLFI+Q +DR + G F +PPA L + + + ++I+V IY+
Sbjct: 344 IPIWFFTLAFGVCATQSSTLFIKQAIIMDRHITGTSFIVPPASLFSLIALSIIITVTIYE 403
Query: 284 RVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDTI 343
++ VP++RR T N RGI++LQR+G+G+V + MI+A LIE+KRL A+E+++ T+
Sbjct: 404 KLLVPLLRRATGNERGISILQRIGVGMVFSLFAMIIAALIEKKRLDYAKEHHM---NKTM 460
Query: 344 PLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLL 403
L+ L PQF ++G+AD F + E+FYDQ P+SM+SLG ++ + L +F++ L+
Sbjct: 461 TLSAIWLAPQFLVLGVADAFTLVGLQEYFYDQVPDSMRSLGIAFYLSVLGAASFVNNLLI 520
Query: 404 SIVADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVY 455
++ L + KGW +LN SR+D +Y LA L+A N CF ++A + Y
Sbjct: 521 TVSDHLAEEISGKGWFGKDLNSSRLDRFYWMLAALTAANICCFVIVAMRYTY 572
>At2g02040 histidine transport protein (PTR2-B)
Length = 585
Score = 323 bits (828), Expect = 1e-88
Identities = 165/405 (40%), Positives = 262/405 (63%), Gaps = 8/405 (1%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G + P S+ GADQFD+ + +ER +K SF+NW+ F I IG + + ++LV+IQ+N G+ LG
Sbjct: 172 GGIKPCVSSFGADQFDDTDSRERVRKASFFNWFYFSINIGALVSSSLLVWIQENRGWGLG 231
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
+GIPT+ + ++I F GTPLYR + P GSP+TR+ QV VA+ RK + VP D+ L+E
Sbjct: 232 FGIPTVFMGLAIASFFFGTPLYRFQKPGGSPITRISQVVVASFRKSSVKVPEDATLLYET 291
Query: 173 SIEEYTSKGRYRINHSSSLRFLDKAAV------KTGQTS-PWMLCTVTQIEETKQMTKML 225
+ G +I H+ ++LDKAAV K+G S W LCTVTQ+EE K + +M
Sbjct: 292 QDKNSAIAGSRKIEHTDDCQYLDKAAVISEEESKSGDYSNSWRLCTVTQVEELKILIRMF 351
Query: 226 PVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRV 285
P+ + I S + +Q +T+F++QG ++ +G+ F++PPA L F ++I V +YDR
Sbjct: 352 PIWASGIIFSAVYAQMSTMFVQQGRAMNCKIGS-FQLPPAALGTFDTASVIIWVPLYDRF 410
Query: 286 FVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDTIPL 345
VP+ R++T +G T +QR+GIGL + V+ M A ++E RL +A + L+ +P+
Sbjct: 411 IVPLARKFTGVDKGFTEIQRMGIGLFVSVLCMAAAAIVEIIRLHMANDLGLVESGAPVPI 470
Query: 346 TIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSI 405
++ +PQ+ ++G A+ F I +LEFFYDQ+P++M+SL ++ A T ++GN+LS+ +L++
Sbjct: 471 SVLWQIPQYFILGAAEVFYFIGQLEFFYDQSPDAMRSLCSALALLTNALGNYLSSLILTL 530
Query: 406 VADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIA 450
V T++NG +GWI +NLN +DY++ LA LS VN +F A
Sbjct: 531 VTYFTTRNGQEGWISDNLNSGHLDYFFWLLAGLSLVNMAVYFFSA 575
>At3g54450 oligopeptide transporter -like protein
Length = 488
Score = 314 bits (804), Expect = 8e-86
Identities = 156/414 (37%), Positives = 257/414 (61%), Gaps = 12/414 (2%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G P T ADQF E +E++ K SF+N+W I++ + A L++IQ+ V ++LG
Sbjct: 62 GGHKPCVMTFAADQFGEANAEEKAAKTSFFNYWYMAIVLASSIAVLALIFIQERVSWSLG 121
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVP-----IDSK 167
+ I +V++I++F++G P YR ++P GSP TR+ QV VAA++KW+L+ + +
