

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147517.5 + phase: 0
(167 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
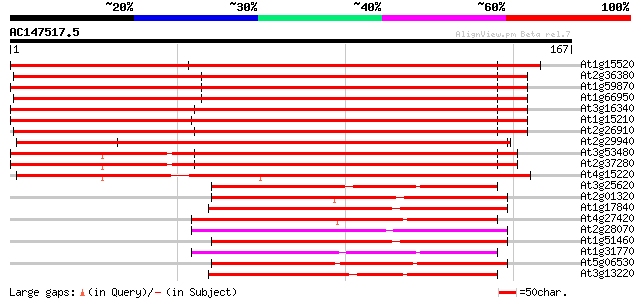
Score E
Sequences producing significant alignments: (bits) Value
At1g15520 hypothetical protein 233 5e-62
At2g36380 putative ABC transporter 202 9e-53
At1g59870 putative ABC transporter (At1g59870) 200 3e-52
At1g66950 ABC transporter, putative 198 1e-51
At3g16340 putative ABC transporter 197 3e-51
At1g15210 putative ABC transporter 196 5e-51
At2g26910 putative ABC transporter 181 2e-46
At2g29940 putative ABC transporter 180 4e-46
At3g53480 ABC transporter - like protein 165 1e-41
At2g37280 putative ABC transporter 157 2e-39
At4g15220 ABC transporter homolog 146 6e-36
At3g25620 membrane transporter, putative 75 1e-14
At2g01320 putative ABC transporter 74 5e-14
At1g17840 putative ABC transporter (At1g17840) 72 1e-13
At4g27420 putative protein 72 2e-13
At2g28070 putative ABC transporter 69 2e-12
At1g51460 ATP-dependent transmembrane transporter like protein 68 3e-12
At1g31770 unknown protein 67 3e-12
At5g06530 ABC transporter like protein 64 3e-11
At3g13220 ABC transporter 64 3e-11
>At1g15520 hypothetical protein
Length = 1423
Score = 233 bits (593), Expect = 5e-62
Identities = 111/158 (70%), Positives = 133/158 (83%)
Query: 1 MDRVGIDLPTIEVRFEHLNIEAEAHVGSISLPTFTNFMVNIVESLLNSLHVLPSRKQRLN 60
+DRVGIDLPTIEVRF+HL +EAE HVG +LPTF NF+ N + LN+LH++P+RK++
Sbjct: 109 IDRVGIDLPTIEVRFDHLKVEAEVHVGGRALPTFVNFISNFADKFLNTLHLVPNRKKKFT 168
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQR 120
IL DVSGI+KP RM LLLGPPSSGKTTLLLALAGKLD +LK +GRVTYNGH M+EFVPQR
Sbjct: 169 ILNDVSGIVKPGRMALLLGPPSSGKTTLLLALAGKLDQELKQTGRVTYNGHGMNEFVPQR 228
Query: 121 TAAYVDQNDLHIGELTVRETLAFSARVQGVGPQYGWST 158
TAAY+ QND+HIGE+TVRET A++AR QGVG +Y T
Sbjct: 229 TAAYIGQNDVHIGEMTVRETFAYAARFQGVGSRYDMLT 266
Score = 60.1 bits (144), Expect = 5e-10
Identities = 32/92 (34%), Positives = 54/92 (57%), Gaps = 1/92 (1%)
Query: 54 SRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEM 113
+++ RL +LK V+G +P +T L+G +GKTTL+ LAG+ G +T +G+
Sbjct: 845 TQEDRLVLLKGVNGAFRPGVLTALMGVSGAGKTTLMDVLAGRKTGGY-IDGNITISGYPK 903
Query: 114 SEFVPQRTAAYVDQNDLHIGELTVRETLAFSA 145
++ R + Y +Q D+H +TV E+L +SA
Sbjct: 904 NQQTFARISGYCEQTDIHSPHVTVYESLVYSA 935
>At2g36380 putative ABC transporter
Length = 1450
Score = 202 bits (513), Expect = 9e-53
Identities = 97/153 (63%), Positives = 122/153 (79%)
Query: 2 DRVGIDLPTIEVRFEHLNIEAEAHVGSISLPTFTNFMVNIVESLLNSLHVLPSRKQRLNI 61
DRVGI++P IEVR+E+L++E + S +LPT N +N +ES+L H+LPS+K+++ I
