

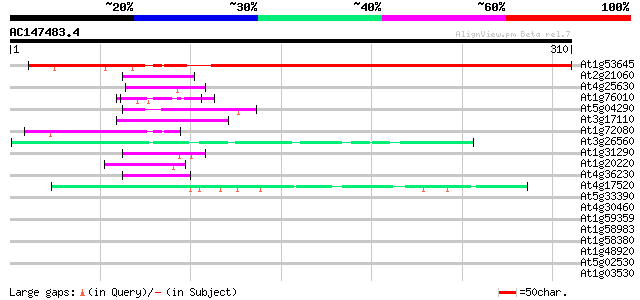
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147483.4 + phase: 0
(310 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g53645 unknown protein 291 3e-79
At2g21060 glycine-rich protein (AtGRP2) 44 9e-05
At4g25630 fibrillarin - like protein 43 2e-04
At1g76010 unknown protein 43 2e-04
At5g04290 glycine-rich protein 43 2e-04
At3g17110 hypothetical protein 43 2e-04
At1g72080 hypothetical protein 42 3e-04
At3g26560 ATP-dependent RNA helicase, putative 42 4e-04
At1g31290 hypothetical protein 42 4e-04
At1g20220 unknown protein 42 6e-04
At4g36230 putative glycine-rich cell wall protein 41 7e-04
At4g17520 nuclear RNA binding protein A-like protein 41 7e-04
At5g33390 unknown protein (At5g33390) 41 0.001
At4g30460 glycine-rich protein 41 0.001
At1g59359 ribosomal protein S2, putative 41 0.001
At1g58983 ribosomal protein S2, putative 41 0.001
At1g58380 unknown protein 41 0.001
At1g48920 unknown protein 41 0.001
At5g02530 unknown protein 40 0.001
At1g03530 unknown protein 40 0.001
>At1g53645 unknown protein
Length = 523
Score = 291 bits (745), Expect = 3e-79
Identities = 162/326 (49%), Positives = 205/326 (62%), Gaps = 44/326 (13%)
Query: 11 GRGRGKPLEEAAQ---------EAPQAPVVNRHIRVRQTPADAESDNVPR---------R 52
G GRGKPL E+A P P + ++ +Q A D P+ R
Sbjct: 216 GAGRGKPLVESAPIRQEDNRQIRRPPPPPQQQRVQPQQKRAPTVKDGTPKPQLSAEEAGR 275
Query: 53 QPMNRFVRDDGDGS------GRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGD 106
+ + R + +GS GRGRGRGRG ARGRG RGRGG G R D +
Sbjct: 276 RARSELSRGEAEGSSVGGRGGRGRGRGRG----ARGRG-RGRGGDGWRDDKK-------- 322
Query: 107 DRTSHQDIARSNADGLYVGDNADGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYV 166
++ A ++ GD+ADGEK A+K+GPE+M + E +EEI E+ LPS D +
Sbjct: 323 -----EEEGEQEAMRIFAGDSADGEKFAEKMGPELMKTLAEGFEEICEKALPSTTHDAII 377
Query: 167 EAMDINCAIEFEPEYAV-EF-DNPDIDEKEPIALRDALEKMKPFLMTYEGIRSQEEWEEV 224
+A D N IE EPEY + +F NPDIDEK P++LR+ LEK+KPF++ YEGI+ QEEWEE
Sbjct: 378 DAYDTNLMIECEPEYIMPDFGSNPDIDEKPPMSLRECLEKVKPFIVAYEGIKDQEEWEEA 437
Query: 225 IEELMQRVPLLKKIVDHYSGPDRVTAKKQQEELERVAKTLPTSAPSSVKEFTNRAVVSLQ 284
I E M + PL+K+IVDHYSGPDRVTAKKQ EEL+R+A TLP SAP SVK F +RA ++L+
Sbjct: 438 INEAMTQAPLMKEIVDHYSGPDRVTAKKQNEELDRIATTLPASAPDSVKRFADRAALTLK 497
Query: 285 SNPGWGFDKKCQFMDKLVFEVSQHHK 310
SNPGWGFDKK QFMDKLV EVSQ +K
Sbjct: 498 SNPGWGFDKKYQFMDKLVLEVSQSYK 523
Score = 32.7 bits (73), Expect = 0.26
Identities = 22/49 (44%), Positives = 22/49 (44%), Gaps = 13/49 (26%)
Query: 64 DGSGRGRGRGRGRD--VYARGRGD-----------RGRGGRGGRGDGRG 99
D SGRGRGRG G D A GRG R GG G GRG
Sbjct: 39 DSSGRGRGRGSGEDGGFPAAGRGQFGVNREPVVPGREPSSAGGYGHGRG 87
>At2g21060 glycine-rich protein (AtGRP2)
Length = 201
Score = 44.3 bits (103), Expect = 9e-05
Identities = 22/40 (55%), Positives = 22/40 (55%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFK 102
G G G GRG GRG Y G G RG GGRGG G FK
Sbjct: 101 GFGGGGGRGGGRGGGSYGGGYGGRGSGGRGGGGGDNSCFK 140
Score = 31.6 bits (70), Expect = 0.57
Identities = 23/58 (39%), Positives = 26/58 (44%), Gaps = 7/58 (12%)
Query: 43 DAESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
D E DN R + + D G G G + GRG G GG GGRG GRGG
Sbjct: 64 DVEVDNSGRPKAIEVSGPDGAPVQGNSGGGGS-----SGGRG--GFGGGGGRGGGRGG 114
>At4g25630 fibrillarin - like protein
Length = 320
Score = 43.1 bits (100), Expect = 2e-04
Identities = 27/47 (57%), Positives = 28/47 (59%), Gaps = 3/47 (6%)
Query: 65 GSGRGRGRGRGRDVYARGRGDRG-RGGR--GGRGDGRGGFKRYGDDR 108
GSG G GRGR Y+ GRGD G GGR GGRG GRG R G R
Sbjct: 7 GSGGGFSGGRGRGGYSGGRGDGGFSGGRGGGGRGGGRGFSDRGGRGR 53
Score = 39.7 bits (91), Expect = 0.002
Identities = 24/46 (52%), Positives = 26/46 (56%), Gaps = 9/46 (19%)
Query: 63 GDGSGRGRGR---GRGRDVYARGRGDRGRGG------RGGRGDGRG 99
G GRGRG GRG ++ GRG GRGG RGGRG GRG
Sbjct: 11 GFSGGRGRGGYSGGRGDGGFSGGRGGGGRGGGRGFSDRGGRGRGRG 56
Score = 37.4 bits (85), Expect = 0.010
Identities = 29/81 (35%), Positives = 35/81 (42%), Gaps = 2/81 (2%)
Query: 61 DDGDGSGRGRG-RGRGRDVYARGRGDRGRGG-RGGRGDGRGGFKRYGDDRTSHQDIARSN 118