Sbjct: 122 FSIIAGSVVIAIVIFLIGIPKYRKQVPVGSPFTRVAQVMVAALKKWRLSSTRHHYGLCYE 181
Query: 168 ELHEVSIEEYTSKGRYRINHSSSLRFLDKAAV-----KTGQTSPWMLCTVTQIEETKQMT 222
E E +E S Y + ++ RFLDKA + +PW LCTV Q+EE K +
Sbjct: 182 EEDEHKLESTNSNQVYLLARTNQFRFLDKATIIDEIDHNKNRNPWRLCTVNQVEEVKLIL 241
Query: 223 KMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIY 282
+++P+ I+ + ++Q T F++QG+ +DR +G HF IPPA + V + +LI + +Y
Sbjct: 242 RLIPIWISLIMFCATLTQLNTFFLKQGSMMDRTIGNHFTIPPAAFQSIVGVTILILIPLY 301
Query: 283 DRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLL-GPLD 341
DRVFVP++R+ T + GIT LQR+G+GL + M++ L+E KRL VAR++ L+ P +
Sbjct: 302 DRVFVPMVRKITNHHSGITSLQRIGVGLFVATFNMVICGLVEAKRLKVARDHGLIDSPKE 361
Query: 342 TIPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTF 401
+P++ L+PQ+ L+GI D F + E FYDQ PE+M+S+G + + + +G+F+ST
Sbjct: 362 VVPMSSLWLLPQYILVGIGDVFTIVGMQELFYDQMPETMRSIGAAIFISVVGVGSFVSTG 421
Query: 402 LLSIVADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVY 455
++S V ++ +G + W++NNLN + +DYYY +A L+AV+ + IA F+Y
Sbjct: 422 IISTVQTISKSHGEE-WLVNNLNRAHLDYYYWIIASLNAVSLCFYLFIANHFLY 474
>At2g37900 putative peptide/amino acid transporter
Length = 575
Score = 308 bits (788), Expect = 6e-84
Identities = 158/406 (38%), Positives = 247/406 (59%), Gaps = 18/406 (4%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G P+ + GADQFD+ +ER K+SF+NWW + G ++A T + YI+D VG+ +
Sbjct: 167 GGHKPSLESFGADQFDDDHVEERKMKMSFFNWWNVSLCAGILTAVTAVAYIEDRVGWGVA 226
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
I T+++ +S+++F +G P YR+R PSGSPLT ++QVFVAA+ K L P D LHEV
Sbjct: 227 GIILTVVMAISLIIFFIGKPFYRYRTPSGSPLTPILQVFVAAIAKRNLPYPSDPSLLHEV 286
Query: 173 SIEEYTSKGRYRINHSSSLRFLDKAAV-------KTGQTSPWMLCTVTQIEETKQMTKML 225
S E+TS GR + H+ L+FLDKAA+ + SPW L T+T++EETK + ++
Sbjct: 287 SKTEFTS-GRL-LCHTEHLKFLDKAAIIEDKNPLALEKQSPWRLLTLTKVEETKLIINVI 344
Query: 226 PVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRV 285
P+ +T +Q +T FI+Q T+DR +G F +PPA + + ++IS+ +Y+++
Sbjct: 345 PIWFSTLAFGICATQASTFFIKQAITMDRHIGG-FTVPPASMFTLTALTLIISLTVYEKL 403
Query: 286 FVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDTIPL 345
VP++R T+N RGI +LQR+G G++ +I MI+A L+E++RL N P+
Sbjct: 404 LVPLLRSITRNQRGINILQRIGTGMIFSLITMIIAALVEKQRLDRTNNNK--------PM 455
Query: 346 TIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSI 405
++ L PQF ++G AD F + E+FY Q P+SM+SLG ++ + + +FL+ L++
Sbjct: 456 SVIWLAPQFMVIGFADAFTLVGLQEYFYHQVPDSMRSLGIAFYLSVIGAASFLNNLLITA 515
Query: 406 VADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAK 451
V L K W +LN SR+D +Y FLA + A N F ++AK
Sbjct: 516 VDTLAENFSGKSWFGKDLNSSRLDRFYWFLAGVIAANICVFVIVAK 561
>At1g68570 peptide transporter like
Length = 596
Score = 303 bits (776), Expect = 1e-82
