

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147517.11 + phase: 0
(83 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
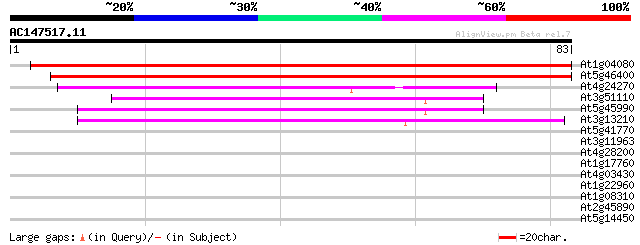
Sequences producing significant alignments: (bits) Value
At1g04080 unknown protein 145 3e-36
At5g46400 putative protein 99 5e-22
At4g24270 unknown protein 40 3e-04
At3g51110 crooked neck-like protein 39 4e-04
At5g45990 CRN (crooked neck) protein 38 8e-04
At3g13210 crn-like protein 38 8e-04
At5g41770 cell cycle control crn (crooked neck) protein-like 37 0.002
At3g11963 pre-rRNA processing protein like 36 0.004
At4g28200 unknown protein 34 0.011
At1g17760 unknown protein 32 0.073
At4g03430 putative pre-mRNA splicing factor 29 0.47
At1g22960 putative salt-inducible protein 26 3.1
At1g08310 unknown protein 26 4.0
At2g45890 hypothetical protein 25 5.2
At5g14450 early nodule-specific protein - like 25 9.0
>At1g04080 unknown protein
Length = 768
Score = 145 bits (366), Expect = 3e-36
Identities = 63/80 (78%), Positives = 75/80 (93%)
Query: 4 LLKNNILKMRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAVQGVTYSVDM 63
+ ++NI K+R+VYDAFLAEFPLCYGYWKK+ADHEAR+G+ DKVVEVYERAV GVTYSVD+
Sbjct: 113 IAQDNIAKIRKVYDAFLAEFPLCYGYWKKFADHEARVGAMDKVVEVYERAVLGVTYSVDI 172
Query: 64 WLHYCIFAISTYGDPDTVRR 83
WLHYC FAI+TYGDP+T+RR
Sbjct: 173 WLHYCTFAINTYGDPETIRR 192
Score = 31.2 bits (69), Expect = 0.096
Identities = 14/38 (36%), Positives = 19/38 (49%)
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHY 67
W Y D R G +KVV++YER V + W+ Y
Sbjct: 364 WHNYLDFIERDGDFNKVVKLYERCVVTCANYPEYWIRY 401
Score = 24.6 bits (52), Expect = 9.0
Identities = 14/47 (29%), Positives = 26/47 (54%), Gaps = 3/47 (6%)
Query: 32 KYADHEARLGSADKVVEVYERAV---QGVTYSVDMWLHYCIFAISTY 75
K+A+ E RLG+ D +YE+ + +G +S + L Y ++ +Y
Sbjct: 470 KHANMEYRLGNLDDAFSLYEQVIAVEKGKEHSTILPLLYAQYSRFSY 516
>At5g46400 putative protein
Length = 1022
Score = 98.6 bits (244), Expect = 5e-22
Identities = 44/77 (57%), Positives = 57/77 (73%)
Query: 7 NNILKMRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLH 66
++I K+ VYDAFL EFPLC+GYW+KYA H+ +L + + VEV+ERAVQ TYSV +WL
Sbjct: 62 DDIEKLCLVYDAFLLEFPLCHGYWRKYAYHKIKLCTLEDAVEVFERAVQAATYSVAVWLD 121
Query: 67 YCIFAISTYGDPDTVRR 83
YC FA++ Y DP V R
Sbjct: 122 YCAFAVAAYEDPHDVSR 138
Score = 25.0 bits (53), Expect = 6.9
Identities = 12/47 (25%), Positives = 16/47 (33%)
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHYCIFAISTYG 76
W Y G D + +YER + + W Y F S G
Sbjct: 301 WHAYLSFGETYGDFDWAINLYERCLIPCANYTEFWFRYVDFVESKGG 347
>At4g24270 unknown protein
Length = 816
Score = 39.7 bits (91), Expect = 3e-04
Identities = 23/68 (33%), Positives = 36/68 (52%), Gaps = 4/68 (5%)
Query: 8 NILKMRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEV---YERAVQGVTYSVDMW 64