D G GRG G RG GR RG RGRG RGG GRG R G S +
Sbjct: 27 DGGFSGGRGGGGRGGGRGFSDRGGRGRGRGPPRGGARGGRGPAGRGGMKGGSKVIVEPHR 86
Query: 119 ADGLYVGDNADGEKLAKKLGP 139
G+++ + + K L P
Sbjct: 87 HAGVFIAKGKEDALVTKNLVP 107
Score = 35.8 bits (81), Expect = 0.030
Identities = 25/51 (49%), Positives = 25/51 (49%), Gaps = 8/51 (15%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGR-------GGRGGRGD-GRGGFK 102
R DG SG G GRG RG RGR G RGGRG GRGG K
Sbjct: 25 RGDGGFSGGRGGGGRGGGRGFSDRGGRGRGRGPPRGGARGGRGPAGRGGMK 75
Score = 35.0 bits (79), Expect = 0.052
Identities = 33/78 (42%), Positives = 38/78 (48%), Gaps = 9/78 (11%)
Query: 63 GDGSGRGRGRGRG---RDVYARGRGD-RG--RGGRG--GRGDGRGGFKRYGD-DRTSHQD 113
G G GRG GRG R RGRG RG RGGRG GRG +GG K + R +
Sbjct: 32 GGRGGGGRGGGRGFSDRGGRGRGRGPPRGGARGGRGPAGRGGMKGGSKVIVEPHRHAGVF 91
Query: 114 IARSNADGLYVGDNADGE 131
IA+ D L + GE
Sbjct: 92 IAKGKEDALVTKNLVPGE 109
Score = 34.3 bits (77), Expect = 0.088
Identities = 18/34 (52%), Positives = 18/34 (52%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGG 93
R D GRGRGRG R GRG GRGG G
Sbjct: 43 RGFSDRGGRGRGRGPPRGGARGGRGPAGRGGMKG 76
>At1g76010 unknown protein
Length = 350
Score = 43.1 bits (100), Expect = 2e-04
Identities = 30/72 (41%), Positives = 34/72 (46%), Gaps = 20/72 (27%)
Query: 60 RDDGDGSGRGRGRGRG------------------RDVYARGRGDRGRGGRGGRGDGRGGF 101
R G G GRGRGRGRG Y RGRG RGR GR RG GRGG+
Sbjct: 151 RGRGRGRGRGRGRGRGGRGNAYVNVEHEDGGWEREQSYGRGRG-RGR-GRSSRGRGRGGY 208
Query: 102 KRYGDDRTSHQD 113
++ + QD
Sbjct: 209 NGPPNEYDAPQD 220
Score = 42.7 bits (99), Expect = 2e-04
Identities = 28/50 (56%), Positives = 29/50 (58%), Gaps = 11/50 (22%)
Query: 62 DGDGSGRG-----RGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGD 106
DG GRG +GRGRGR RGRG RGRGG GRG G GGF D
Sbjct: 299 DGPSQGRGGYDGPQGRGRGR---GRGRGGRGRGG--GRG-GDGGFNNRSD 342
Score = 40.8 bits (94), Expect = 0.001
Identities = 29/66 (43%), Positives = 32/66 (47%), Gaps = 23/66 (34%)
Query: 53 QPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRG-------------------G 93
+PM + +GS GRGRGRGR RGRG RGRGGRG G
Sbjct: 134 KPMGDIDYEGREGSPGGRGRGRGR---GRGRG-RGRGGRGNAYVNVEHEDGGWEREQSYG 189
Query: 94 RGDGRG 99
RG GRG
Sbjct: 190 RGRGRG 195
Score = 39.3 bits (90), Expect = 0.003
Identities = 19/31 (61%), Positives = 19/31 (61%)
Query: 67 GRGRGRGRGRDVYARGRGDRGRGGRGGRGDG 97
GRGRGRGRGR RG G G GG R DG
Sbjct: 313 GRGRGRGRGRGGRGRGGGRGGDGGFNNRSDG 343
Score = 32.0 bits (71), Expect = 0.44
Identities = 19/43 (44%), Positives = 24/43 (55%), Gaps = 8/43 (18%)
Query: 64 DGSGRGRGRGRGRDVYARGRG-----DRGRGGRGGRGDGRGGF 101
DG +GRG G D ++GRG +GRGG G GRGG+
Sbjct: 269 DGPPQGRG---GYDGPSQGRGGYDGPSQGRGGYDGPSQGRGGY 308
Score = 29.3 bits (64), Expect = 2.8
Identities = 18/44 (40%), Positives = 21/44 (46%), Gaps = 5/44 (11%)
Query: 63 GDGSGRGRGRGRGRDVYARGRG-----DRGRGGRGGRGDGRGGF 101
G G G RG D +GRG +GRGG G GRGG+
Sbjct: 255 GRGGYDGPQGRRGYDGPPQGRGGYDGPSQGRGGYDGPSQGRGGY 298
>At5g04290 glycine-rich protein
Length = 1493
Score = 42.7 bits (99), Expect = 2e-04
Identities = 27/76 (35%), Positives = 35/76 (45%), Gaps = 10/76 (13%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNADGL 122
G+GSG G+G G G G +G G RGG+G+G FK + GL
Sbjct: 401 GEGSGGGKGEGSG--------GGKGEGSRGGKGEGSSDFKSESSYELYNLVCFSRKDFGL 452
Query: 123 YVG--DNADGEKLAKK 136
VG D DG K+ K+
Sbjct: 453 IVGVDDKGDGYKVLKE 468
Score = 33.9 bits (76), Expect = 0.12
Identities = 18/53 (33%), Positives = 23/53 (42%)
Query: 61 DDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQD 113
++ G GRG G RG + GR GRGGR G +K + T D
Sbjct: 1326 NEHSGGGRGFGERRGGGGFRGGRNQSGRGGRSFDGGRSSSWKTDNQENTWKSD 1378
Score = 30.8 bits (68), Expect = 0.98
Identities = 28/83 (33%), Positives = 31/83 (36%), Gaps = 3/83 (3%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGR--GGRGDGRGGFKRYGDDRTSHQDIARS 117
+DDG GS G+ G RG RG GGR GGR GR F R S S
Sbjct: 1066 KDDG-GSSWGKKDEGGYSEQTFDRGGRGFGGRRGGGRRGGRDQFGRGSSFGNSEDPAPWS 1124
Query: 118 NADGLYVGDNADGEKLAKKLGPE 140
G DG+ G E
Sbjct: 1125 KPSGGSSWGKQDGDGGGSSWGKE 1147