Identities = 170/439 (38%), Positives = 262/439 (58%), Gaps = 20/439 (4%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G + P GADQFDE + + ++ +++NW+ F + + A T+LV+IQDNVG+ LG
Sbjct: 155 GGIRPCVVAFGADQFDESDPNQTTKTWNYFNWYYFCMGAAVLLAVTVLVWIQDNVGWGLG 214
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELH-E 171
GIPT+ + +S++ FV G LYRH +P+GSP TR++QV VAA RK KL + D L+
Sbjct: 215 LGIPTVAMFLSVIAFVGGFQLYRHLVPAGSPFTRLIQVGVAAFRKRKLRMVSDPSLLYFN 274
Query: 172 VSIEEYTSKGRYRINHSSSLRFLDKAAV-------KTGQ-TSPWMLCTVTQIEETKQMTK 223
I+ S G ++ H+ + FLDKAA+ K GQ + W L TV ++EE K + +
Sbjct: 275 DEIDAPISLGG-KLTHTKHMSFLDKAAIVTEEDNLKPGQIPNHWRLSTVHRVEELKSVIR 333
Query: 224 MLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYD 283
M P+ + + T +Q T ++Q T++R + F+IP + F + ML +++ YD
Sbjct: 334 MGPIGASGILLITAYAQQGTFSLQQAKTMNRHLTNSFQIPAGSMSVFTTVAMLTTIIFYD 393
Query: 284 RVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLG-PLDT 342
RVFV V R++T RGIT L R+GIG V+ +I +VA +E KR SVA E+ LL P
Sbjct: 394 RVFVKVARKFTGLERGITFLHRMGIGFVISIIATLVAGFVEVKRKSVAIEHGLLDKPHTI 453
Query: 343 IPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFL 402
+P++ L+PQ+ L G+A+ F+ I LEFFYDQAPESM+S T+ +SIGN++ST L
Sbjct: 454 VPISFLWLIPQYGLHGVAEAFMSIGHLEFFYDQAPESMRSTATALFWMAISIGNYVSTLL 513
Query: 403 LSIVADLTSKNGHKGWIL-NNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVY------ 455
+++V ++K W+ NNLN R++Y+Y + +L AVN + + AK + Y
Sbjct: 514 VTLVHKFSAKPDGSNWLPDNNLNRGRLEYFYWLITVLQAVNLVYYLWCAKIYTYKPVQVH 573
Query: 456 --NDDVTRTKMDLEMNPNS 472
+D + K +L+++ S
Sbjct: 574 HSKEDSSPVKEELQLSNRS 592
>At1g22540 Similar to peptide transporter
Length = 557
Score = 301 bits (771), Expect = 5e-82
Identities = 157/407 (38%), Positives = 244/407 (59%), Gaps = 18/407 (4%)
Query: 52 RGELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFAL 111
+G P GADQFDE E +E K SF+NWW F + GT++ +L YIQDN+ +AL
Sbjct: 158 QGGHKPCVQAFGADQFDEKEPEECKAKSSFFNWWYFGMCFGTLTTLWVLNYIQDNLSWAL 217
Query: 112 GYGIPTILLVVSILVFVLGTPLYRH--RLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKEL 169
G+GIP I +VV+++V +LGT YR R SP R+ V+VAA++ W ++ L
Sbjct: 218 GFGIPCIAMVVALVVLLLGTCTYRFSIRREDQSPFVRIGNVYVAAVKNWSVSA------L 271
Query: 170 HEVSIEEYTSKGRYRINHSSSLRFLDKAAV-KTGQTSPWMLCTVTQIEETKQMTKMLPVL 228
+ EE G + S FL+KA V K G C++ ++EE K + ++ P+
Sbjct: 272 DVAAAEERL--GLVSCSSSQQFSFLNKALVAKNGS------CSIDELEEAKSVLRLAPIW 323
Query: 229 ITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRVFVP 288
+T + + + +Q+ T F +QG T++R + ++I PA L +F+++ ++I + IYDRV +P
Sbjct: 324 LTCLVYAVVFAQSPTFFTKQGATMERSITPGYKISPATLQSFISLSIVIFIPIYDRVLIP 383
Query: 289 VIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLL-GPLDTIPLTI 347
+ R +T P GITMLQR+G G+ + + M+VA L+E KRL A + L+ P T+P+++
