

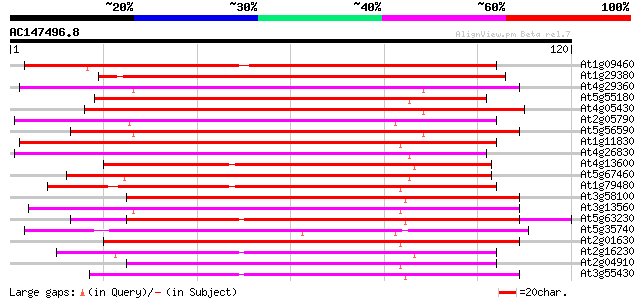
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147496.8 + phase: 1 /partial
(120 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g09460 unknown protein 134 7e-33
At1g29380 beta-1,3 glucanase, putative 127 7e-31
At4g29360 beta-1,3-glucanase-like protein 100 2e-22
At5g55180 beta-1,3-glucanase-like protein 99 4e-22
At4g05430 putative protein 99 4e-22
At2g05790 putative beta-1,3-glucanase 99 4e-22
At5g56590 beta-1,3-glucanase-like protein 98 6e-22
At1g11830 putative beta-1,3-glucanase, predicted GPI-anchored pr... 97 2e-21
At4g26830 putative beta-1,3-glucanase 96 4e-21
At4g13600 putative protein 93 2e-20
At5g67460 putative protein 92 3e-20
At1g79480 hypothetical protein 92 3e-20
At3g58100 unknown protein 92 4e-20
At3g13560 glucan endo-1,3-beta-glucosidase precursor, putative 89 5e-19
At5g63230 unknown protein 88 8e-19
At5g35740 unknown protein 87 1e-18
At2g01630 beta-1,3-glucanase like protein 87 1e-18
At2g16230 putative beta-1,3-glucanase 86 2e-18
At2g04910 hypothetical protein 86 2e-18
At3g55430 beta-1,3-glucanase - like protein 86 3e-18
>At1g09460 unknown protein
Length = 213
Score = 134 bits (337), Expect = 7e-33
Identities = 66/106 (62%), Positives = 74/106 (69%), Gaps = 7/106 (6%)
Query: 4 PSSTVPITAPST-----SNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQP 58
PS TVP+ P SNSP SG SWC+A P ASQ SLQ ALDYACG DCS +Q
Sbjct: 110 PSGTVPVPVPVVAPPVVSNSPSVSGQSWCVAKPGASQVSLQQALDYACGIA--DCSQLQQ 167
Query: 59 GGSCYNPNSVHDHASFAFNKYYQKNPVPNSCNFGGNAVLTNTNPSK 104
GG+CY+P S+ HASFAFN YYQKNP P SC+FGG A L NTNPS+
Sbjct: 168 GGNCYSPISLQSHASFAFNSYYQKNPSPQSCDFGGAASLVNTNPSE 213
>At1g29380 beta-1,3 glucanase, putative
Length = 228
Score = 127 bits (320), Expect = 7e-31
Identities = 59/87 (67%), Positives = 67/87 (76%), Gaps = 1/87 (1%)
Query: 20 VSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNKY 79
V SG WCIA +AS SLQVALDYACGYGG DC IQ G +CY PN++ DHASFAFN Y
Sbjct: 143 VGSG-QWCIAKANASPTSLQVALDYACGYGGADCGQIQQGAACYEPNTIRDHASFAFNSY 201
Query: 80 YQKNPVPNSCNFGGNAVLTNTNPSKAS 106
YQK+P +SCNFGG A LT+T+PSK S
Sbjct: 202 YQKHPGSDSCNFGGAAQLTSTDPSKTS 228
>At4g29360 beta-1,3-glucanase-like protein
Length = 534
Score = 99.8 bits (247), Expect = 2e-22
Identities = 51/122 (41%), Positives = 73/122 (59%), Gaps = 15/122 (12%)
Query: 3 SPSSTVPITAPSTSNSPVSSGAS--------------WCIASPSASQRSLQVALDYACGY 48
SP+++ T+PS S+SP+ +G S WCIAS AS LQ ALD+ACG
Sbjct: 354 SPTNSTTGTSPSPSSSPIINGNSTVTIGGGGGGGTKKWCIASSQASVTELQTALDWACGP 413