Score = 28.5 bits (62), Expect = 4.9
Identities = 18/60 (30%), Positives = 28/60 (46%), Gaps = 1/60 (1%)
Query: 47 DNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGR-GDRGRGGRGGRGDGRGGFKRYG 105
D + + P++R D + G R + + R G +G G GG+G+G GG K G
Sbjct: 360 DELLKFTPVDRKETGDVEWISEIYGEERKKKILPTCREGGKGEGSGGGKGEGSGGGKGEG 419
>At3g17110 hypothetical protein
Length = 175
Score = 42.7 bits (99), Expect = 2e-04
Identities = 25/62 (40%), Positives = 25/62 (40%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNA 119
R G GSG G G G G G G G GG GG GDG G YG S N
Sbjct: 31 RGSGSGSGYGSGSGGGNGGGGGGGGGGGGGGGGGGGDGSGSGSGYGSGYGSGSGYGGGNN 90
Query: 120 DG 121
G
Sbjct: 91 GG 92
Score = 39.3 bits (90), Expect = 0.003
Identities = 20/39 (51%), Positives = 21/39 (53%)
Query: 62 DGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
DG GSG G G G G G + G GG GGRG G GG
Sbjct: 67 DGSGSGSGYGSGYGSGSGYGGGNNGGGGGGGGRGGGGGG 105
Score = 37.7 bits (86), Expect = 0.008
Identities = 21/43 (48%), Positives = 23/43 (52%), Gaps = 3/43 (6%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYG 105
G+GSG G G G G GRG G GG GG G G GG + G
Sbjct: 111 GNGSGYGSGSGYGS---GAGRGGGGGGGGGGGGGGGGGSRGNG 150
Score = 37.4 bits (85), Expect = 0.010
Identities = 22/45 (48%), Positives = 24/45 (52%), Gaps = 1/45 (2%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGG-RGDGRGGFKRYGD 106
G GSG G G G GR G G G GG GG RG+G G YG+
Sbjct: 115 GYGSGSGYGSGAGRGGGGGGGGGGGGGGGGGSRGNGSGYGSGYGE 159
Score = 37.0 bits (84), Expect = 0.014
Identities = 24/63 (38%), Positives = 25/63 (39%), Gaps = 1/63 (1%)
Query: 65 GSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNADGLYV 124
GSG G G G G Y G G G GG GG G G GG G+ G
Sbjct: 109 GSGNGSGYGSGSG-YGSGAGRGGGGGGGGGGGGGGGGGSRGNGSGYGSGYGEGYGSGYGG 167
Query: 125 GDN 127
GDN
Sbjct: 168 GDN 170
Score = 36.6 bits (83), Expect = 0.018
Identities = 21/39 (53%), Positives = 21/39 (53%), Gaps = 4/39 (10%)
Query: 63 GDGSGRGRGRGRGRDVYARGRG-DRGRGGRGGRGDGRGG 100
GDGSG G G G G Y G G G G GG G GRGG
Sbjct: 66 GDGSGSGSGYGSG---YGSGSGYGGGNNGGGGGGGGRGG 101
Score = 36.2 bits (82), Expect = 0.023
Identities = 19/38 (50%), Positives = 20/38 (52%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
G GSG G G G G G G GRGG GG G+G G
Sbjct: 74 GYGSGYGSGSGYGGGNNGGGGGGGGRGGGGGGGNGGSG 111
Score = 35.4 bits (80), Expect = 0.040
Identities = 20/43 (46%), Positives = 21/43 (48%), Gaps = 1/43 (2%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYG 105
G G G G G G G + G G RG GG GG G G G YG
Sbjct: 76 GSGYGSGSGYGGGNNGGGGGGGGRGGGGGGGNG-GSGNGSGYG 117
Score = 35.0 bits (79), Expect = 0.052
Identities = 20/38 (52%), Positives = 21/38 (54%), Gaps = 4/38 (10%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
G G+G G G G G Y G G RG GG GG G G GG
Sbjct: 109 GSGNGSGYGSGSG---YGSGAG-RGGGGGGGGGGGGGG 142
Score = 34.3 bits (77), Expect = 0.088
Identities = 24/68 (35%), Positives = 25/68 (36%), Gaps = 8/68 (11%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNADGL 122
G GSG G G G G G G GG GG G G GG GD S G
Sbjct: 32 GSGSGSGYGSGSG--------GGNGGGGGGGGGGGGGGGGGGGDGSGSGSGYGSGYGSGS 83
Query: 123 YVGDNADG 130
G +G
Sbjct: 84 GYGGGNNG 91
Score = 34.3 bits (77), Expect = 0.088
Identities = 22/55 (40%), Positives = 22/55 (40%), Gaps = 11/55 (20%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRG-----------GRGDGRGGFKRYGD 106
G G G G GRG G G G G G RG G G G GG YGD
Sbjct: 119 GSGYGSGAGRGGGGGGGGGGGGGGGGGSRGNGSGYGSGYGEGYGSGYGGGDNYGD 173
Score = 32.3 bits (72), Expect = 0.34
Identities = 19/44 (43%), Positives = 19/44 (43%), Gaps = 6/44 (13%)
Query: 63 GDGSGRGRGRGRGR------DVYARGRGDRGRGGRGGRGDGRGG 100
G G GRG G G G Y G G GRGG G G GG
Sbjct: 94 GGGGGRGGGGGGGNGGSGNGSGYGSGSGYGSGAGRGGGGGGGGG 137
>At1g72080 hypothetical protein
Length = 243
Score = 42.4 bits (98), Expect = 3e-04
Identities = 34/95 (35%), Positives = 43/95 (44%), Gaps = 13/95 (13%)
Query: 9 GGGRGRGKPLEEA--------AQEAPQAPV-VNRHIRVRQTPADAESDNVPRRQPMNRFV 59
G GRGRG+ + +E P A + H+ ++ + RR +