Sbjct: 384 IARSFTHKPGGITMLQRIGTGIFLSFLAMVVAALVEMKRLKTAADYGLVDSPDATVPMSV 443
Query: 348 FILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSIVA 407
+ LVPQ+ L GI D F + EFFYDQ P ++S+G + + IGNFLS+F++SI+
Sbjct: 444 WWLVPQYVLFGITDVFAMVGLQEFFYDQVPNELRSVGLALYLSIFGIGNFLSSFMISIIE 503
Query: 408 DLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFV 454
TS++G W NNLN + +DY+Y LA LS + + +AK +V
Sbjct: 504 KATSQSGQASWFANNLNQAHLDYFYWLLACLSFIGLASYLYVAKSYV 550
>At2g02020 putative peptide/amino acid transporter
Length = 545
Score = 295 bits (756), Expect = 3e-80
Identities = 164/395 (41%), Positives = 243/395 (61%), Gaps = 35/395 (8%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G + P S+ GADQFD+ + ER +K SF+NW+ F I IG + T+LV+IQ+N G+ LG
Sbjct: 173 GGIKPCVSSFGADQFDKTDPSERVRKASFFNWFYFTINIGAFVSSTVLVWIQENYGWELG 232
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
+ IPT+ + ++ + F GTPLYR + P GSP+T + QV VAA RK L VP DS
Sbjct: 233 FLIPTVFMGLATMSFFFGTPLYRFQKPRGSPITSVCQVLVAAYRKSNLKVPEDS------ 286
Query: 173 SIEEYTSKGRYRINHSSSLRFLDKAAVKTGQTSPWMLCTVTQIEETKQMTKMLPVLITTC 232
T +G T+PW LCTVTQ+EE K + +++P+ +
Sbjct: 287 -----TDEG-------------------DANTNPWKLCTVTQVEEVKILLRLVPIWASGI 322
Query: 233 IPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRVFVPVIRR 292
I S + SQ TLF++QG + R +G FEIPPA L F +LISV IYDRV VP++RR
Sbjct: 323 IFSVLHSQIYTLFVQQGRCMKRTIGL-FEIPPATLGMFDTASVLISVPIYDRVIVPLVRR 381
Query: 293 YTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDTIPLTIFILVP 352
+T +G T LQR+GIGL + V+ + A ++E RL +AR+ +L+ D +PL IF +P
Sbjct: 382 FTGLAKGFTELQRMGIGLFVSVLSLTFAAIVETVRLQLARDLDLVESGDIVPLNIFWQIP 441
Query: 353 QFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSIVADLTSK 412
Q+ LMG A F + ++EFFY+Q+P+SM+SL +++A T ++GN+LS+ ++++VA L+ K
Sbjct: 442 QYFLMGTAGVFFFVGRIEFFYEQSPDSMRSLCSAWALLTTTLGNYLSSLIITLVAYLSGK 501
Query: 413 NGHKGWI-LNNLNVSRIDYYYAFLALLSAVNFICF 446
+ WI +N+N +DY++ L L +VN F
Sbjct: 502 D---CWIPSDNINNGHLDYFFWLLVSLGSVNIPVF 533
>At2g26690 putative nitrate transporter
Length = 586
Score = 290 bits (743), Expect = 9e-79
Identities = 158/412 (38%), Positives = 251/412 (60%), Gaps = 23/412 (5%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G L + S G+DQFD+ + KE++ F+N + F+I +GT+ A T+LVY+QD VG +
Sbjct: 165 GGLKSSISGFGSDQFDDKDPKEKAHMAFFFNRFFFFISMGTLLAVTVLVYMQDEVGRSWA 224
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
YGI T+ + ++I++F+ GT YR++ GSP+ ++ QV AA RK K+ +P L+E
Sbjct: 225 YGICTVSMAIAIVIFLCGTKRYRYKKSQGSPVVQIFQVIAAAFRKRKMELPQSIVYLYED 284
Query: 173 SIEEYTSKGRYRINHSSSLRFLDKAAVKTG----QT-------SPWMLCTVTQIEETKQM 221
+ E RI H+ LDKAA+ QT +PW L +VT++EE K M
Sbjct: 285 NPEGI------RIEHTDQFHLLDKAAIVAEGDFEQTLDGVAIPNPWKLSSVTKVEEVKMM 338