Sbjct: 129 DRVGIEVPKIEVRYENLSVEGDVRSASRALPTLFNVTLNTIESILGLFHLLPSKKRKIEI 188
Query: 62 LKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQRT 121
LKD+SGIIKPSRMTLLLGPPSSGKTTLL ALAGKLD L+ SGR+TY GHE EFVPQ+T
Sbjct: 189 LKDISGIIKPSRMTLLLGPPSSGKTTLLQALAGKLDDTLQMSGRITYCGHEFREFVPQKT 248
Query: 122 AAYVDQNDLHIGELTVRETLAFSARVQGVGPQY 154
AY+ Q+DLH GE+TVRE+L FS R GVG +Y
Sbjct: 249 CAYISQHDLHFGEMTVRESLDFSGRCLGVGTRY 281
Score = 60.8 bits (146), Expect = 3e-10
Identities = 32/88 (36%), Positives = 51/88 (57%), Gaps = 1/88 (1%)
Query: 58 RLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFV 117
RL +L+DV G +P +T L+G +GKTTL+ LAG+ G + +G+ ++
Sbjct: 873 RLQLLRDVGGAFRPGVLTALVGVSGAGKTTLMDVLAGRKTGGY-VEGSINISGYPKNQAT 931
Query: 118 PQRTAAYVDQNDLHIGELTVRETLAFSA 145
R + Y +QND+H +TV E+L +SA
Sbjct: 932 FARVSGYCEQNDIHSPHVTVYESLIYSA 959
>At1g59870 putative ABC transporter (At1g59870)
Length = 1469
Score = 200 bits (508), Expect = 3e-52
Identities = 96/154 (62%), Positives = 125/154 (80%)
Query: 1 MDRVGIDLPTIEVRFEHLNIEAEAHVGSISLPTFTNFMVNIVESLLNSLHVLPSRKQRLN 60
+DRVGI LPT+EVR+EHL I+A+ + G+ SLPT N + N+ ES L + + ++K +L
Sbjct: 126 IDRVGIKLPTVEVRYEHLTIKADCYTGNRSLPTLLNVVRNMGESALGMIGIQFAKKAQLT 185
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQR 120
ILKD+SG+IKP RMTLLLGPPSSGKTTLLLALAGKLD L+ SG +TYNG+++ EFVP++
Sbjct: 186 ILKDISGVIKPGRMTLLLGPPSSGKTTLLLALAGKLDKSLQVSGDITYNGYQLDEFVPRK 245
Query: 121 TAAYVDQNDLHIGELTVRETLAFSARVQGVGPQY 154
T+AY+ QNDLH+G +TV+ETL FSAR QGVG +Y
Sbjct: 246 TSAYISQNDLHVGIMTVKETLDFSARCQGVGTRY 279
Score = 59.3 bits (142), Expect = 9e-10
Identities = 34/88 (38%), Positives = 50/88 (56%), Gaps = 1/88 (1%)
Query: 58 RLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFV 117
RL +LK V+G +P +T L+G +GKTTL+ LAG+ G V +G +
Sbjct: 891 RLQLLKGVTGAFRPGVLTALMGVSGAGKTTLMDVLAGRKTGGY-IEGDVRISGFPKVQET 949
Query: 118 PQRTAAYVDQNDLHIGELTVRETLAFSA 145
R + Y +Q D+H ++TVRE+L FSA
Sbjct: 950 FARISGYCEQTDIHSPQVTVRESLIFSA 977
>At1g66950 ABC transporter, putative
Length = 1434
Score = 198 bits (503), Expect = 1e-51
Identities = 94/153 (61%), Positives = 121/153 (78%)
Query: 2 DRVGIDLPTIEVRFEHLNIEAEAHVGSISLPTFTNFMVNIVESLLNSLHVLPSRKQRLNI 61
DRVGI++P IEVR+E++++E + S +LPT N +N +ES+L H+LPS+++++ I
Sbjct: 131 DRVGIEVPKIEVRYENISVEGDVRSASRALPTLFNVTLNTLESILGFFHLLPSKRKKIQI 190
Query: 62 LKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQRT 121
LKD+SGI+KPSRMTLLLGPPSSGKTTLL ALAGKLD L+ SGR+TY GHE EFVPQ+T
Sbjct: 191 LKDISGIVKPSRMTLLLGPPSSGKTTLLQALAGKLDDTLQMSGRITYCGHEFREFVPQKT 250
Query: 122 AAYVDQNDLHIGELTVRETLAFSARVQGVGPQY 154
AY+ Q+DLH GE+TVRE L FS R GVG +Y
Sbjct: 251 CAYISQHDLHFGEMTVREILDFSGRCLGVGSRY 283
