

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147496.3 - phase: 0
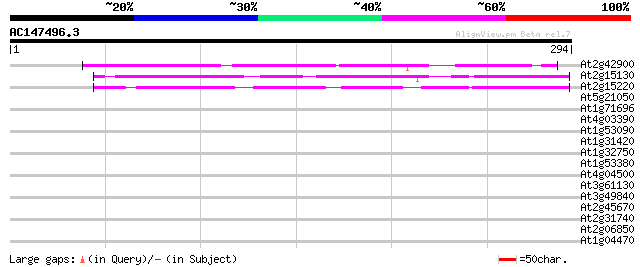
(294 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g42900 unknown protein 164 7e-41
At2g15130 unknown protein 76 2e-14
At2g15220 hypothetical protein 69 2e-12
At5g21050 putative protein 30 1.5
At1g71696 carboxypeptidase, putative 29 2.6
At4g03390 SRF3 (LRR receptor-like protein kinase like protein) 28 4.5
At1g53090 phytochrome A supressor spa1, putative 28 4.5
At1g31420 unknown protein 28 4.5
At1g32750 unknown protein 28 5.9
At1g53380 hypothetical protein; similar to EST gb|R30194.1 28 7.6
At4g04500 putative receptor-like protein kinase 27 10.0
At3g61130 unknown protein 27 10.0
At3g49840 putative protein 27 10.0
At2g45670 unknown protein 27 10.0
At2g31740 unknown protein 27 10.0
At2g06850 putative endoxyloglucan glycosyltransferase 27 10.0
At1g04470 unknown protein 27 10.0
>At2g42900 unknown protein
Length = 276
Score = 164 bits (414), Expect = 7e-41
Identities = 98/252 (38%), Positives = 141/252 (55%), Gaps = 26/252 (10%)
Query: 39 NNNNIIHRILLILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRI 98
+++ II R+ IL V IS+WAN+EASK F I I+ND KDS +G+RF LF+ S+D A RI
Sbjct: 26 SDSGIILRLFSILLVGAISLWANHEASKGFSISIINDAKDSPSGKRFALFFESDDTAVRI 85
Query: 99 LLNTSSFVEQILYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITADE-KNINSYVI 157
LL+ S FVE+ LY + + + +K + VT++ + D + + + YVI
Sbjct: 86 LLDASFFVERFLY-----EGVPHRLRKPVNHVTVQFCGNSSDRVDRFSVTSGASHGEYVI 140
Query: 158 EISPMLLEDENFNNMAIVGAIQRAMARVWLWDGRSKASPRLLDGMVEYIA--ELAGFHRE 215
+SP L E + F+N A+ A++R+M R+WLW S ASP L+ GMVEY+A H E
Sbjct: 141 RLSPSLTERKKFSN-AVESALRRSMVRIWLWGDESGASPELVAGMVEYLAVESRKRRHFE 199
Query: 216 RLSGGVGESPECEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVG 275
+ G W D + +V LL YCE+ +GFI+RLN M+ W DR V
Sbjct: 200 KFGGN-------------WKDKEKSVYVVSLLDYCERRSEGFIRRLNHGMRLRWDDRTV- 245
Query: 276 DVLGLKATKLCG 287
L ++ CG
Sbjct: 246 ---DLASSGACG 254
>At2g15130 unknown protein
Length = 225
Score = 76.3 bits (186), Expect = 2e-14
Identities = 61/252 (24%), Positives = 102/252 (40%), Gaps = 35/252 (13%)
Query: 45 HRILLILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSS 104
H+I L+ IS+ D +V++T DS GRRF + L + +
Sbjct: 4 HKIFLV-----ISLMLAVSLVSAVDFSVVDNTGDSPGGRRFRNEIGGVSYGEQSLRDATD 58
Query: 105 FVEQILYPNGNNDNIDIKNKKIIKSVTLRLARQNLDTITTITADEKNINSYVIEISPMLL 164
F ++ +D D+ + L +N + I + DE + N+ L+
Sbjct: 59 FTWRLFQQTNPSDRKDVTK--------ITLFMENSNGIAYSSQDEIHYNA------GSLV 104