Query: 222 TKMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVI 281
++LP+ TT I T +Q T + Q +T+ R +G+ F+IP L F +LI++ +
Sbjct: 339 VRLLPIWATTIIFWTTYAQMITFSVEQASTMRRNIGS-FKIPAGSLTVFFVAAILITLAV 397
Query: 282 YDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLD 341
YDR +P +++ P G + LQR+ IGLV+ M A L+E+KRLSVA+ ++
Sbjct: 398 YDRAIMPFWKKWKGKP-GFSSLQRIAIGLVLSTAGMAAAALVEQKRLSVAKSSSQ----K 452
Query: 342 TIPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTF 401
T+P+++F+LVPQF L+G + F+ +L+FF Q+P+ MK++ T TTLS+G F+S+F
Sbjct: 453 TLPISVFLLVPQFFLVGAGEAFIYTGQLDFFITQSPKGMKTMSTGLFLTTLSLGFFVSSF 512
Query: 402 LLSIVADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFF 453
L+SIV +TS + GW+ +N+N R+DY+Y L +LS +NF+ + + A +F
Sbjct: 513 LVSIVKRVTSTSTDVGWLADNINHGRLDYFYWLLVILSGINFVVYIICALWF 564
>At1g22570 Similar to LeOPT1 [Lycopersicon esculentum
Length = 565
Score = 285 bits (730), Expect = 3e-77
Identities = 152/411 (36%), Positives = 245/411 (58%), Gaps = 18/411 (4%)
Query: 52 RGELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFAL 111
+G P GADQFD + KE + SF+NWW + G + ++ Y+Q+NV +A
Sbjct: 160 QGGHKPCVQAFGADQFDAEDPKEVIARGSFFNWWFLSLSAGISISIIVVAYVQENVNWAF 219
Query: 112 GYGIPTILLVVSILVFVLGTPLYRH------RLPSGSPLTRMVQVFVAAMRKWKLNVPID 165
G+GIP + +V+++ +F+LG +YR+ + S + R+ +VFV A + KL +
Sbjct: 220 GFGIPCLFMVMALAIFLLGRKIYRYPKGHHEEVNSSNTFARIGRVFVIAFKNRKLRLEHS 279
Query: 166 SKELHEVSIEEYTSKGRYRINHSSSLRFLDKAAVKTGQTSPWMLCTVTQIEETKQMTKML 225
S EL + +E+ S+ R L FL KA + P C+ +++ K + +++
Sbjct: 280 SLELDQGLLEDGQSEKR-----KDRLNFLAKAMISREGVEP---CSGRDVDDAKALVRLI 331
Query: 226 PVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRV 285
P+ IT + + +Q T F +QG T+DR + EIP A L++FV + +LISV +Y+RV
Sbjct: 332 PIWITYVVSTIPYAQYITFFTKQGVTVDRRILPGVEIPAASLLSFVGVSILISVPLYERV 391
Query: 286 FVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLD-TIP 344
F+P+ R+ TK P GITMLQR+G G+V+ V M++A L+E KRL +ARE+ L+ D T+P
Sbjct: 392 FLPIARKITKKPFGITMLQRIGAGMVLSVFNMMLAALVESKRLKIAREHGLVDKPDVTVP 451
Query: 345 LTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLS 404
++I+ VPQ+ L+G+ D F + EFFYDQ P ++S+G S + + + + +FLS FL+S
Sbjct: 452 MSIWWFVPQYLLLGMIDLFSMVGTQEFFYDQVPTELRSIGLSLSLSAMGLSSFLSGFLIS 511
Query: 405 IVADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVY 455
++ T K+ GW +NLN + +DY+Y LA +A+ F F I+K +VY
Sbjct: 512 LIDWATGKD---GWFNSNLNRAHVDYFYWLLAAFTAIAFFAFLFISKMYVY 559
>At1g12110 nitrate/chlorate transporter CHL1
Length = 590
Score = 285 bits (730), Expect = 3e-77
Identities = 162/416 (38%), Positives = 252/416 (59%), Gaps = 17/416 (4%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G + + S G+DQFDE E KERS+ F+N + F I +G++ A T+LVY+QD+VG G
Sbjct: 161 GGVKASVSGFGSDQFDETEPKERSKMTYFFNRFFFCINVGSLLAVTVLVYVQDDVGRKWG 220