Sbjct: 59 GRGRGRGRSRRRSRSPSHSNGGREPPFATINTEEHLTMQIESPSTIINTEERRAGDDDGS 118
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGR 94
R G G GRGRGRGRGR RGRG RGRG R
Sbjct: 119 RSRGRGRGRGRGRGRGR---GRGRG-RGRGRNRSR 149
Score = 32.3 bits (72), Expect = 0.34
Identities = 16/32 (50%), Positives = 17/32 (53%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGR 91
R G G GRGRGRGRGR+ GGR
Sbjct: 129 RGRGRGRGRGRGRGRGRNRSRSRSPSHSNGGR 160
Score = 32.0 bits (71), Expect = 0.44
Identities = 18/39 (46%), Positives = 20/39 (51%), Gaps = 3/39 (7%)
Query: 65 GSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKR 103
G GR G R R +RGR D GGRG GRG +R
Sbjct: 34 GLGRRGGNNRNR---SRGRDDNQSPSNGGRGRGRGRSRR 69
Score = 29.6 bits (65), Expect = 2.2
Identities = 62/254 (24%), Positives = 87/254 (33%), Gaps = 70/254 (27%)
Query: 17 PLEEAAQEAPQAPVV-------NRHIRVRQTPADAESDNVPRRQPMNRFVRDDGDG-SGR 68
P +++QE PQ VV N +R R +N R + RDD S
Sbjct: 4 PSNQSSQEQPQRCVVLALPAISNTSLRHRYGLGRRGGNNRNRSRG-----RDDNQSPSNG 58
Query: 69 GRGRGRGR---------------------------------------DVYARGRGD---- 85
GRGRGRGR + R GD
Sbjct: 59 GRGRGRGRSRRRSRSPSHSNGGREPPFATINTEEHLTMQIESPSTIINTEERRAGDDDGS 118
Query: 86 --RGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNADGLYVGDNADGEKLAKKLGPEIMD 143
RGRG GRG GRG + G R ++ +RS + N E L E D
Sbjct: 119 RSRGRGRGRGRGRGRGRGRGRGRGRGRNRSRSRSPSH-----SNGGREPPTTTLNTE--D 171
Query: 144 QIT---EAYEEIIERVLPSPLQDEYVEAMDINCAIEFEPEYAVEFDNPDIDEKEPIALRD 200
+T E II R + D V +I + + + D +++++ I R
Sbjct: 172 HLTMQIEPPSTIINRENGTDANDNVVAVNNIESDCKIDETVPPKTDEEILEDEDDI--RS 229
Query: 201 ALEKMKPFLMTYEG 214
L+K+ M Y G
Sbjct: 230 LLDKLPIPSMFYLG 243
>At3g26560 ATP-dependent RNA helicase, putative
Length = 1168
Score = 42.0 bits (97), Expect = 4e-04
Identities = 58/255 (22%), Positives = 97/255 (37%), Gaps = 24/255 (9%)
Query: 2 TVPKVLPGGGRGRGKPLEEAAQEAPQAPVVNRHIRVRQTPADAESDNVPRRQPMNRFVRD 61
T+ + P + K E Q+ + + +V++ + E + RR+ +R
Sbjct: 72 TIHGIYPPKPKSEKKKEEGDDQKFKGLAIKDTKDKVKELEKEIEREAEERRREEDRNRDR 131
Query: 62 DGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNADG 121
D SGR R R R RD R DR R RGD G D R+ + R DG
Sbjct: 132 DRRESGRDRDRDRNRD--RDDRRDRHRDRERNRGDEEG-----EDRRSDRRHRERGRGDG 184
Query: 122 LYVGDNADGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYVEAMDINCAIEFEPEY 181
G+ D + + + + A E + +V + MD C ++F+
Sbjct: 185 ---GEGEDRRRDRRAKDEYVEEDKGGANEPELYQV----YKGRVTRVMDAGCFVQFDKFR 237
Query: 182 AVEFDNPDIDEKEPIALRDALEKMKPFLMTYEGIRSQEEWEEVIEELMQRVPLLKKIVDH 241
E + +A R ++K K F+ R E + +VI + L + VD
Sbjct: 238 GKE----GLVHVSQMATR-RVDKAKEFVK-----RDMEVYVKVISISSDKYSLSMRDVDQ 287
Query: 242 YSGPDRVTAKKQQEE 256
+G D + +K +E
Sbjct: 288 NTGRDLIPLRKPSDE 302
>At1g31290 hypothetical protein
Length = 1194
Score = 42.0 bits (97), Expect = 4e-04
Identities = 29/61 (47%), Positives = 30/61 (48%), Gaps = 15/61 (24%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGD-RGRGGRG-----GRGDGRG---------GFKRYGDD 107
G G GRGRG G R GRGD RGRGG G GRGDG G G+ GD
Sbjct: 9 GRGDGRGRGGGGDRGRGYSGRGDGRGRGGDGDRGYSGRGDGHGRGGGGDRGRGYSGRGDG 68
Query: 108 R 108
R
Sbjct: 69 R 69
Score = 40.4 bits (93), Expect = 0.001
Identities = 23/44 (52%), Positives = 23/44 (52%), Gaps = 4/44 (9%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRG----GRGDGRG 99
R DG G G G RGRG GRG G G RG GRGDG G
Sbjct: 46 RGDGHGRGGGGDRGRGYSGRGDGRGRGGGGDRGRGYSGRGDGHG 89
Score = 39.7 bits (91), Expect = 0.002
Identities = 23/37 (62%), Positives = 23/37 (62%), Gaps = 7/37 (18%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRG 99
G GRG GRGRG G GDRGRG GRGDGRG
Sbjct: 5 GYRGGRGDGRGRG------GGGDRGRG-YSGRGDGRG 34
Score = 39.3 bits (90), Expect = 0.003
Identities = 22/39 (56%), Positives = 22/39 (56%), Gaps = 1/39 (2%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGD-RGRGGRGGRGDGRGG 100
G G GRGRG G R GRGD GRGG G RG G G
Sbjct: 64 GRGDGRGRGGGGDRGRGYSGRGDGHGRGGGGDRGRGYSG 102
Score = 39.3 bits (90), Expect = 0.003
Identities = 22/39 (56%), Positives = 22/39 (56%), Gaps = 1/39 (2%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGD-RGRGGRGGRGDGRGG 100