Query: 165 EDENFNNMAIVGAIQRAMARVWLWDGRSKASPRLLDGMVEYIAELAGF---HRERLSGGV 221
+D+ + G + + W W+G +A L++G+ +Y+ AG+ H R GG
Sbjct: 105 DDKGYVRRGFTGVVYHEVVHSWQWNGAGRAPGGLIEGIADYVRLKAGYVASHWVRPGGG- 163
Query: 222 GESPECEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLK 281
+ W D T AR L YC GF+ LN+ M+ ++D D+LG
Sbjct: 164 ----------DRWDQGYDVT--ARFLEYCNDLRNGFVAELNKKMRSDYNDGFFVDLLGKD 211
Query: 282 ATKLCGLYNATY 293
+L Y A Y
Sbjct: 212 VNQLWREYKANY 223
>At2g15220 hypothetical protein
Length = 225
Score = 69.3 bits (168), Expect = 2e-12
Identities = 61/251 (24%), Positives = 104/251 (41%), Gaps = 33/251 (13%)
Query: 45 HRILLILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSS 104
H+I ++ + ++ N D +V+++ DS GRRF ++ L + +
Sbjct: 4 HKIFFVISLMLVVSLVN-----AVDYSVVDNSGDSTGGRRFRGEIGGISYGTQTLRSATD 58
Query: 105 FVEQILYPNGNNDNIDIKNKKIIKSVT-LRLARQNLDTITTITADEKNIN-SYVIEISPM 162
FV ++ +D KSVT + L +N D + +A+E + N Y+ +S
Sbjct: 59 FVWRLFQQTNPSDR---------KSVTKITLFMENGDGVAYNSANEIHFNVGYLAGVSGD 109
Query: 163 LLEDENFNNMAIVGAIQRAMARVWLWDGRSKASPRLLDGMVEYIAELAGFHRERLSGGVG 222
+ + G + + W W+G +A L++G+ +Y+ RL G
Sbjct: 110 VKRE-------FTGVVYHEVVHSWQWNGAGRAPGGLIEGIADYV---------RLKAGYA 153
Query: 223 ESPECEDGRELWWDNKDPTHVARLLHYCEKYDKGFIQRLNEAMKDTWHDRMVGDVLGLKA 282
S GR WD AR L YC GF+ LN+ M++ + D D+LG
Sbjct: 154 PSHWVGPGRGDRWDQGYDV-TARFLDYCNGLRNGFVAELNKKMRNGYSDGFFVDLLGKDV 212
Query: 283 TKLCGLYNATY 293
+L Y A Y
Sbjct: 213 NQLWREYKAKY 223
>At5g21050 putative protein
Length = 355
Score = 30.0 bits (66), Expect = 1.5
Identities = 24/103 (23%), Positives = 49/103 (47%), Gaps = 11/103 (10%)
Query: 115 NNDNIDIKNKKIIKSVTLRLARQNLDTITTITADEKNINSYVIEISPMLLEDENFNNMAI 174
N + K + I+S++ + N+ + TI D++ +++ IS +LL ++
Sbjct: 27 NESESNTKCQTAIQSLSTIVTNTNIPSTITILLDDEAVST---AISSLLLRPDS------ 77
Query: 175 VGAIQRAMARVWLWDGRSKASPRLLDGMVEYIAELAGFHRERL 217
GA + R WL+D A P L ++ ++ +AG + R+
Sbjct: 78 -GAGDNNLCR-WLYDTFQSAEPSLQLLVLRFVPLIAGLYLSRV 118
>At1g71696 carboxypeptidase, putative
Length = 367
Score = 29.3 bits (64), Expect = 2.6
Identities = 13/50 (26%), Positives = 26/50 (52%), Gaps = 4/50 (8%)
Query: 13 LLPHFNPSSYEINQQNDYNNSKFFIPNNNNIIHRILLILFVAIISIWANY 62
++P NP + I ++N+ NN + N + +I L+LF+ + +Y
Sbjct: 163 IMPSLNPDGFSIRKRNNANN----VDLNRDFPDQIRLVLFIVTFDLLNSY 208
>At4g03390 SRF3 (LRR receptor-like protein kinase like protein)
Length = 776
Score = 28.5 bits (62), Expect = 4.5
Identities = 18/61 (29%), Positives = 30/61 (48%), Gaps = 2/61 (3%)
Query: 118 NIDIKNKKIIKSVTLRLARQNLDTITTITADEKNINSYVIEISPMLLEDENFNNMAIVGA 177