Query: 49 GGTDCSAIQPGGSCYNPNSVHDHASFAFNKYYQKNPVPN-SCNFGGNAVLTNTNPSKAST 107
G DCSA+QP C+ P++V HAS+AFN YYQ++ + C+F G +V + +PS +
Sbjct: 414 GNVDCSAVQPDQPCFEPDTVLSHASYAFNTYYQQSGASSIDCSFNGASVEVDKDPSYGNC 473
Query: 108 IY 109
+Y
Sbjct: 474 LY 475
>At5g55180 beta-1,3-glucanase-like protein
Length = 460
Score = 98.6 bits (244), Expect = 4e-22
Identities = 44/85 (51%), Positives = 55/85 (63%), Gaps = 1/85 (1%)
Query: 19 PVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNK 78
P G +WC+A+ ++ LQ LDYACG GG DC IQPG +CYNP S+ HAS+AFN
Sbjct: 365 PSLVGQTWCVANGKTTKEKLQEGLDYACGEGGADCRPIQPGATCYNPESLEAHASYAFNS 424
Query: 79 YYQKNP-VPNSCNFGGNAVLTNTNP 102
YYQKN +CNFGG A + + P
Sbjct: 425 YYQKNARGVGTCNFGGAAYVVSQPP 449
>At4g05430 putative protein
Length = 119
Score = 98.6 bits (244), Expect = 4e-22
Identities = 46/95 (48%), Positives = 58/95 (60%), Gaps = 1/95 (1%)
Query: 17 NSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAF 76
N+ G +WC+A P A+Q LQ ALD+ACG G DCS I+ G CY P+++ HASFAF
Sbjct: 14 NTTFLEGTTWCVARPGATQAELQRALDWACGIGRVDCSVIERHGDCYEPDTIVSHASFAF 73
Query: 77 NKYYQKNPVPN-SCNFGGNAVLTNTNPSKASTIYD 110
N YYQ N +C FGG A T NP++ S D
Sbjct: 74 NAYYQTNGNNRIACYFGGTATFTKINPNRKSPPQD 108
>At2g05790 putative beta-1,3-glucanase
Length = 473
Score = 98.6 bits (244), Expect = 4e-22
Identities = 49/112 (43%), Positives = 61/112 (53%), Gaps = 9/112 (8%)
Query: 2 YSPSSTVPITAPSTSNSPVSSGA--------SWCIASPSASQRSLQVALDYACGYGGTDC 53
Y P+T P SG +WC+A+ A + LQ LDYACG GG DC
Sbjct: 352 YRDGGHTPVTGGDQVTKPPMSGGVSKSLNGYTWCVANGDAGEERLQGGLDYACGEGGADC 411
Query: 54 SAIQPGGSCYNPNSVHDHASFAFNKYYQ-KNPVPNSCNFGGNAVLTNTNPSK 104
IQPG +CY+P+++ HASFAFN YYQ K SC FGG A + + PSK
Sbjct: 412 RPIQPGANCYSPDTLEAHASFAFNSYYQKKGRAGGSCYFGGAAYVVSQPPSK 463
>At5g56590 beta-1,3-glucanase-like protein
Length = 506
Score = 98.2 bits (243), Expect = 6e-22
Identities = 49/99 (49%), Positives = 65/99 (65%), Gaps = 3/99 (3%)
Query: 14 STSNSPVSSGAS--WCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDH 71
S S+ SSG+S WCIAS AS+R L+ ALD+ACG G DC+AIQP C+ P+++ H
Sbjct: 355 SNSSGTNSSGSSNSWCIASSKASERDLKGALDWACGPGNVDCTAIQPSQPCFQPDTLVSH 414
Query: 72 ASFAFNKYYQKNPVPN-SCNFGGNAVLTNTNPSKASTIY 109
ASF FN Y+Q+N + +C+FGG V N +PS IY
Sbjct: 415 ASFVFNSYFQQNRATDVACSFGGAGVKVNKDPSYDKCIY 453
>At1g11830 putative beta-1,3-glucanase, predicted GPI-anchored
protein
Length = 477
Score = 96.7 bits (239), Expect = 2e-21
Identities = 47/103 (45%), Positives = 66/103 (63%), Gaps = 1/103 (0%)
Query: 3 SPSSTVPITAPSTSNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSC 62
+P + ++ T + ++ ++CIA ++LQ ALD+ACG G ++CS IQPG SC
Sbjct: 358 TPVYLLHVSGSGTFLANDTTNQTYCIAMDGVDAKTLQAALDWACGPGRSNCSEIQPGESC 417
Query: 63 YNPNSVHDHASFAFNKYYQK-NPVPNSCNFGGNAVLTNTNPSK 104
Y PN+V HASFAFN YYQK SC+F G A++T T+PSK
Sbjct: 418 YQPNNVKGHASFAFNSYYQKEGRASGSCDFKGVAMITTTDPSK 460
>At4g26830 putative beta-1,3-glucanase
Length = 448
Score = 95.5 bits (236), Expect = 4e-21
Identities = 44/102 (43%), Positives = 59/102 (57%), Gaps = 1/102 (0%)
Query: 2 YSPSSTVPITAPSTSNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGS 61
++ ST P+ G +WC+++ ++ LQ ALDYACG GG DC IQPG +
Sbjct: 343 FTKKSTTPVNGNRGKVPVTHEGHTWCVSNGEVAKEKLQEALDYACGEGGADCRPIQPGAT 402
Query: 62 CYNPNSVHDHASFAFNKYYQKNP-VPNSCNFGGNAVLTNTNP 102
CY+P S+ HAS+AFN YYQKN +C FGG A + P
Sbjct: 403 CYHPESLEAHASYAFNSYYQKNSRRVGTCFFGGAAHVVTQPP 444
>At4g13600 putative protein
Length = 256
Score = 92.8 bits (229), Expect = 2e-20
Identities = 42/84 (50%), Positives = 56/84 (66%), Gaps = 2/84 (2%)
Query: 21 SSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNKYY 80
SS A WC+A + ++LQ ALDYAC G DC+ IQP G C+ PN+V HAS+AFN Y+
Sbjct: 56 SSAAMWCVARFDVTSQALQAALDYACA-AGADCAPIQPNGLCFLPNTVQAHASYAFNSYF 114
Query: 81 QKNPV-PNSCNFGGNAVLTNTNPS 103
Q+ + P SCNF G + + T+PS
Sbjct: 115 QRAAMAPGSCNFAGTSTIAKTDPS 138
>At5g67460 putative protein
Length = 380
Score = 92.4 bits (228), Expect = 3e-20
Identities = 44/95 (46%), Positives = 60/95 (62%), Gaps = 4/95 (4%)
Query: 13 PSTSNSPVSSG---ASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVH 69
P+ S PV WC+A PS + +LQ +LD+ACG GG +C I+P G CY P++V
Sbjct: 277 PAPSPQPVKKKNVEGLWCVAKPSVAAETLQQSLDFACGQGGANCDEIKPHGICYYPDTVM 336
Query: 70 DHASFAFNKYYQKNP-VPNSCNFGGNAVLTNTNPS 103
HAS+AFN Y+QK +C+FGG A+L T+PS
Sbjct: 337 AHASYAFNSYWQKTKRNGGTCSFGGTAMLITTDPS 371
>At1g79480 hypothetical protein
Length = 356
Score = 92.4 bits (228), Expect = 3e-20
Identities = 44/97 (45%), Positives = 60/97 (61%), Gaps = 4/97 (4%)
Query: 9 PITAPSTSNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSV 68
P+ PS S +P S A WC+A PS +Q A+++ACG G DC +IQP G C+ PN++
Sbjct: 252 PMAPPSPSVTPTS--AYWCVAKPSVPDPIIQEAMNFACG-SGADCHSIQPNGPCFKPNTL 308
Query: 69 HDHASFAFNKYYQK-NPVPNSCNFGGNAVLTNTNPSK 104
HASFA+N Y+Q+ SC FGG +L +PSK
Sbjct: 309 WAHASFAYNSYWQRTKSTGGSCTFGGTGMLVTVDPSK 345
>At3g58100 unknown protein
Length = 180
Score = 92.0 bits (227), Expect = 4e-20
Identities = 42/85 (49%), Positives = 52/85 (60%), Gaps = 1/85 (1%)