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
YGI +V+++ VF+ GT YR + GSP+T++ V VAA R KL +P D L++V
Sbjct: 221 YGICAFAIVLALSVFLAGTNRYRFKKLIGSPMTQVAAVIVAAWRNRKLELPADPSYLYDV 280
Query: 173 S---IEEYTSKGRYRINHSSSLRFLDKAAVK---TGQTS----PWMLCTVTQIEETKQMT 222
E + KG+ ++ H+ R LDKAA++ G TS W L T+T +EE KQ+
Sbjct: 281 DDIIAAEGSMKGKQKLPHTEQFRSLDKAAIRDQEAGVTSNVFNKWTLSTLTDVEEVKQIV 340
Query: 223 KMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIY 282
+MLP+ T + T+ +Q TTL + Q TLDR +G+ FEIPPA + F +L++ +Y
Sbjct: 341 RMLPIWATCILFWTVHAQLTTLSVAQSETLDRSIGS-FEIPPASMAVFYVGGLLLTTAVY 399
Query: 283 DRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGP-LD 341
DRV + + ++ P G+ LQR+G+GL + M VA L+E KRL A + GP +
Sbjct: 400 DRVAIRLCKKLFNYPHGLRPLQRIGLGLFFGSMAMAVAALVELKRLRTAHAH---GPTVK 456
Query: 342 TIPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTF 401
T+PL ++L+PQ+ ++GI + + +L+FF + P+ MK + T +TL++G F S+
Sbjct: 457 TLPLGFYLLIPQYLIVGIGEALIYTGQLDFFLRECPKGMKGMSTGLLLSTLALGFFFSSV 516
Query: 402 LLSIVADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVYND 457
L++IV T K H WI ++LN R+ +Y +A+L A+NF+ F V +K++VY +
Sbjct: 517 LVTIVEKFTGK-AHP-WIADDLNKGRLYNFYWLVAVLVALNFLIFLVFSKWYVYKE 570
>At1g22550 Similar to LeOPT1 [Lycopersicon esculentum
Length = 564
Score = 277 bits (708), Expect = 1e-74
Identities = 150/410 (36%), Positives = 238/410 (57%), Gaps = 20/410 (4%)
Query: 52 RGELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFAL 111
+G P GADQFD + KER + SF+NWW + G + ++VY+QDNV +AL
Sbjct: 163 QGGHKPCVQAFGADQFDVGDPKERISRGSFFNWWFLSLSAGITLSIIVVVYVQDNVNWAL 222
Query: 112 GYGIPTILLVVSILVFVLGTPLYRH----RLPSGSPLTRMVQVFVAAMRKWKLNVPIDSK 167
G+GIP + +V+++ +F+ G YR+ R + R+ +VF+ A + KL K
Sbjct: 223 GFGIPCLFMVMALALFLFGRKTYRYPRGDREGKNNAFARIGRVFLVAFKNRKL------K 276
Query: 168 ELHEVSIEEYTSKGRYRINHSSSLRFLDKAAVK-TGQTSPWMLCTVTQIEETKQMTKMLP 226
H +E G Y+ L FL KA + G P C+ +E+ + +++P
Sbjct: 277 LTHSGQLEV----GSYK-KCKGQLEFLAKALLPGEGGVEP---CSSRDVEDAMALVRLIP 328
Query: 227 VLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRVF 286
+ IT+ + + +Q T F +QG T+DR + FEIPPA A + + + ISV Y+RVF
Sbjct: 329 IWITSVVSTIPYAQYATFFTKQGVTVDRKILPGFEIPPASFQALIGLSIFISVPTYERVF 388
Query: 287 VPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLG-PLDTIPL 345
+P+ R TK P GITMLQR+G G+V+ + M+VA L+E KRL A+E+ L+ P TIP+
Sbjct: 389 LPLARLITKKPSGITMLQRIGAGMVLSSLNMVVAALVEMKRLETAKEHGLVDRPDATIPM 448
Query: 346 TIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSI 405
+I+ VPQ+ L+G+ D F + EFFYDQ P ++S+G + + + + + +FLS FL+++
Sbjct: 449 SIWWFVPQYLLLGMIDVFSLVGTQEFFYDQVPTELRSIGLALSLSAMGLASFLSGFLITV 508
Query: 406 VADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVY 455