Score = 61.6 bits (148), Expect = 2e-10
Identities = 32/88 (36%), Positives = 52/88 (58%), Gaps = 1/88 (1%)
Query: 58 RLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFV 117
RL +L+DV G +P +T L+G +GKTTL+ LAG+ G ++ +G+ ++
Sbjct: 857 RLQLLRDVGGAFRPGILTALVGVSGAGKTTLMDVLAGRKTGGY-IEGSISISGYPKNQTT 915
Query: 118 PQRTAAYVDQNDLHIGELTVRETLAFSA 145
R + Y +QND+H +TV E+L +SA
Sbjct: 916 FARVSGYCEQNDIHSPHVTVYESLIYSA 943
>At3g16340 putative ABC transporter
Length = 1416
Score = 197 bits (500), Expect = 3e-51
Identities = 99/154 (64%), Positives = 120/154 (77%)
Query: 1 MDRVGIDLPTIEVRFEHLNIEAEAHVGSISLPTFTNFMVNIVESLLNSLHVLPSRKQRLN 60
+DRV I LPT+EVRFE + IEA H+G +LPT N +NI E L L ++ ++
Sbjct: 102 IDRVRIKLPTVEVRFEKVTIEANCHIGKRALPTLPNAALNIAERGLRLLGFNFTKTTKVT 161
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQR 120
IL+DVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLD LK +GRVTYNGH + EFVPQ+
Sbjct: 162 ILRDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDQSLKVTGRVTYNGHGLEEFVPQK 221
Query: 121 TAAYVDQNDLHIGELTVRETLAFSARVQGVGPQY 154
T+AY+ QND+H+G +TV+ETL FSAR QGVG +Y
Sbjct: 222 TSAYISQNDVHVGVMTVQETLDFSARCQGVGTRY 255
Score = 62.8 bits (151), Expect = 8e-11
Identities = 32/90 (35%), Positives = 54/90 (59%), Gaps = 1/90 (1%)
Query: 56 KQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSE 115
K +L +LK+V+G+ +P +T L+G +GKTTL+ LAG+ G + +G +
Sbjct: 836 KDKLQLLKEVTGVFRPGVLTALMGVSGAGKTTLMDVLAGRKTGGY-IEGDIRISGFPKRQ 894
Query: 116 FVPQRTAAYVDQNDLHIGELTVRETLAFSA 145
R + Y +QND+H ++TV+E+L +SA
Sbjct: 895 ETFARISGYCEQNDIHSPQVTVKESLIYSA 924
>At1g15210 putative ABC transporter
Length = 1442
Score = 196 bits (498), Expect = 5e-51
Identities = 95/154 (61%), Positives = 123/154 (79%)
Query: 1 MDRVGIDLPTIEVRFEHLNIEAEAHVGSISLPTFTNFMVNIVESLLNSLHVLPSRKQRLN 60
+DRVGI LPT+EVR++HL ++A+ + G SLP+ N + N+ E+ L + + ++K +L
Sbjct: 124 IDRVGIQLPTVEVRYDHLTVKADCYTGDRSLPSLLNAVRNMGEAALGMIGIRLAKKAQLT 183
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQR 120
ILKDVSGI+KPSRMTLLLGPPSSGKTTLLLALAGKLD L SG VTYNG+ ++EFVP +
Sbjct: 184 ILKDVSGIVKPSRMTLLLGPPSSGKTTLLLALAGKLDKSLDVSGEVTYNGYRLNEFVPIK 243
Query: 121 TAAYVDQNDLHIGELTVRETLAFSARVQGVGPQY 154
T+AY+ QNDLH+G +TV+ETL FSAR QGVG +Y
Sbjct: 244 TSAYISQNDLHVGIMTVKETLDFSARCQGVGTRY 277
Score = 58.2 bits (139), Expect = 2e-09
Identities = 33/91 (36%), Positives = 51/91 (55%), Gaps = 1/91 (1%)
Query: 55 RKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMS 114
++ RL +LK V+ +P +T L+G +GKTTL+ LAG+ G V +G
Sbjct: 861 QETRLQLLKGVTSAFRPGVLTALMGVSGAGKTTLMDVLAGRKTGGY-IEGDVRVSGFPKK 919
Query: 115 EFVPQRTAAYVDQNDLHIGELTVRETLAFSA 145