N+ K+R+ +A A FPL W ++A EA L +++ V E+ YER + SV +W
Sbjct: 81 NLEKLRQAREAMSAIFPLSPSLWLEWARDEASLAASENVPEIVMLYERGLSDY-QSVSLW 139
Query: 65 LHYCIFAI 72
Y F +
Sbjct: 140 CDYLSFML 147
Score = 33.5 bits (75), Expect = 0.019
Identities = 15/62 (24%), Positives = 31/62 (49%), Gaps = 5/62 (8%)
Query: 11 KMRRVYDAFLAEFPLCYGYWKKYA---DHEARLGSADKVVEVYERAVQGVTYSVDMWLHY 67
+++ +Y+ +AE+P+ W Y D ++G A + Y RA + ++ D+W Y
Sbjct: 312 RVQAIYERAVAEYPVSSDLWIDYTVYLDKTLKVGKA--ITHAYSRATRSCPWTGDLWARY 369
Query: 68 CI 69
+
Sbjct: 370 LL 371
Score = 31.2 bits (69), Expect = 0.096
Identities = 14/42 (33%), Positives = 20/42 (47%)
Query: 33 YADHEARLGSADKVVEVYERAVQGVTYSVDMWLHYCIFAIST 74
Y E G +V +YERAV S D+W+ Y ++ T
Sbjct: 300 YIKFEKTSGDPTRVQAIYERAVAEYPVSSDLWIDYTVYLDKT 341
Score = 26.9 bits (58), Expect = 1.8
Identities = 13/40 (32%), Positives = 18/40 (44%)
Query: 13 RRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYER 52
R V+D+FL + W Y D E LG + +Y R
Sbjct: 473 RGVWDSFLKKSGGMLAAWHAYIDMEVHLGHIKEARSIYRR 512
>At3g51110 crooked neck-like protein
Length = 599
Score = 39.3 bits (90), Expect = 4e-04
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 9/64 (14%)
Query: 16 YDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAVQGVTYS---------VDMWLH 66
Y+ + + PL Y W Y E LG D++ EVYERA+ V + + +W+
Sbjct: 305 YEGEVRKNPLNYDSWFDYISLEETLGDKDRIREVYERAIANVPLAEEKRYWQRYIYLWID 364
Query: 67 YCIF 70
Y +F
Sbjct: 365 YALF 368
Score = 32.7 bits (73), Expect = 0.033
Identities = 23/81 (28%), Positives = 38/81 (46%), Gaps = 8/81 (9%)
Query: 8 NILKMRRVYDAFLAEFP-LCYGYWKKYADHEARLGSADKVVEVYERAVQ-----GVTYSV 61
NI + R++Y +L P CY W K+A+ E L ++ ++E A+ T
Sbjct: 418 NIDRCRKLYARYLEWSPESCYA-WTKFAEFERSLAETERARAIFELAISQPRLLDRTKHY 476
Query: 62 DMWLHYCIF-AISTYGDPDTV 81
+WL + F A + G DT+
Sbjct: 477 KVWLSFAKFEASAAQGQEDTI 497
Score = 30.4 bits (67), Expect = 0.16
Identities = 15/42 (35%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGVTY-SVDMWLHYCIF 70
W +YAD E D+ V+ERA++ +Y + +WL Y F
Sbjct: 68 WVRYADWEESQKDHDRARSVWERALEDESYRNHTLWLKYAEF 109
Score = 27.3 bits (59), Expect = 1.4
Identities = 13/40 (32%), Positives = 20/40 (49%)
Query: 13 RRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYER 52
R V+D + P +W KY E LG+ D +++ER
Sbjct: 120 RNVWDRAVKILPRVDQFWYKYIHMEEILGNIDGARKIFER 159
Score = 26.9 bits (58), Expect = 1.8
Identities = 13/38 (34%), Positives = 19/38 (49%)
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHY 67
W KYA+ E R S + V++RAV+ + W Y
Sbjct: 103 WLKYAEFEMRNKSVNHARNVWDRAVKILPRVDQFWYKY 140
Score = 26.9 bits (58), Expect = 1.8
Identities = 16/63 (25%), Positives = 30/63 (47%), Gaps = 1/63 (1%)
Query: 8 NILKMRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHY 67