N+DI + I S TL + +NL T+TT+ ++ + + + L+D N N G
Sbjct: 172 NLDISSNNI--SGTLPPSMENLLTLTTLRVQNNQLSGTLDVLQGLPLQDLNIENNLFSGP 229
Query: 178 I 178
I
Sbjct: 230 I 230
>At1g53090 phytochrome A supressor spa1, putative
Length = 794
Score = 28.5 bits (62), Expect = 4.5
Identities = 15/43 (34%), Positives = 21/43 (47%), Gaps = 6/43 (13%)
Query: 203 VEYIAELAGFHRERLSGGVGESP-----ECED-GRELWWDNKD 239
++Y+ L G H+E GG+ + ECED W DN D
Sbjct: 32 IDYVRSLLGSHKEANLGGLDDDSIVRALECEDVSLRQWLDNPD 74
>At1g31420 unknown protein
Length = 592
Score = 28.5 bits (62), Expect = 4.5
Identities = 19/72 (26%), Positives = 33/72 (45%), Gaps = 3/72 (4%)
Query: 18 NPSSYEINQQNDYNNSKFFIPNNNNIIHRILLILFVAIISIWANYEASKTFDIHIVNDTK 77
NPSS+ + QN NS + + + + +LL VA++ W + K + I + K
Sbjct: 221 NPSSHSQSGQNQKKNSGKLLISASATVGALLL---VALMCFWGCFLYKKLGKVEIKSLAK 277
Query: 78 DSLAGRRFTLFY 89
D G +F+
Sbjct: 278 DVGGGASIVMFH 289
>At1g32750 unknown protein
Length = 1919
Score = 28.1 bits (61), Expect = 5.9
Identities = 18/48 (37%), Positives = 25/48 (51%), Gaps = 5/48 (10%)
Query: 195 SPRLLDGMVEYIAELAGFHRE---RLSGGVGESPECEDGRELWWDNKD 239
SPR + +AELAGF R+ RLS E ED +WW+ ++
Sbjct: 1643 SPRFESRKTKRMAELAGFQRQQSFRLSENSLERRPKED--RVWWEEEE 1688
>At1g53380 hypothetical protein; similar to EST gb|R30194.1
Length = 453
Score = 27.7 bits (60), Expect = 7.6
Identities = 11/28 (39%), Positives = 17/28 (60%)
Query: 239 DPTHVARLLHYCEKYDKGFIQRLNEAMK 266
+PTH LH+ K +GF++ + E MK
Sbjct: 207 NPTHFVTYLHHTVKSTRGFVKLMIEQMK 234
>At4g04500 putative receptor-like protein kinase
Length = 646
Score = 27.3 bits (59), Expect = 10.0
Identities = 31/133 (23%), Positives = 53/133 (39%), Gaps = 13/133 (9%)
Query: 30 YNNSKFFIPNNNNIIHRILLILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFY 89
+ N+ F + NN +H VA+ +A KT H++ DTK ++ + +
Sbjct: 60 FYNASFGRDSKNNRVH------VVALCRRGYEKQACKTCLEHVIEDTKSKCPRQKESFSW 113
Query: 90 VSND----KASRILLNTSSFVEQILYPNGNNDN---IDIKNKKIIKSVTLRLARQNLDTI 142
V+++ S N S+ + L PN N N ID K + +A N
Sbjct: 114 VTDEFDDVSCSLRYTNHSTLGKLELLPNTINPNPNSIDSKFNNMAMFSQEWIAMVNRTLE 173
Query: 143 TTITADEKNINSY 155
TA+ ++ Y
Sbjct: 174 AASTAENSSVLKY 186
>At3g61130 unknown protein
Length = 673
Score = 27.3 bits (59), Expect = 10.0
Identities = 20/77 (25%), Positives = 37/77 (47%), Gaps = 18/77 (23%)
Query: 105 FVEQILYPNGNND--------NIDIKNKKIIKSVTLRLARQNLDTITTITADEKNINSYV 156
FV + +Y + +ND N+D + + ++SV +++ LD I T TAD
Sbjct: 39 FVGRGVYIDSSNDYSIVSVKQNLDWRERLAMQSVRSLFSKEILDVIATSTAD-------- 90
Query: 157 IEISPMLLEDENFNNMA 173