+ R + Y +Q D+H ++TVRE+L FSA
Sbjct: 920 QETFARISGYCEQTDIHSPQVTVRESLIFSA 950
>At2g26910 putative ABC transporter
Length = 1420
Score = 181 bits (459), Expect = 2e-46
Identities = 84/153 (54%), Positives = 117/153 (75%)
Query: 2 DRVGIDLPTIEVRFEHLNIEAEAHVGSISLPTFTNFMVNIVESLLNSLHVLPSRKQRLNI 61
D V + P IEVRF++L +E+ HVGS +LPT NF++N+ E LL ++HV+ ++ +L I
Sbjct: 91 DAVDLKFPKIEVRFQNLMVESFVHVGSRALPTIPNFIINMAEGLLRNIHVIGGKRNKLTI 150
Query: 62 LKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQRT 121
L +SG+I+PSR+TLLLGPPSSGKTTLLLALAG+L L+ SG++TYNG+++ E + RT
Sbjct: 151 LDGISGVIRPSRLTLLLGPPSSGKTTLLLALAGRLGTNLQTSGKITYNGYDLKEIIAPRT 210
Query: 122 AAYVDQNDLHIGELTVRETLAFSARVQGVGPQY 154
+AYV Q D H+ E+TVR+TL F+ R QGVG +Y
Sbjct: 211 SAYVSQQDWHVAEMTVRQTLEFAGRCQGVGFKY 243
Score = 54.7 bits (130), Expect = 2e-08
Identities = 33/90 (36%), Positives = 50/90 (54%), Gaps = 1/90 (1%)
Query: 56 KQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSE 115
+ RL +L +++G +P +T L+G +GKTTL+ LAG+ G V +G +
Sbjct: 840 EDRLQLLVNITGAFRPGVLTALVGVSGAGKTTLMDVLAGRKTGG-TIEGDVYISGFPKRQ 898
Query: 116 FVPQRTAAYVDQNDLHIGELTVRETLAFSA 145
R + Y +QND+H LTV E+L FSA
Sbjct: 899 ETFARISGYCEQNDVHSPCLTVVESLLFSA 928
>At2g29940 putative ABC transporter
Length = 1443
Score = 180 bits (456), Expect = 4e-46
Identities = 89/147 (60%), Positives = 113/147 (76%)
Query: 3 RVGIDLPTIEVRFEHLNIEAEAHVGSISLPTFTNFMVNIVESLLNSLHVLPSRKQRLNIL 62
+VG+++P IEVRFE+LNIEA+ G+ +LPT N + E L+SL ++ RK +LNIL
Sbjct: 134 KVGMEVPKIEVRFENLNIEADVQAGTRALPTLVNVSRDFFERCLSSLRIIKPRKHKLNIL 193
Query: 63 KDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQRTA 122
KD+SGIIKP RMTLLLGPP SGK+TLLLALAGKLD LK +G +TYNG +++F +RT+
Sbjct: 194 KDISGIIKPGRMTLLLGPPGSGKSTLLLALAGKLDKSLKKTGNITYNGENLNKFHVKRTS 253
Query: 123 AYVDQNDLHIGELTVRETLAFSARVQG 149
AY+ Q D HI ELTVRETL F+AR QG
Sbjct: 254 AYISQTDNHIAELTVRETLDFAARCQG 280
Score = 65.9 bits (159), Expect = 1e-11
Identities = 39/116 (33%), Positives = 61/116 (51%), Gaps = 1/116 (0%)
Query: 33 TFTNFMVNIVESLLNSLHVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLAL 92
T T VN + + + RL +L +VSG+ P +T L+G +GKTTL+ L
Sbjct: 842 TMTFHNVNYYVDMPKEMRSQGVPETRLQLLSNVSGVFSPGVLTALVGSSGAGKTTLMDVL 901
Query: 93 AGKLDPKLKFSGRVTYNGHEMSEFVPQRTAAYVDQNDLHIGELTVRETLAFSARVQ 148
AG+ G + +GH + R + YV+QND+H ++TV E+L FSA ++
Sbjct: 902 AGRKTGGYT-EGDIRISGHPKEQQTFARISGYVEQNDIHSPQVTVEESLWFSASLR 956
>At3g53480 ABC transporter - like protein
Length = 1450
Score = 165 bits (417), Expect = 1e-41
Identities = 85/152 (55%), Positives = 111/152 (72%), Gaps = 2/152 (1%)
Query: 1 MDRVGIDLPTIEVRFEHLNIEAEAHV-GSISLPTFTNFMVNIVESLLNSLHVLPSRKQRL 59