+ T KNG W NLN + +DY+Y LA +A+ F+ F ++++ +VY
Sbjct: 509 INWATGKNGGDSWFNTNLNRAHVDYFYWLLAAFTAIGFLAFLLLSRLYVY 558
>At1g72140 peptide transporter like protein (PTR2-B)
Length = 555
Score = 272 bits (696), Expect = 3e-73
Identities = 141/403 (34%), Positives = 236/403 (57%), Gaps = 32/403 (7%)
Query: 63 GADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALGYGIPTILLVV 122
GADQFDE + E K S++NW F I IG ++ + + Y+Q+N+ +ALGY IP + +++
Sbjct: 173 GADQFDEQDPNESKAKSSYFNWLYFAISIGILTTRLVTNYVQENLSWALGYAIPCLSMML 232
Query: 123 SILVFVLGTPLYRHRLPS--------GSPLTRMVQVFVAAMRKWKLNVPIDSKELHEVSI 174
++ +F+LG YR +P R+ +VFVAA R + P D+ L
Sbjct: 233 ALFLFLLGIKTYRFSTGGEGRQGKKHDNPFVRIGRVFVAAARN-RRQTPSDTCLLLP--- 288
Query: 175 EEYTSKGRYRINHSSSLRFLDKAAVKTGQTSPWMLCTVTQIEETKQMTKMLPVLITTCIP 234
E T K RFLD+A + C ++EE K + ++P+ + + +
Sbjct: 289 NESTKK----------FRFLDRAVIS---------CDSYEVEEAKAVLSLIPIWLCSLVF 329
Query: 235 STIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRVFVPVIRRYT 294
+ +Q+ T F +QG+T+DR + + ++P A L F+++ +L+ + IYDR+FVP+ R T
Sbjct: 330 GIVFAQSPTFFTKQGSTMDRSISSTLQVPAATLQCFISLAILVFIPIYDRLFVPIARSIT 389
Query: 295 KNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLL-GPLDTIPLTIFILVPQ 353
+ P GIT LQR+ G+ + +I M++A L+E KRL AR++ L+ P T+P+++ L+PQ
Sbjct: 390 RKPAGITTLQRISTGIFLSIISMVIAALVEMKRLKTARDHGLVDSPKATVPMSVCWLIPQ 449
Query: 354 FALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSIVADLTSKN 413
+ L G++D F + EFFY + P ++S+G + + + IGNFLS+F++S++ + TS++
Sbjct: 450 YILFGVSDVFTMVGLQEFFYGEVPPQLRSMGLALYLSIIGIGNFLSSFMVSVIEEATSQS 509
Query: 414 GHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVYN 456
G W NNLN + +DY+Y LA LS++ FI AK ++YN
Sbjct: 510 GQVSWFSNNLNQAHLDYFYWLLACLSSLAFIFTVYFAKSYLYN 552
>At3g21670 nitrate transporter
Length = 590
Score = 271 bits (694), Expect = 5e-73
Identities = 158/417 (37%), Positives = 238/417 (56%), Gaps = 30/417 (7%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G + S G+DQFD + KE Q + F+N + F I +G++ A LVY+QDNVG G
Sbjct: 165 GGIKSNVSGFGSDQFDTSDPKEEKQMIFFFNRFYFSISVGSLFAVIALVYVQDNVGRGWG 224
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
YGI +VV+ +V + GT YR + P GSP T + +V A +K K + P H
Sbjct: 225 YGISAATMVVAAIVLLCGTKRYRFKKPKGSPFTTIWRVGFLAWKKRKESYP-----AHPS 279
Query: 173 SIEEYTSKGRYRINHSSSLRFLDKAAVKTGQTS----------PWMLCTVTQIEETKQMT 222
+ Y + + H+ L+ LDKAA+ ++S PW++ TVTQ+EE K +
Sbjct: 280 LLNGYDNT---TVPHTEMLKCLDKAAISKNESSPSSKDFEEKDPWIVSTVTQVEEVKLVM 336
Query: 223 KMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIY 282
K++P+ T + TI SQ TT + Q T +DR +G+ F +P AF+ + +L+ +
Sbjct: 337 KLVPIWATNILFWTIYSQMTTFTVEQATFMDRKLGS-FTVPAGSYSAFLILTILLFTSLN 395
Query: 283 DRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDT 342
+RVFVP+ RR TK P+GIT LQR+G+GLV + M VA +IE R A N+
