

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147484.13 + phase: 0 /pseudo
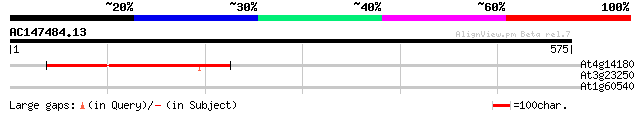
(575 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g14180 hypothetical protein 169 3e-42
At3g23250 myb-related transcription factor like protein 31 1.6
At1g60540 30 4.8
>At4g14180 hypothetical protein
Length = 1268
Score = 169 bits (429), Expect = 3e-42
Identities = 98/197 (49%), Positives = 129/197 (64%), Gaps = 9/197 (4%)
Query: 38 RGLGNMNYQIHYSREAEEIIFQLINNGEWDLLSARIHTVSLKWLFQQENIINSLCHQILK 97
R L N YQI YS EAE IIF L+N EWDL S IH SLKWLFQQE+I SL +QI K
Sbjct: 661 RSLQNERYQISYSLEAERIIFHLLNEYEWDLGSINIHLESLKWLFQQESISKSLIYQIQK 720
Query: 98 FCRNNNLEGDDM--IIGNSNQTVNVQTLAELVSSEDNYGARLFICLLAQLAEEEGQEHDI 155
R NNL G+++ + G+ Q A+L+S DNY A L + LL QLAE+E QE+D+
Sbjct: 721 ISR-NNLIGNEVHNVYGDGRQRSLTYWFAKLISEGDNYAATLLVNLLTQLAEKEEQENDV 779
Query: 156 ISVLNLMATMIHICPAASDQLSLHGIGTTIRTCC--FSTA----TFMSILVLVFNTLSSV 209
IS+LNLM T++ I P AS+ LS++GIG+ I FS + +F ++L+LVFN L+SV
Sbjct: 780 ISILNLMNTIVSIFPTASNNLSMNGIGSVIHRLVSGFSNSSLGTSFRTLLLLVFNILTSV 839
Query: 210 HPKTLSTDQSWVAVTME 226
P L D+SW AV+++
Sbjct: 840 QPAVLMIDESWYAVSIK 856
Score = 37.7 bits (86), Expect = 0.017
Identities = 23/83 (27%), Positives = 44/83 (52%), Gaps = 2/83 (2%)
Query: 484 LTS*VLSKLQL*VKCCRKQMYTNNAQDGLPSEPIKKVLATSYDLGDVCWFLIDLMSPDTL 543
L S ++KL + CRKQM+ N +D E +++ + + +D + C +L+ LM ++
Sbjct: 1159 LNSQQVTKLDRAFQECRKQMHRNGTRDETVEEQVQRKIPSIHDHSEFCNYLVHLMVSNSF 1218
Query: 544 --DMDSWGIHMDGKRLLEEIELF 564
+S K++L+E+E F
Sbjct: 1219 GHPSESETYTQKKKQILDEMEQF 1241
>At3g23250 myb-related transcription factor like protein
Length = 285
Score = 31.2 bits (69), Expect = 1.6
Identities = 19/61 (31%), Positives = 32/61 (52%), Gaps = 3/61 (4%)
Query: 12 LNSSDSHNWTTDSLTVTRLVNLYSLLRGLGNMNYQIHYSREAEEIIFQLI--NNGEWDLL 69
L S D HN+ ++ L V LV + + LG+ N ++ + E + F ++ GE DLL
Sbjct: 224 LYSQDEHNYVSNDLEVAGLVEIQQEFQNLGSANNEMIFDSEM-DFWFDVLARTGGEQDLL 282
Query: 70 S 70
+
Sbjct: 283 A 283
>At1g60540
Length = 648
Score = 29.6 bits (65), Expect = 4.8
Identities = 39/200 (19%), Positives = 81/200 (40%), Gaps = 19/200 (9%)
Query: 63 NGEWDLLSARIHTVSLKWLFQQENIINSLCHQILKFCRNNNLEGDDMIIGNSNQTVNVQT 122
NG+ + + +I + +K++ QE+II ++ + F ++ + +T+ V T
Sbjct: 182 NGQPENIYEQISRMVMKYIEPQESIILNVLSATVDFTTCESIRMSKQVDKTGERTLAVVT 241
Query: 123 LAEL--------VSSEDNYGARLFICLLAQLAEEEGQE-----------HDIISVLNLMA 163
A++ V+++D ++C+ ++ EE +E H ++S++N
Sbjct: 242 KADMAPEGLLQKVTADDVSIGLGYVCVRNRIGEETYEEARMQEELLFRTHPMLSMINDEI 301
Query: 164 TMIHICPAASDQLSLHGIGTTIRTCCFSTATFMSILVLVFNTLSSVHPKTLSTDQSWVAV 223
I + Q+ I + M I VL FN L V T S + +
Sbjct: 302 VGIPVLAQKLTQIQGMMISRCLPEIERKINVKMEISVLEFNKLPIVMASTGEALMSLMDI 361
Query: 224 TMESRKSLRHWHSFLDFTSF 243
+++SL DF+ +
Sbjct: 362 IGSAKESLLRILVHGDFSEY 381
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.349 0.152 0.538
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,322,016
Number of Sequences: 26719
Number of extensions: 422180
Number of successful extensions: 1646
Number of sequences better than 10.0: 3
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 1641
Number of HSP's gapped (non-prelim): 4
length of query: 575
length of database: 11,318,596
effective HSP length: 105
effective length of query: 470
effective length of database: 8,513,101
effective search space: 4001157470
effective search space used: 4001157470
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC147484.13