+DRVG++LPTIEVR+E L + AE V +LPT N ++ L+ L + + ++
Sbjct: 129 IDRVGMELPTIEVRYESLKVVAECEVVEGKALPTLWNTAKRVLSELVK-LTGAKTHEAKI 187
Query: 60 NILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQ 119
NI+ DV+GIIKP R+TLLLGPPS GKTTLL AL+G L+ LK SG ++YNGH + EFVPQ
Sbjct: 188 NIINDVNGIIKPGRLTLLLGPPSCGKTTLLKALSGNLENNLKCSGEISYNGHRLDEFVPQ 247
Query: 120 RTAAYVDQNDLHIGELTVRETLAFSARVQGVG 151
+T+AY+ Q DLHI E+TVRET+ FSAR QGVG
Sbjct: 248 KTSAYISQYDLHIAEMTVRETVDFSARCQGVG 279
Score = 55.8 bits (133), Expect = 1e-08
Identities = 29/90 (32%), Positives = 50/90 (55%), Gaps = 1/90 (1%)
Query: 56 KQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSE 115
+++L +L D++G +P +T L+G +GKTTLL LAG+ G + +G +
Sbjct: 872 QKKLQLLSDITGAFRPGILTALMGVSGAGKTTLLDVLAGRKTSGY-IEGDIRISGFPKVQ 930
Query: 116 FVPQRTAAYVDQNDLHIGELTVRETLAFSA 145
R + Y +Q D+H +TV E++ +SA
Sbjct: 931 ETFARVSGYCEQTDIHSPNITVEESVIYSA 960
>At2g37280 putative ABC transporter
Length = 1413
Score = 157 bits (398), Expect = 2e-39
Identities = 81/152 (53%), Positives = 108/152 (70%), Gaps = 2/152 (1%)
Query: 1 MDRVGIDLPTIEVRFEHLNIEAEAHV-GSISLPTFTNFMVNIVESLLNSLHVLPSRKQRL 59
M+RVG++ P+IEVR+EHL +EA V +LPT N + ++ LL L + + + +
Sbjct: 94 MERVGVEFPSIEVRYEHLGVEAACEVVEGKALPTLWNSLKHVFLDLLK-LSGVRTNEANI 152
Query: 60 NILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQ 119
IL DVSGII P R+TLLLGPP GKTTLL AL+G L+ LK G ++YNGH ++E VPQ
Sbjct: 153 KILTDVSGIISPGRLTLLLGPPGCGKTTLLKALSGNLENNLKCYGEISYNGHGLNEVVPQ 212
Query: 120 RTAAYVDQNDLHIGELTVRETLAFSARVQGVG 151
+T+AY+ Q+DLHI E+T RET+ FSAR QGVG
Sbjct: 213 KTSAYISQHDLHIAEMTTRETIDFSARCQGVG 244
Score = 55.1 bits (131), Expect = 2e-08
Identities = 29/90 (32%), Positives = 50/90 (55%), Gaps = 1/90 (1%)
Query: 56 KQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSE 115
+++L +L +++G +P +T L+G +GKTTLL LAG+ G + +G +
Sbjct: 835 EKKLQLLSEITGAFRPGVLTALMGISGAGKTTLLDVLAGRKTSGY-IEGEIRISGFLKVQ 893
Query: 116 FVPQRTAAYVDQNDLHIGELTVRETLAFSA 145
R + Y +Q D+H +TV E+L +SA
Sbjct: 894 ETFARVSGYCEQTDIHSPSITVEESLIYSA 923
>At4g15220 ABC transporter homolog
Length = 290
Score = 146 bits (368), Expect = 6e-36
Identities = 79/164 (48%), Positives = 107/164 (65%), Gaps = 16/164 (9%)
Query: 3 RVGIDLPTIEVRFEHLNIEAEAHV-GSISLPTFTNFMVNIVESLLNSLHVLPSRKQRLNI 61
RVGI+LPT+EVRF +L++EAE V +PT N + ++ + S ++ ++ I
Sbjct: 123 RVGIELPTVEVRFNNLSVEAECQVIHGKPIPTLWNTIKGLLSEFICS-----KKETKIGI 177
Query: 62 LKDVSGIIKPSR----------MTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGH 111
LK VSGI++P R MTLLLGPP GKTTLL AL+GK +K G V YNG
Sbjct: 178 LKGVSGIVRPGRVQKRCWFSCRMTLLLGPPGCGKTTLLQALSGKFSDSVKVGGEVCYNGC 237