+ P+ L+ NN++
Sbjct: 91 --LGPLSLDSFKKNNLS 105
>At3g49840 putative protein
Length = 651
Score = 27.3 bits (59), Expect = 10.0
Identities = 23/93 (24%), Positives = 43/93 (45%), Gaps = 7/93 (7%)
Query: 47 ILLILFVAIISIWANYEASKTFDIHIVNDTKDSLAGRRFTLFYVSNDKASRILLNTSSFV 106
ILL++FV ++ W+ + K F + TKD ++F + +A + L S +
Sbjct: 312 ILLVVFVFMLGAWSGFWTVKKFVV-----TKDGSVDISTSIFVSWSIRAFAVALILQSSL 366
Query: 107 EQILYPNGNNDNIDIKNKKIIKSVTLRLARQNL 139
+ +L I I I+KS++ +L Q +
Sbjct: 367 DPLLAGGALITGILI--SLILKSISRKLLAQRI 397
>At2g45670 unknown protein
Length = 539
Score = 27.3 bits (59), Expect = 10.0
Identities = 11/33 (33%), Positives = 21/33 (63%)
Query: 155 YVIEISPMLLEDENFNNMAIVGAIQRAMARVWL 187
Y E+SP ++ E+ +++ VG I RAM +++
Sbjct: 187 YFYELSPTIVASESHDSLPFVGTIIRAMQVIYV 219
>At2g31740 unknown protein
Length = 760
Score = 27.3 bits (59), Expect = 10.0
Identities = 25/92 (27%), Positives = 39/92 (42%), Gaps = 17/92 (18%)
Query: 101 NTSSFVEQILYPNGNNDNI----------DIKNKKIIKSVTLRLARQNLDTITTITADEK 150
++SS QIL P N + DI N K V + R+N+ T
Sbjct: 64 SSSSDSLQILVPGCGNSRLTEHLYDAGFRDITNVDFSKVVISDMLRRNIRT-------RP 116
Query: 151 NINSYVIEISPMLLEDENFNNMAIVGAIQRAM 182
+ V++I+ M L DE+F+ + GA+ M
Sbjct: 117 ELRWRVMDITKMQLADESFDTVLDKGALDALM 148
>At2g06850 putative endoxyloglucan glycosyltransferase
Length = 296
Score = 27.3 bits (59), Expect = 10.0
Identities = 11/35 (31%), Positives = 17/35 (48%), Gaps = 1/35 (2%)
Query: 205 YIAELAGFHRERLSGGVGESPECEDGRELWWDNKD 239
++A GFH + V E+ C +WWD K+
Sbjct: 219 FVASYKGFHIDGCQASV-EAKYCATQGRMWWDQKE 252
>At1g04470 unknown protein
Length = 1035
Score = 27.3 bits (59), Expect = 10.0
Identities = 23/70 (32%), Positives = 35/70 (49%), Gaps = 9/70 (12%)
Query: 81 AGRRFTLFYVSNDKASRIL-LNTSSFVEQILYPNGNNDNIDIKNKKIIKSVTLRLARQNL 139
AG +VS A R++ L++ S + LYP D+ N +I LR+ +QNL
Sbjct: 825 AGIESACQHVSEVAAYRLIFLDSYSVFYESLYPG------DVANGRI--KPALRILKQNL 876
Query: 140 DTITTITADE 149
+T I AD+
Sbjct: 877 TLMTAILADK 886
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,797,379
Number of Sequences: 26719
Number of extensions: 287134
Number of successful extensions: 718
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 705
Number of HSP's gapped (non-prelim): 17
length of query: 294
length of database: 11,318,596
effective HSP length: 99
effective length of query: 195
effective length of database: 8,673,415
effective search space: 1691315925
effective search space used: 1691315925
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC147496.3