G G G GRG G R GRGD RGRGG G RG G G
Sbjct: 45 GRGDGHGRGGGGDRGRGYSGRGDGRGRGGGGDRGRGYSG 83
Score = 38.5 bits (88), Expect = 0.005
Identities = 27/50 (54%), Positives = 27/50 (54%), Gaps = 11/50 (22%)
Query: 61 DDGDG-SGRGRGRGRGRDVY--------ARGRGDRGRGGRG--GRGDGRG 99
D G G SGRG GRGRG D GRG G GRG GRGDGRG
Sbjct: 21 DRGRGYSGRGDGRGRGGDGDRGYSGRGDGHGRGGGGDRGRGYSGRGDGRG 70
Score = 37.0 bits (84), Expect = 0.014
Identities = 27/52 (51%), Positives = 28/52 (52%), Gaps = 10/52 (19%)
Query: 62 DGD----GSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDG--RG-GFKRYGD 106
DGD G G G GRG G D RGRG GRG GRG G RG G+ GD
Sbjct: 38 DGDRGYSGRGDGHGRGGGGD---RGRGYSGRGDGRGRGGGGDRGRGYSGRGD 86
Score = 36.6 bits (83), Expect = 0.018
Identities = 26/55 (47%), Positives = 26/55 (47%), Gaps = 14/55 (25%)
Query: 63 GDGSGRGRGRGRGRDVYARGRG---------DRGRGGRGGRGDGRGGFKRYGDDR 108
GDG GRG G RGR RG G RG GRG G GRGG G DR
Sbjct: 47 GDGHGRGGGGDRGRGYSGRGDGRGRGGGGDRGRGYSGRGD-GHGRGG----GGDR 96
Score = 35.0 bits (79), Expect = 0.052
Identities = 25/57 (43%), Positives = 26/57 (44%), Gaps = 5/57 (8%)
Query: 60 RDDGDGSGRGRG-RGRGRDVYARGRGDRGRG----GRGGRGDGRGGFKRYGDDRTSH 111
R G G RGRG GRG G GDRGRG GRG D GG+ G H
Sbjct: 69 RGRGGGGDRGRGYSGRGDGHGRGGGGDRGRGYSGRGRGFVQDRDGGWVNPGQSSGGH 125
Score = 31.6 bits (70), Expect = 0.57
Identities = 19/42 (45%), Positives = 20/42 (47%)
Query: 59 VRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
V D G G GR DV G GD GRGG G RG + G
Sbjct: 204 VGDVGQGGVGDVGRDGVGDVGRDGVGDVGRGGVGDRGQSQSG 245
Score = 29.3 bits (64), Expect = 2.8
Identities = 18/44 (40%), Positives = 18/44 (40%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKR 103
R DG G G G RGRG RG GG G GG R
Sbjct: 84 RGDGHGRGGGGDRGRGYSGRGRGFVQDRDGGWVNPGQSSGGHVR 127
Score = 27.7 bits (60), Expect = 8.3
Identities = 37/129 (28%), Positives = 49/129 (37%), Gaps = 15/129 (11%)
Query: 9 GGGRGRGKPLEEAAQEAPQAPVVNRHIRVRQTPADAESDNVPRRQPMNRFVRDDGDGSGR 68
G RGRG L++ + + P + +V Q A + V D G
Sbjct: 124 GHVRGRGTQLQQPPPQ--EVPPSSSQAQVSQGVAPGDVGQGGVGDVGRDGVGDVGRDGVG 181
Query: 69 GRGRGRGRDVYARGRGDRGRG-----GRGGRGD-GRGGFKRYGDDRTSHQDIARSNADGL 122
G+G DV G GD G+G G+GG GD GR G G D D+ R
Sbjct: 182 DVGQGGVGDVGQVGVGDVGQGGVGDVGQGGVGDVGRDGVGDVGRDGVG--DVGRGG---- 235
Query: 123 YVGDNADGE 131
VGD +
Sbjct: 236 -VGDRGQSQ 243
>At1g20220 unknown protein
Length = 315
Score = 41.6 bits (96), Expect = 6e-04
Identities = 25/58 (43%), Positives = 29/58 (49%), Gaps = 13/58 (22%)
Query: 53 QPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRG-------------GRGGRGDG 97
+P+ + DGS RGRG RGR RGRG GRG GRGGRG+G
Sbjct: 134 KPLAEIDYEGQDGSPRGRGGRRGRGGRGRGRGRGGRGNGPANVEYDDGGRGRGGRGNG 191
Score = 39.7 bits (91), Expect = 0.002
Identities = 31/78 (39%), Positives = 38/78 (47%), Gaps = 20/78 (25%)
Query: 41 PADAESDNVPRRQPM--NRFVRDDGDGSGRGRGRGRGRDV---------------YARGR 83
PA+ E D+ R + N +V ++ D GRGRG GRG Y RGR
Sbjct: 173 PANVEYDDGGRGRGGRGNGYVNNEYDDGGRGRG-GRGSGYVNNEYNDGGMEQDRSYGRGR 231
Query: 84 GDRGRGGRGGRGDGRGGF 101
G GGRGGR GRGG+
Sbjct: 232 GRGRGGGRGGR--GRGGY 247
Score = 37.4 bits (85), Expect = 0.010
Identities = 28/63 (44%), Positives = 28/63 (44%), Gaps = 19/63 (30%)
Query: 54 PMNRFVRDDGDGSGRGRGRG-RGR-------DVYARGRGDRGRG-----------GRGGR 94
P R R G GRGRGRG RG D RGRG RG G GRGGR
Sbjct: 148 PRGRGGRRGRGGRGRGRGRGGRGNGPANVEYDDGGRGRGGRGNGYVNNEYDDGGRGRGGR 207
Query: 95 GDG 97
G G
Sbjct: 208 GSG 210
Score = 37.0 bits (84), Expect = 0.014
Identities = 34/94 (36%), Positives = 38/94 (40%), Gaps = 28/94 (29%)
Query: 10 GGRGRGKPLEEAAQEAPQAPVVNRHIRVRQTPADAESDNVPRRQPMNRFVRDDGDGSGRG 69
GGRGRG P P + +Q D +NV D G
Sbjct: 240 GGRGRG------GYNGPPPP----YYEAQQDGGDYGYNNVAPPA-------DHGYDGPPP 282
Query: 70 RGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKR 103
+GRGRGR GRGGRG RG GRGGF R
Sbjct: 283 QGRGRGR----------GRGGRG-RGGGRGGFNR 305
Score = 33.9 bits (76), Expect = 0.12
Identities = 17/25 (68%), Positives = 17/25 (68%), Gaps = 3/25 (12%)