N+ RR+ + + P + +KKY + E LG+ D+ ++Y R ++ S W +
Sbjct: 385 NLSGARRILGNAIGKAPK-HKIFKKYIEIELHLGNIDRCRKLYARYLEWSPESCYAWTKF 443
Query: 68 CIF 70
F
Sbjct: 444 AEF 446
>At5g45990 CRN (crooked neck) protein
Length = 673
Score = 38.1 bits (87), Expect = 8e-04
Identities = 19/69 (27%), Positives = 36/69 (51%), Gaps = 9/69 (13%)
Query: 11 KMRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAVQGVTYS---------V 61
K R Y+ +++ PL Y W Y E +G+ D++ E+YERA+ V + +
Sbjct: 312 KKRFEYEDEVSKNPLNYDSWFDYVRLEESVGNKDRIREIYERAIANVPPAQEKRFWQRYI 371
Query: 62 DMWLHYCIF 70
+W++Y ++
Sbjct: 372 YLWINYALY 380
Score = 34.7 bits (78), Expect = 0.009
Identities = 21/69 (30%), Positives = 34/69 (48%), Gaps = 4/69 (5%)
Query: 8 NILKMRRVYDAFLAEFPL-CYGYWKKYADHEARLGSADKVVEVYERAVQGVTYSVD--MW 64
NI + R++Y+ FL P CY W+ YA+ E L ++ ++E A+ + +W
Sbjct: 458 NIDRCRKLYERFLEWSPENCYA-WRNYAEFEISLAETERARAIFELAISQPALDMPELLW 516
Query: 65 LHYCIFAIS 73
Y F IS
Sbjct: 517 KTYIDFEIS 525
Score = 29.3 bits (64), Expect = 0.36
Identities = 14/44 (31%), Positives = 25/44 (56%)
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHYCIFAIS 73
+KKY + E +L + D+ ++YER ++ + W +Y F IS
Sbjct: 446 FKKYIEMELKLVNIDRCRKLYERFLEWSPENCYAWRNYAEFEIS 489
Score = 28.1 bits (61), Expect = 0.81
Identities = 15/51 (29%), Positives = 25/51 (48%), Gaps = 1/51 (1%)
Query: 3 FLLKNNILK-MRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYER 52
F +KN + R V+D + P W+KY E +LG+ +++ER
Sbjct: 121 FEMKNKFVNNARNVWDRSVTLLPRVDQLWEKYIYMEEKLGNVTGARQIFER 171
Score = 28.1 bits (61), Expect = 0.81
Identities = 20/66 (30%), Positives = 32/66 (48%), Gaps = 2/66 (3%)
Query: 7 NNILKMRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVV-EVYERAVQGVTYSVDMWL 65
N I + R +Y+ F+ P + + YA E + G K+ EVYERAV + + +
Sbjct: 193 NEIERARSIYERFVLCHPKVSAFIR-YAKFEMKRGGQVKLAREVYERAVDKLANDEEAEI 251
Query: 66 HYCIFA 71
+ FA
Sbjct: 252 LFVSFA 257
Score = 26.9 bits (58), Expect = 1.8
Identities = 13/41 (31%), Positives = 20/41 (48%)
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHYCIF 70
W KYA E + V+ERA++G + +W+ Y F
Sbjct: 81 WVKYAKWEESQMDYARARSVWERALEGEYRNHTLWVKYAEF 121
Score = 26.9 bits (58), Expect = 1.8
Identities = 14/44 (31%), Positives = 20/44 (44%), Gaps = 1/44 (2%)
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHYCIFAIS 73
WK Y D E G +K +YER + T +W+ + F S
Sbjct: 516 WKTYIDFEISEGEFEKTRALYERLLDR-TKHCKVWISFAKFEAS 558
>At3g13210 crn-like protein
Length = 657
Score = 38.1 bits (87), Expect = 8e-04
Identities = 23/83 (27%), Positives = 38/83 (45%), Gaps = 11/83 (13%)
Query: 11 KMRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAVQGV-----------TY 59
K R Y+ + + PL Y W + E +G+ D++ E+YERAV V
Sbjct: 280 KRRCQYEDEVRKNPLNYDSWFDFVRLEETVGNKDRIREIYERAVANVPPPEAQEKRYWQR 339