Query: 112 EMSEFVPQRTAAYVDQNDLHIGELTVRETLAFSARVQGVGPQYG 155
+SEF+P++T++Y+ QNDLHI EL+VRETL FSA QG+G + G
Sbjct: 238 SLSEFIPEKTSSYISQNDLHIPELSVRETLDFSACCQGIGSRMG 281
>At3g25620 membrane transporter, putative
Length = 646
Score = 75.5 bits (184), Expect = 1e-14
Identities = 42/85 (49%), Positives = 57/85 (66%), Gaps = 3/85 (3%)
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQR 120
+LK VSGI+KP + +LGP SGKTTL+ ALAG+L KL SG V+YNG + V ++
Sbjct: 98 VLKCVSGIVKPGELLAMLGPSGSGKTTLVTALAGRLQGKL--SGTVSYNGEPFTSSVKRK 155
Query: 121 TAAYVDQNDLHIGELTVRETLAFSA 145
T +V Q+D+ LTV ETL ++A
Sbjct: 156 T-GFVTQDDVLYPHLTVMETLTYTA 179
>At2g01320 putative ABC transporter
Length = 725
Score = 73.6 bits (179), Expect = 5e-14
Identities = 43/90 (47%), Positives = 56/90 (61%), Gaps = 4/90 (4%)
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGK--LDPKLKFSGRVTYNGHEMSEFVP 118
+LK+VSG KP R+ ++GP SGKTTLL LAG+ L P+L SG + NG S
Sbjct: 90 LLKNVSGEAKPGRLLAIMGPSGSGKTTLLNVLAGQLSLSPRLHLSGLLEVNGKPSSS--K 147
Query: 119 QRTAAYVDQNDLHIGELTVRETLAFSARVQ 148
A+V Q DL +LTVRETL+F+A +Q
Sbjct: 148 AYKLAFVRQEDLFFSQLTVRETLSFAAELQ 177
>At1g17840 putative ABC transporter (At1g17840)
Length = 703
Score = 72.0 bits (175), Expect = 1e-13
Identities = 41/89 (46%), Positives = 57/89 (63%), Gaps = 2/89 (2%)
Query: 60 NILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQ 119
N+L+ ++G +P +T L+GP SGK+T+L ALA +L SG V NG + +
Sbjct: 68 NVLEGLTGYAEPGSLTALMGPSGSGKSTMLDALASRLAANAFLSGTVLLNGRKTK--LSF 125
Query: 120 RTAAYVDQNDLHIGELTVRETLAFSARVQ 148
TAAYV Q+D IG LTVRET+ +SARV+
Sbjct: 126 GTAAYVTQDDNLIGTLTVRETIWYSARVR 154
>At4g27420 putative protein
Length = 635
Score = 71.6 bits (174), Expect = 2e-13
Identities = 39/92 (42%), Positives = 59/92 (63%), Gaps = 2/92 (2%)
Query: 55 RKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKL-DPKLKFSGRVTYNGHEM 113
+ + ILK ++GI+KP + +LGP SGKT+LL AL G++ + K K +G ++YN +
Sbjct: 60 KTEERTILKGLTGIVKPGEILAMLGPSGSGKTSLLTALGGRVGEGKGKLTGNISYNNKPL 119
Query: 114 SEFVPQRTAAYVDQNDLHIGELTVRETLAFSA 145
S+ V +RT +V Q+D LTV ETL F+A
Sbjct: 120 SKAV-KRTTGFVTQDDALYPNLTVTETLVFTA 150
>At2g28070 putative ABC transporter
Length = 705
Score = 68.6 bits (166), Expect = 2e-12
Identities = 39/94 (41%), Positives = 55/94 (58%), Gaps = 2/94 (2%)
Query: 55 RKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMS 114
RK ++K +G P MT+++GP SGK+TLL ALAG+L P K G V NG +
Sbjct: 127 RKYSDKVVKSSNGYAFPGTMTVIMGPAKSGKSTLLRALAGRLPPSAKMYGEVFVNGSK-- 184
Query: 115 EFVPQRTAAYVDQNDLHIGELTVRETLAFSARVQ 148
+P + +V++ IG LTVRE L +SA +Q
Sbjct: 185 SHMPYGSYGFVERETQLIGSLTVREFLYYSALLQ 218