Query: 69 GRGRGRGRDVYARGRGDRGRGGRGG 93
GRGRGRGR GRG RGRGG G
Sbjct: 228 GRGRGRGR---GGGRGGRGRGGYNG 249
>At4g36230 putative glycine-rich cell wall protein
Length = 221
Score = 41.2 bits (95), Expect = 7e-04
Identities = 21/38 (55%), Positives = 21/38 (55%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
G GSG G GRGRG G G G GG GG G G GG
Sbjct: 141 GGGSGNGSGRGRGGGGGGGGGGGGGGGGGGGGGGGGGG 178
Score = 40.8 bits (94), Expect = 0.001
Identities = 22/45 (48%), Positives = 22/45 (48%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDD 107
G G G G G GRGR G G G GG GG G G GG G D
Sbjct: 139 GGGGGSGNGSGRGRGGGGGGGGGGGGGGGGGGGGGGGGGGGGGSD 183
Score = 38.1 bits (87), Expect = 0.006
Identities = 23/54 (42%), Positives = 25/54 (45%), Gaps = 3/54 (5%)
Query: 47 DNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
DN +R + G G G G G G GR RG G G GG GG G G GG
Sbjct: 121 DNSHKRNKSSGGGGGGGGGGGGGSGNGSGR---GRGGGGGGGGGGGGGGGGGGG 171
Score = 35.4 bits (80), Expect = 0.040
Identities = 21/47 (44%), Positives = 24/47 (50%), Gaps = 3/47 (6%)
Query: 62 DGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRG---DGRGGFKRYG 105
+G G GRG G G G G G G GG GG G DG+GG +G
Sbjct: 146 NGSGRGRGGGGGGGGGGGGGGGGGGGGGGGGGGGGGSDGKGGGWGFG 192
Score = 33.5 bits (75), Expect = 0.15
Identities = 24/54 (44%), Positives = 26/54 (47%), Gaps = 10/54 (18%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRG---------GRGDGRG-GFKRYGD 106
G+GSGRGRG G G G G G GG G G+G G G GF GD
Sbjct: 145 GNGSGRGRGGGGGGGGGGGGGGGGGGGGGGGGGGGGGSDGKGGGWGFGFGWGGD 198
Score = 32.7 bits (73), Expect = 0.26
Identities = 19/46 (41%), Positives = 22/46 (47%), Gaps = 9/46 (19%)
Query: 63 GDGSGRGRGRGRG---------RDVYARGRGDRGRGGRGGRGDGRG 99
G GSG G G G G R+ + G G G GG GG G+G G
Sbjct: 104 GQGSGGGGGEGNGGNGKDNSHKRNKSSGGGGGGGGGGGGGSGNGSG 149
Score = 31.6 bits (70), Expect = 0.57
Identities = 19/56 (33%), Positives = 26/56 (45%), Gaps = 4/56 (7%)
Query: 45 ESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
E +N Q +N+ + + G G G G G G G +G GG GG G+G G
Sbjct: 68 EFENALDDQNVNKTEKCESRAGGGGGGGGGG----GGGGGGQGSGGGGGEGNGGNG 119
Score = 30.4 bits (67), Expect = 1.3
Identities = 18/46 (39%), Positives = 20/46 (43%), Gaps = 1/46 (2%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDR 108
G G G G G G G+D + G GG GG G G G G R
Sbjct: 108 GGGGGEGNG-GNGKDNSHKRNKSSGGGGGGGGGGGGGSGNGSGRGR 152
Score = 30.4 bits (67), Expect = 1.3
Identities = 18/51 (35%), Positives = 22/51 (42%), Gaps = 5/51 (9%)
Query: 63 GDGSGRGRGRGRGRDVYARGRG-----DRGRGGRGGRGDGRGGFKRYGDDR 108
G G G+G G G G G+ ++ GG GG G G GG G R
Sbjct: 100 GGGGGQGSGGGGGEGNGGNGKDNSHKRNKSSGGGGGGGGGGGGGSGNGSGR 150
Score = 28.9 bits (63), Expect = 3.7
Identities = 16/39 (41%), Positives = 19/39 (48%), Gaps = 5/39 (12%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGR 98
R G G G G G G G G+G G GG G G+G+
Sbjct: 87 RAGGGGGGGGGGGGGGG-----GQGSGGGGGEGNGGNGK 120
Score = 27.7 bits (60), Expect = 8.3
Identities = 17/38 (44%), Positives = 18/38 (46%), Gaps = 4/38 (10%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
G G G G G G G D G+G G G GDG GG
Sbjct: 169 GGGGGGGGGGGGGSD----GKGGGWGFGFGWGGDGPGG 202
>At4g17520 nuclear RNA binding protein A-like protein
Length = 360
Score = 41.2 bits (95), Expect = 7e-04
Identities = 68/289 (23%), Positives = 112/289 (38%), Gaps = 40/289 (13%)
Query: 24 EAPQAPVVNRHIRVRQTPADAESDNVPRRQPM-NRFVRDDGDGSGRGRGRGRGRDVYARG 82
E P V ++ ++V A+S +P + P ++ VR+ + G GRG GRG G
Sbjct: 15 EDPSQIVASKPLKVVAPVQTAKSGKMPTKPPPPSQAVREARNAPGGGRGAGRGGSYGRGG 74
Query: 83 RGDRGRGGRGGRGDGR-----GGFKR--YGDDRTSHQDIA--RSNADGLYV--GDNADGE 131
RG R R G GG++R GD + R + G Y GD+ D E
Sbjct: 75 RGGNNRDSRNNDGPANENGYGGGYRRSEEGDGARRGGPVGGYRGDRRGSYSNGGDSGDSE 134
Query: 132 KLAKKL---------GPEIMDQITEAYEEIIERVLPSPLQDEYVEAMDINCAIEFEPEYA 182
+ K + D A + + +P+ +E +D N +E +
Sbjct: 135 RPRKNYDRHSRTAYGNEDKRDGAGRANWGTTQDDI-TPVTEESTAVVDKNLTVEKQ---- 189
Query: 183 VEFDNPDIDEKEPIALRDALEKMKPFLMTYEGIRSQEEWEEVIEE---LMQRVPLLKKIV 239