Query: 60 SVDMWLHYCIFAISTYGDPDTVR 82
+ +W++Y FA D ++ R
Sbjct: 340 YIYLWINYAFFAEMVTEDVESTR 362
Score = 35.8 bits (81), Expect = 0.004
Identities = 23/75 (30%), Positives = 37/75 (48%), Gaps = 4/75 (5%)
Query: 2 IFLLKNNILKMRRVYDAFLAEFP-LCYGYWKKYADHEARLGSADKVVEVYERAVQGVTYS 60
I L NI + R++Y+ +L P CY W+KYA+ E L ++ ++E A+
Sbjct: 422 IELQLRNIDRCRKLYERYLEWSPGNCYA-WRKYAEFEMSLAETERTRAIFELAISQPALD 480
Query: 61 VD--MWLHYCIFAIS 73
+ +W Y F IS
Sbjct: 481 MPELLWKTYIDFEIS 495
Score = 28.1 bits (61), Expect = 0.81
Identities = 15/58 (25%), Positives = 28/58 (47%)
Query: 10 LKMRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHY 67
L+ R+ ++ + L W KYAD E + S ++ V++RAV + +W +
Sbjct: 74 LRRRKEFEDQIRRARLNTQVWVKYADFEMKNKSVNEARNVWDRAVSLLPRVDQLWYKF 131
Score = 26.6 bits (57), Expect = 2.4
Identities = 13/44 (29%), Positives = 24/44 (54%)
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHYCIFAIS 73
+KKY + E +L + D+ ++YER ++ + W Y F +S
Sbjct: 416 FKKYIEIELQLRNIDRCRKLYERYLEWSPGNCYAWRKYAEFEMS 459
Score = 26.6 bits (57), Expect = 2.4
Identities = 17/65 (26%), Positives = 31/65 (47%), Gaps = 1/65 (1%)
Query: 7 NNILKMRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLH 66
N I R +Y+ F+ P Y + YA E + G + ++V+ERA + + + +
Sbjct: 172 NEIECARSIYERFVLCHPKVSAYIR-YAKFEMKHGQVELAMKVFERAKKELADDEEAEIL 230
Query: 67 YCIFA 71
+ FA
Sbjct: 231 FVAFA 235
Score = 25.8 bits (55), Expect = 4.0
Identities = 13/44 (29%), Positives = 20/44 (44%), Gaps = 1/44 (2%)
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHYCIFAIS 73
WK Y D E G ++ +YER + T +W+ + F S
Sbjct: 486 WKTYIDFEISEGELERTRALYERLLDR-TKHCKVWVDFAKFEAS 528
Score = 24.6 bits (52), Expect = 9.0
Identities = 14/51 (27%), Positives = 26/51 (50%), Gaps = 1/51 (1%)
Query: 3 FLLKN-NILKMRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYER 52
F +KN ++ + R V+D ++ P W K+ E +LG+ ++ ER
Sbjct: 100 FEMKNKSVNEARNVWDRAVSLLPRVDQLWYKFIHMEEKLGNIAGARQILER 150
>At5g41770 cell cycle control crn (crooked neck) protein-like
Length = 705
Score = 36.6 bits (83), Expect = 0.002
Identities = 19/69 (27%), Positives = 34/69 (48%), Gaps = 9/69 (13%)
Query: 11 KMRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAVQGVTYS---------V 61
K R Y+ + + P Y W Y E +G+ D++ E+YERA+ V + +
Sbjct: 325 KRRFQYEDEVRKSPSNYDSWFDYVRLEESVGNKDRIREIYERAIANVPPAEEKRYWQRYI 384
Query: 62 DMWLHYCIF 70
+W++Y +F
Sbjct: 385 YLWINYALF 393
Score = 32.0 bits (71), Expect = 0.056
Identities = 22/75 (29%), Positives = 36/75 (47%), Gaps = 4/75 (5%)
Query: 2 IFLLKNNILKMRRVYDAFLAEFPL-CYGYWKKYADHEARLGSADKVVEVYERAVQGVTYS 60
I L N+ + R++Y+ +L P CY W KYA+ E L ++ ++E A+
Sbjct: 465 IELQLGNMDRCRKLYERYLEWSPENCYA-WSKYAELERSLVETERARAIFELAISQPALD 523
Query: 61 VD--MWLHYCIFAIS 73
+ +W Y F IS