>At1g51460 ATP-dependent transmembrane transporter like protein
Length = 678
Score = 67.8 bits (164), Expect = 3e-12
Identities = 37/88 (42%), Positives = 58/88 (65%), Gaps = 2/88 (2%)
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQR 120
+L V+G +P+R+ ++GP SGK+TLL ALAG+L + SG+V NG + +
Sbjct: 30 LLNGVNGCGEPNRILAIMGPSGSGKSTLLDALAGRLAGNVVMSGKVLVNGKKRR--LDFG 87
Query: 121 TAAYVDQNDLHIGELTVRETLAFSARVQ 148
AAYV Q D+ +G LTVRE++++SA ++
Sbjct: 88 AAAYVTQEDVLLGTLTVRESISYSAHLR 115
>At1g31770 unknown protein
Length = 648
Score = 67.4 bits (163), Expect = 3e-12
Identities = 39/91 (42%), Positives = 54/91 (58%), Gaps = 3/91 (3%)
Query: 55 RKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMS 114
+ + IL ++G++ P +LGP SGKTTLL AL G+L FSG+V YNG S
Sbjct: 75 KSKEKTILNGITGMVCPGEFLAMLGPSGSGKTTLLSALGGRLSK--TFSGKVMYNGQPFS 132
Query: 115 EFVPQRTAAYVDQNDLHIGELTVRETLAFSA 145
+ +RT +V Q+D+ LTV ETL F+A
Sbjct: 133 GCIKRRT-GFVAQDDVLYPHLTVWETLFFTA 162
>At5g06530 ABC transporter like protein
Length = 627
Score = 64.3 bits (155), Expect = 3e-11
Identities = 35/88 (39%), Positives = 54/88 (60%), Gaps = 2/88 (2%)
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQR 120
IL +SG + P + L+GP SGKTTLL LAG++ + G VTYN S+++ +
Sbjct: 55 ILTGISGSVNPGEVLALMGPSGSGKTTLLSLLAGRIS-QSSTGGSVTYNDKPYSKYLKSK 113
Query: 121 TAAYVDQNDLHIGELTVRETLAFSARVQ 148
+V Q+D+ LTV+ETL ++AR++
Sbjct: 114 -IGFVTQDDVLFPHLTVKETLTYAARLR 140
>At3g13220 ABC transporter
Length = 647
Score = 64.3 bits (155), Expect = 3e-11
Identities = 37/86 (43%), Positives = 54/86 (62%), Gaps = 3/86 (3%)
Query: 60 NILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQ 119
+ILK ++G P + L+GP SGKTTLL + G+L +K G++TYN S V +
Sbjct: 67 HILKGITGSTGPGEILALMGPSGSGKTTLLKIMGGRLTDNVK--GKLTYNDIPYSPSV-K 123
Query: 120 RTAAYVDQNDLHIGELTVRETLAFSA 145
R +V Q+D+ + +LTV ETLAF+A
Sbjct: 124 RRIGFVTQDDVLLPQLTVEETLAFAA 149
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.138 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,545,835
Number of Sequences: 26719
Number of extensions: 139987
Number of successful extensions: 784
Number of sequences better than 10.0: 197
Number of HSP's better than 10.0 without gapping: 140
Number of HSP's successfully gapped in prelim test: 57
Number of HSP's that attempted gapping in prelim test: 545
Number of HSP's gapped (non-prelim): 245
length of query: 167
length of database: 11,318,596
effective HSP length: 92
effective length of query: 75
effective length of database: 8,860,448
effective search space: 664533600
effective search space used: 664533600
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC147517.5