Query: 26 WCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNKYYQKN-P 84
WC+A +A SLQ A+++ACG GG DC IQ GG C +P V ASF FN YY KN
Sbjct: 41 WCVAKNNAEDSSLQTAIEWACGQGGADCGPIQQGGPCNDPTDVQKMASFVFNNYYLKNGE 100
Query: 85 VPNSCNFGGNAVLTNTNPSKASTIY 109
+CNF NA LT+ NPS+ + Y
Sbjct: 101 EDEACNFNNNAALTSLNPSQGTCKY 125
>At3g13560 glucan endo-1,3-beta-glucosidase precursor, putative
Length = 505
Score = 88.6 bits (218), Expect = 5e-19
Identities = 45/107 (42%), Positives = 62/107 (57%), Gaps = 2/107 (1%)
Query: 5 SSTVPITAPSTSNSPVSSGAS-WCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCY 63
+S P++ S+S +G+S +C+A A L L++ACG G +C+AIQPG CY
Sbjct: 340 TSVYPLSLSGGSSSAALNGSSMFCVAKADADDDKLVDGLNWACGQGRANCAAIQPGQPCY 399
Query: 64 NPNSVHDHASFAFNKYYQK-NPVPNSCNFGGNAVLTNTNPSKASTIY 109
PN V HASFAFN YYQK +C+F G A+ T +PS + Y
Sbjct: 400 LPNDVKSHASFAFNDYYQKMKSAGGTCDFDGTAITTTRDPSYRTCAY 446
>At5g63230 unknown protein
Length = 178
Score = 87.8 bits (216), Expect = 8e-19
Identities = 40/85 (47%), Positives = 53/85 (62%), Gaps = 2/85 (2%)
Query: 26 WCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNKYYQKN-P 84
WC+A P+A+ LQ +DYAC DC+ IQPGG+CY PN++ DHASFA N YYQ +
Sbjct: 93 WCMAMPNATGEQLQANIDYACSQN-VDCTPIQPGGTCYEPNTLLDHASFAMNAYYQSHGR 151
Query: 85 VPNSCNFGGNAVLTNTNPSKASTIY 109
+ ++C FG +PS S IY
Sbjct: 152 IEDACRFGRTGCFVFIDPSNGSCIY 176
Score = 79.3 bits (194), Expect = 3e-16
Identities = 39/108 (36%), Positives = 56/108 (51%), Gaps = 2/108 (1%)
Query: 14 STSNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHAS 73
ST + PV + WC P+ S LQ +D+AC G DC+ IQPGG+CYNPN++ DHAS
Sbjct: 14 STVSIPVVTCRQWCTPMPNTSDEQLQANIDFACS-NGVDCTPIQPGGNCYNPNTLFDHAS 72
Query: 74 FAFNKYYQKN-PVPNSCNFGGNAVLTNTNPSKASTIYDHGFKLQSDTT 120
+ N YY + V ++C+ + N + D+ D T
Sbjct: 73 YVMNAYYHSHGRVEDACSRQWCMAMPNATGEQLQANIDYACSQNVDCT 120
>At5g35740 unknown protein
Length = 119
Score = 87.4 bits (215), Expect = 1e-18
Identities = 48/111 (43%), Positives = 61/111 (54%), Gaps = 7/111 (6%)
Query: 4 PSSTVPITAPSTSNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGS-C 62
P + +T P S + WCIA LQ ALD+ACG GG DCS +Q C
Sbjct: 11 PLLIIAMTLPRRSEA---ESEQWCIADEQTPDDELQAALDWACGKGGADCSKMQQENQPC 67
Query: 63 YNPNSVHDHASFAFNKYYQ--KNPVPNSCNFGGNAVLTNTNPSKASTIYDH 111
+ PN++ DHASFAFN YYQ KN SC F G A++T +PS S Y++
Sbjct: 68 FLPNTIRDHASFAFNSYYQTYKNK-GGSCYFKGAAMITELDPSHGSCQYEY 117
>At2g01630 beta-1,3-glucanase like protein
Length = 501
Score = 87.4 bits (215), Expect = 1e-18
Identities = 39/90 (43%), Positives = 58/90 (64%), Gaps = 1/90 (1%)