Sbjct: 524 MPELLWKAYIDFEIS 538
Score = 29.3 bits (64), Expect = 0.36
Identities = 14/41 (34%), Positives = 20/41 (48%)
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHYCIF 70
W KYA E + V+ERA++G + +WL Y F
Sbjct: 95 WVKYAQWEESQKDYARARSVWERAIEGDYRNHTLWLKYAEF 135
Score = 28.5 bits (62), Expect = 0.62
Identities = 18/65 (27%), Positives = 30/65 (45%), Gaps = 1/65 (1%)
Query: 7 NNILKMRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLH 66
N I + R +Y+ F+ P Y + YA E + G + VYERA + + + +
Sbjct: 207 NEIERARTIYERFVLCHPKVSAYIR-YAKFEMKGGEVARCRSVYERATEKLADDEEAEIL 265
Query: 67 YCIFA 71
+ FA
Sbjct: 266 FVAFA 270
Score = 27.3 bits (59), Expect = 1.4
Identities = 12/38 (31%), Positives = 22/38 (57%)
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHY 67
+KKY + E +LG+ D+ ++YER ++ + W Y
Sbjct: 459 FKKYIEIELQLGNMDRCRKLYERYLEWSPENCYAWSKY 496
Score = 25.8 bits (55), Expect = 4.0
Identities = 13/44 (29%), Positives = 20/44 (44%), Gaps = 1/44 (2%)
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWLHYCIFAIS 73
WK Y D E G ++ +YER + T +W+ + F S
Sbjct: 529 WKAYIDFEISEGELERTRALYERLLDR-TKHYKVWVSFAKFEAS 571
Score = 25.4 bits (54), Expect = 5.2
Identities = 20/73 (27%), Positives = 32/73 (43%), Gaps = 6/73 (8%)
Query: 3 FLLKNNILKM-RRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAVQGVTYSV 61
F +KN + R V+D + P W KY E LG+ +++ER + +S
Sbjct: 135 FEMKNKFVNSARNVWDRAVTLLPRVDQLWYKYIHMEEILGNIAGARQIFER---WMDWSP 191
Query: 62 DM--WLHYCIFAI 72
D WL + F +
Sbjct: 192 DQQGWLSFIKFEL 204
>At3g11963 pre-rRNA processing protein like
Length = 1126
Score = 35.8 bits (81), Expect = 0.004
Identities = 15/42 (35%), Positives = 23/42 (54%)
Query: 13 RRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAV 54
R +++ L E+P W Y D E RLG D + ++ERA+
Sbjct: 1040 RSLFEGVLREYPKRTDLWSVYLDQEIRLGEDDVIRSLFERAI 1081
>At4g28200 unknown protein
Length = 648
Score = 34.3 bits (77), Expect = 0.011
Identities = 17/57 (29%), Positives = 30/57 (51%)
Query: 6 KNNILKMRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAVQGVTYSVD 62
K+ + R++YD+ +A + WK Y E +LG+++ VY RA + + S D
Sbjct: 589 KDGLSNARKLYDSAVASYGQDVELWKNYYSLETKLGTSETANGVYWRARKTLNESAD 645
>At1g17760 unknown protein
Length = 734
Score = 31.6 bits (70), Expect = 0.073
Identities = 15/71 (21%), Positives = 32/71 (44%), Gaps = 1/71 (1%)
Query: 5 LKNNILKMRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAVQGVTYSVDMW 64
L + I + +Y+ L+ +P +WK+Y + + + + D +++ R + V +W
Sbjct: 18 LHSPIAQATPIYEQLLSLYPTSARFWKQYVEAQMAVNNDDATKQIFSRCLL-TCLQVPLW 76
Query: 65 LHYCIFAISTY 75
Y F Y
Sbjct: 77 QCYIRFIRKVY 87
Score = 28.1 bits (61), Expect = 0.81
Identities = 9/28 (32%), Positives = 18/28 (64%)
Query: 30 WKKYADHEARLGSADKVVEVYERAVQGV 57
W YA+ + GS D ++V++RA++ +