+ + D K A EK + MT EE+E+V+EE +Q + ++ V
Sbjct: 190 -DGEGEATDAKNETPAEKAEEKPEDKEMTL------EEYEKVLEEKKKALQATKVEERKV 242
Query: 240 D--HYSGPDRVTAKKQQEELERVAKTLPTSAPSSVKEFTNRAVVSLQSN 286
D + ++++KK + V L T + E + SL N
Sbjct: 243 DTKAFEAMQQLSSKKSNN--DEVFIKLGTEKDKRITEREEKTRKSLSIN 289
>At5g33390 unknown protein (At5g33390)
Length = 118
Score = 40.8 bits (94), Expect = 0.001
Identities = 22/41 (53%), Positives = 23/41 (55%), Gaps = 1/41 (2%)
Query: 65 GSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYG 105
GSG G G G G RG G RG G GGRGDGRG + G
Sbjct: 32 GSGSG-GSGSGGRANGRGNGGRGSGRGGGRGDGRGDGRGIG 71
Score = 40.4 bits (93), Expect = 0.001
Identities = 28/64 (43%), Positives = 31/64 (47%), Gaps = 11/64 (17%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRG-------GRGGRGDGRGGFKRYGDDRTSHQ 112
R G G GRG GRG GR + G G RGRG GR GRG GRG + D Q
Sbjct: 52 RGSGRGGGRGDGRGDGRGI---GGGGRGRGRIPAAFMGR-GRGRGRGNHGQMSIDALQAQ 107
Query: 113 DIAR 116
+ R
Sbjct: 108 EARR 111
Score = 32.7 bits (73), Expect = 0.26
Identities = 21/40 (52%), Positives = 23/40 (57%), Gaps = 6/40 (15%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRG 99
R +G G+G GRG GRG GRGD GRG G GRG
Sbjct: 43 RANGRGNG-GRGSGRGG-----GRGDGRGDGRGIGGGGRG 76
Score = 32.3 bits (72), Expect = 0.34
Identities = 24/76 (31%), Positives = 29/76 (37%), Gaps = 1/76 (1%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNADGL 122
G GSG G G G G G GGR +GRG R G R + R + G+
Sbjct: 12 GSGSGGSVSGGSGSGGSGSGGSGSGGSGSGGRANGRGNGGR-GSGRGGGRGDGRGDGRGI 70
Query: 123 YVGDNADGEKLAKKLG 138
G G A +G
Sbjct: 71 GGGGRGRGRIPAAFMG 86
Score = 29.3 bits (64), Expect = 2.8
Identities = 18/40 (45%), Positives = 19/40 (47%), Gaps = 2/40 (5%)
Query: 62 DGDGSGRGRGRGRGRDVYA-RGRGDRGRGGRGGRGDGRGG 100
+G GSG G G G V G G G GG G G G GG
Sbjct: 4 NGGGSGSS-GSGSGGSVSGGSGSGGSGSGGSGSGGSGSGG 42
>At4g30460 glycine-rich protein
Length = 162
Score = 40.8 bits (94), Expect = 0.001
Identities = 20/35 (57%), Positives = 21/35 (59%)
Query: 65 GSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRG 99
GSGRGRG G G G G GRGG GG G+G G
Sbjct: 112 GSGRGRGSGGGGGHGGGGGGGGGRGGGGGSGNGEG 146
Score = 38.5 bits (88), Expect = 0.005
Identities = 20/43 (46%), Positives = 21/43 (48%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYG 105
G G GRG G G G G G RG GG G G+G G YG
Sbjct: 112 GSGRGRGSGGGGGHGGGGGGGGGRGGGGGSGNGEGYGEGGGYG 154
Score = 36.6 bits (83), Expect = 0.018
Identities = 21/44 (47%), Positives = 23/44 (51%), Gaps = 2/44 (4%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGD 106
G GSG G GRGR + G G G GG GG G G GG G+
Sbjct: 104 GSGSGGRSGSGRGRG--SGGGGGHGGGGGGGGGRGGGGGSGNGE 145
Score = 31.2 bits (69), Expect = 0.75
Identities = 20/45 (44%), Positives = 20/45 (44%), Gaps = 4/45 (8%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDD 107
G G G G G G GR G G G G G G G GG GDD
Sbjct: 122 GGGHGGGGGGGGGRG----GGGGSGNGEGYGEGGGYGGGYGGGDD 162
Score = 29.3 bits (64), Expect = 2.8
Identities = 16/40 (40%), Positives = 17/40 (42%)
Query: 61 DDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
D G +G G G R RGRG GG G G GG
Sbjct: 90 DAGSEAGSYAGSHAGSGSGGRSGSGRGRGSGGGGGHGGGG 129
Score = 27.7 bits (60), Expect = 8.3
Identities = 14/33 (42%), Positives = 14/33 (42%)
Query: 67 GRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRG 99
G G G YA G GGR G G GRG
Sbjct: 86 GGGGDAGSEAGSYAGSHAGSGSGGRSGSGRGRG 118
>At1g59359 ribosomal protein S2, putative
Length = 284
Score = 40.8 bits (94), Expect = 0.001
Identities = 22/34 (64%), Positives = 22/34 (64%), Gaps = 5/34 (14%)
Query: 61 DDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGR 94
D G G G GRG GRGRD RG RGRG RGGR
Sbjct: 18 DFGRGFGGGRGGGRGRD-----RGPRGRGRRGGR 46
Score = 33.1 bits (74), Expect = 0.20
Identities = 33/111 (29%), Positives = 44/111 (38%), Gaps = 16/111 (14%)
Query: 67 GRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRG------GFKRYGDDRTSHQDIARSNAD 120
G G RG D RGD GRG GGRG GRG G R G +
Sbjct: 5 GGESGAERGGD-----RGDFGRGFGGGRGGGRGRDRGPRGRGRRGGRASEETKWVPVTKL 59