Query: 21 SSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNKYY 80
++ ++CIA ++ LQ ALD+ACG G DCSA+ G SCY P+ V H+++AFN YY
Sbjct: 355 TTNQTFCIAKEKVDRKMLQAALDWACGPGKVDCSALMQGESCYEPDDVVAHSTYAFNAYY 414
Query: 81 QK-NPVPNSCNFGGNAVLTNTNPSKASTIY 109
QK SC+F G A +T T+PS+ + ++
Sbjct: 415 QKMGKASGSCDFKGVATVTTTDPSRGTCVF 444
>At2g16230 putative beta-1,3-glucanase
Length = 456
Score = 86.3 bits (212), Expect = 2e-18
Identities = 42/98 (42%), Positives = 57/98 (57%), Gaps = 5/98 (5%)
Query: 11 TAPSTSNSPVS---SGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNS 67
T+ TS SP + WC+ A+Q LQ +LD+ CG G DC I PGG C+ PN+
Sbjct: 348 TSSQTSQSPQLGKVTSMGWCVPKEDATQEQLQDSLDWVCGQG-IDCGPIMPGGVCFEPNN 406
Query: 68 VHDHASFAFNKYYQKNPV-PNSCNFGGNAVLTNTNPSK 104
V H ++A N Y+QK+P P C+F A +T+ NPSK
Sbjct: 407 VASHTAYAMNLYFQKSPENPTDCDFSKTARITSENPSK 444
>At2g04910 hypothetical protein
Length = 124
Score = 86.3 bits (212), Expect = 2e-18
Identities = 39/80 (48%), Positives = 48/80 (59%), Gaps = 1/80 (1%)
Query: 26 WCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNKYYQK-NP 84
WC+A +Q A+D+AC GG DCS IQP C+ PN+V DHAS FN YYQ+
Sbjct: 15 WCVADGQIPDNVIQAAVDWACQTGGADCSTIQPNQPCFLPNTVKDHASVVFNNYYQRYKR 74
Query: 85 VPNSCNFGGNAVLTNTNPSK 104
SCNF A +T T+PSK
Sbjct: 75 NGGSCNFNSTAFITQTDPSK 94
>At3g55430 beta-1,3-glucanase - like protein
Length = 449
Score = 85.9 bits (211), Expect = 3e-18
Identities = 37/93 (39%), Positives = 55/93 (58%), Gaps = 2/93 (2%)
Query: 18 SPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFN 77
+P ++G WC+A A+ LQ ++++ CG G DC IQ GGSC+NP+S+ HASF N
Sbjct: 358 APSTAGGKWCVARSGATNTQLQDSINWVCGQG-VDCKPIQAGGSCFNPSSLRTHASFVMN 416
Query: 78 KYYQKN-PVPNSCNFGGNAVLTNTNPSKASTIY 109
Y+Q + +CNF G ++ NPS + Y
Sbjct: 417 AYFQSHGRTDGACNFSGTGMIVGNNPSNGACKY 449
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.125 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,214,564
Number of Sequences: 26719
Number of extensions: 141190
Number of successful extensions: 452
Number of sequences better than 10.0: 81
Number of HSP's better than 10.0 without gapping: 58
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 324
Number of HSP's gapped (non-prelim): 88
length of query: 120
length of database: 11,318,596
effective HSP length: 96
effective length of query: 24
effective length of database: 8,753,572
effective search space: 210085728
effective search space used: 210085728
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 52 (24.6 bits)
Medicago: description of AC147496.8