Sbjct: 269 WYDYAEWHVKSGSTDAAIKVFQRALKAI 296
Score = 27.7 bits (60), Expect = 1.1
Identities = 18/51 (35%), Positives = 25/51 (48%), Gaps = 3/51 (5%)
Query: 29 YWKKYADHEARLGSADKVVEVYERAVQ--GVTYSVDMWLHYCIFAISTYGD 77
Y KYAD RL + ++ERA+ V S ++W + F TYGD
Sbjct: 404 YILKYADFLTRLNDDRNIRALFERALSTLPVEDSAEVWKRFIQFE-QTYGD 453
>At4g03430 putative pre-mRNA splicing factor
Length = 1029
Score = 28.9 bits (63), Expect = 0.47
Identities = 12/58 (20%), Positives = 27/58 (45%)
Query: 8 NILKMRRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWL 65
N+ + RR+ + L +FP + W E R ++ + Y+ ++ + + +WL
Sbjct: 775 NVEEERRLLNEGLKQFPTFFKLWLMLGQLEERFKHLEQARKAYDTGLKHCPHCIPLWL 832
Score = 28.5 bits (62), Expect = 0.62
Identities = 14/53 (26%), Positives = 22/53 (41%)
Query: 13 RRVYDAFLAEFPLCYGYWKKYADHEARLGSADKVVEVYERAVQGVTYSVDMWL 65
R+ YD L P C W AD E ++ +K + A + ++WL
Sbjct: 814 RKAYDTGLKHCPHCIPLWLSLADLEEKVNGLNKARAILTTARKKNPGGAELWL 866
>At1g22960 putative salt-inducible protein
Length = 718
Score = 26.2 bits (56), Expect = 3.1
Identities = 18/65 (27%), Positives = 30/65 (45%), Gaps = 1/65 (1%)
Query: 3 FLLKNNILKMRRVYDAFLAEFPLCYGY-WKKYADHEARLGSADKVVEVYERAVQGVTYSV 61
F+ N+ VYD L + GY + A E RLG +DK ++E V ++
Sbjct: 454 FVKNGNLSMATEVYDEMLRKGIKPDGYAYTTRAVGELRLGDSDKAFRLHEEMVATDHHAP 513
Query: 62 DMWLH 66
D+ ++
Sbjct: 514 DLTIY 518
>At1g08310 unknown protein
Length = 315
Score = 25.8 bits (55), Expect = 4.0
Identities = 9/27 (33%), Positives = 15/27 (55%)
Query: 4 LLKNNILKMRRVYDAFLAEFPLCYGYW 30
+L L+ R V+D +F +C+G W
Sbjct: 209 MLTKEKLRERNVFDTLRDDFMVCFGQW 235
>At2g45890 hypothetical protein
Length = 492
Score = 25.4 bits (54), Expect = 5.2
Identities = 10/34 (29%), Positives = 19/34 (55%)
Query: 46 VVEVYERAVQGVTYSVDMWLHYCIFAISTYGDPD 79
V+E Y R ++G+ Y+V W+ ++ T + D
Sbjct: 458 VLESYSRVLEGLAYNVVSWIDDVLYVDRTVRNRD 491
>At5g14450 early nodule-specific protein - like
Length = 389
Score = 24.6 bits (52), Expect = 9.0
Identities = 11/24 (45%), Positives = 16/24 (65%)
Query: 51 ERAVQGVTYSVDMWLHYCIFAIST 74
ERA V+ +V WL C+FA++T
Sbjct: 6 ERAKLMVSSTVFSWLLLCLFAVTT 29
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.142 0.456
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,868,910
Number of Sequences: 26719
Number of extensions: 59916
Number of successful extensions: 268
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 213
Number of HSP's gapped (non-prelim): 55
length of query: 83
length of database: 11,318,596
effective HSP length: 59
effective length of query: 24
effective length of database: 9,742,175
effective search space: 233812200
effective search space used: 233812200
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 52 (24.6 bits)
Medicago: description of AC147517.11