Sbjct: 396 ERVFVPLTRRLTKKPQGITSLQRIGVGLVFSMAAMAVAAVIENARREAAVNNDK------ 449
Query: 343 IPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFL 402
++ F LVPQ+ L+G + F + +LEFF +APE MKS+ T +T+S+G F+S+ L
Sbjct: 450 -KISAFWLVPQYFLVGAGEAFAYVGQLEFFIREAPERMKSMSTGLFLSTISMGFFVSSLL 508
Query: 403 LSIVADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVYNDDV 459
+S+V +T K+ W+ +NLN +R++Y+Y L +L A+NF+ F V A Y DV
Sbjct: 509 VSLVDRVTDKS----WLRSNLNKARLNYFYWLLVVLGALNFLIFIVFAMKHQYKADV 561
>At1g72125 oligopeptide like transporter
Length = 557
Score = 271 bits (694), Expect = 5e-73
Identities = 144/404 (35%), Positives = 237/404 (58%), Gaps = 16/404 (3%)
Query: 57 PTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALGYGIP 116
P GADQFDE + +E+S + SF+NWW + G A ++VYIQ+ +A G+GIP
Sbjct: 159 PCVQAFGADQFDEKDSQEKSDRSSFFNWWYLSLSAGICFAILVVVYIQEEFSWAFGFGIP 218
Query: 117 TILLVVSILVFVLGTPLYRHRLPSG----SPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
+ +V+S+++FV G +YR+ +P TR+ +VF A++ +L+ S +L +V
Sbjct: 219 CVFMVISLVLFVSGRRIYRYSKRRHEEEINPFTRIGRVFFVALKNQRLS----SSDLCKV 274
Query: 173 SIEEYTSKGRYRINHSSSLRFLDKAAVKTGQTSPWMLCTVTQIEETKQMTKMLPVLITTC 232
+E TS + + + L D + + S + +E+ + +++PV TT
Sbjct: 275 ELEANTSPEKQSFFNKALLVPNDSSQGENASKS-------SDVEDATALIRLIPVWFTTL 327
Query: 233 IPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRVFVPVIRR 292
+ +Q T F +QG T+DR + +IPPA L F+ I +++ V IYDRVFVP+ R
Sbjct: 328 AYAIPYAQYMTFFTKQGVTMDRTILPGVKIPPASLQVFIGISIVLFVPIYDRVFVPIARL 387
Query: 293 YTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLG-PLDTIPLTIFILV 351
TK P GIT L+R+G G+V+ I M++A L+E KRL A+E+ L+ P T+P++I+ L+
Sbjct: 388 ITKEPCGITTLKRIGTGIVLSTITMVIAALVEFKRLETAKEHGLIDQPEATLPMSIWWLI 447
Query: 352 PQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSIVADLTS 411
PQ+ L+G+AD + + EFFY Q P ++S+G + + L +G+ LS+ L+S++ T
Sbjct: 448 PQYLLLGLADVYTLVGMQEFFYSQVPTELRSIGLALYLSALGVGSLLSSLLISLIDLATG 507
Query: 412 KNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVY 455
+ W +NLN + +DY+Y LA++SAV F F I+K ++Y
Sbjct: 508 GDAGNSWFNSNLNRAHLDYFYWLLAIVSAVGFFTFLFISKSYIY 551
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.140 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,133,626
Number of Sequences: 26719
Number of extensions: 362493
Number of successful extensions: 1268
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 1018
Number of HSP's gapped (non-prelim): 64
length of query: 485
length of database: 11,318,596
effective HSP length: 103
effective length of query: 382
effective length of database: 8,566,539
effective search space: 3272417898
effective search space used: 3272417898
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147517.7