Query: 121 GLYVGDNADGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYVEAMDI 171
G V DN KL + + +II+ ++ L+DE ++ M +
Sbjct: 60 GRLVADNK-----ITKLEQIYLHSLPVKEYQIIDHLVGPTLKDEVMKIMPV 105
>At1g58983 ribosomal protein S2, putative
Length = 284
Score = 40.8 bits (94), Expect = 0.001
Identities = 22/34 (64%), Positives = 22/34 (64%), Gaps = 5/34 (14%)
Query: 61 DDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGR 94
D G G G GRG GRGRD RG RGRG RGGR
Sbjct: 18 DFGRGFGGGRGGGRGRD-----RGPRGRGRRGGR 46
Score = 33.1 bits (74), Expect = 0.20
Identities = 33/111 (29%), Positives = 44/111 (38%), Gaps = 16/111 (14%)
Query: 67 GRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRG------GFKRYGDDRTSHQDIARSNAD 120
G G RG D RGD GRG GGRG GRG G R G +
Sbjct: 5 GGESGAERGGD-----RGDFGRGFGGGRGGGRGRDRGPRGRGRRGGRASEETKWVPVTKL 59
Query: 121 GLYVGDNADGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYVEAMDI 171
G V DN KL + + +II+ ++ L+DE ++ M +
Sbjct: 60 GRLVADNK-----ITKLEQIYLHSLPVKEYQIIDHLVGPTLKDEVMKIMPV 105
>At1g58380 unknown protein
Length = 284
Score = 40.8 bits (94), Expect = 0.001
Identities = 22/34 (64%), Positives = 22/34 (64%), Gaps = 5/34 (14%)
Query: 61 DDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGR 94
D G G G GRG GRGRD RG RGRG RGGR
Sbjct: 18 DFGRGFGGGRGGGRGRD-----RGPRGRGRRGGR 46
Score = 32.7 bits (73), Expect = 0.26
Identities = 32/107 (29%), Positives = 43/107 (39%), Gaps = 16/107 (14%)
Query: 71 GRGRGRDVYARGRGDRGRGGRGGRGDGRG------GFKRYGDDRTSHQDIARSNADGLYV 124
G RG D RGD GRG GGRG GRG G R G + G V
Sbjct: 9 GAERGGD-----RGDFGRGFGGGRGGGRGRDRGPRGRGRRGGRASEETKWVPVTKLGRLV 63
Query: 125 GDNADGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYVEAMDI 171
DN KL + + +II+ ++ L+DE ++ M +
Sbjct: 64 ADNK-----ITKLEQIYLHSLPVKEYQIIDHLVGPTLKDEVMKIMPV 105
>At1g48920 unknown protein
Length = 557
Score = 40.8 bits (94), Expect = 0.001
Identities = 22/43 (51%), Positives = 23/43 (53%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYG 105
G G GRGR GRGR GRG G GR G G GRG + G
Sbjct: 499 GSGGGRGRDGGRGRFGSGGGRGRDGGRGRFGSGGGRGSDRGRG 541
Score = 39.7 bits (91), Expect = 0.002
Identities = 26/54 (48%), Positives = 27/54 (49%), Gaps = 8/54 (14%)
Query: 53 QPMNRFVRDDGDGSGRGRGR-----GRGRDVYARGRGDRGRGGRGGRGDGRGGF 101
+P R G G GRG GR GRGRD GRG G GG GR GRG F
Sbjct: 478 EPRPRGDSSGGGGFGRGNGRFGSGGGRGRD---GGRGRFGSGGGRGRDGGRGRF 528
Score = 36.6 bits (83), Expect = 0.018
Identities = 25/50 (50%), Positives = 26/50 (52%), Gaps = 4/50 (8%)
Query: 60 RDDGDGSGRGR-GRGRGRDVYARGRGDRGRGGRGGRGDGR---GGFKRYG 105
R GD SG G GRG GR GRG G GR G G GR GG R+G
Sbjct: 480 RPRGDSSGGGGFGRGNGRFGSGGGRGRDGGRGRFGSGGGRGRDGGRGRFG 529
>At5g02530 unknown protein
Length = 292
Score = 40.4 bits (93), Expect = 0.001
Identities = 24/55 (43%), Positives = 28/55 (50%), Gaps = 7/55 (12%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDI 114
R G GR RG G G + GRG RGRGGRG G GR D+ S +D+
Sbjct: 230 RGRGGFMGRPRGGGFGGGNFRGGRGARGRGGRGSGGRGR-------DENVSAEDL 277
>At1g03530 unknown protein
Length = 801
Score = 40.4 bits (93), Expect = 0.001
Identities = 26/76 (34%), Positives = 36/76 (47%), Gaps = 1/76 (1%)
Query: 17 PLEEAAQEAPQAPVVNRHIRVRQTPADAESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGR 76
P Q Q+ V ++ Q+P+D +S P+ Q + + GRGRGRGR
Sbjct: 727 PANPQFQMQAQSDVRPLQSQIPQSPSDLQSPMEPQSQGFSSGQSSERGRGFHGRGRGRGR 786
Query: 77 DVYARGRGDRGRGGRG 92
+ RGRG RGR G
Sbjct: 787 GRFGRGRG-RGRQQSG 801
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.137 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,801,689
Number of Sequences: 26719
Number of extensions: 404030
Number of successful extensions: 4521
Number of sequences better than 10.0: 236
Number of HSP's better than 10.0 without gapping: 137
Number of HSP's successfully gapped in prelim test: 101
Number of HSP's that attempted gapping in prelim test: 1454
Number of HSP's gapped (non-prelim): 1712
length of query: 310
length of database: 11,318,596
effective HSP length: 99
effective length of query: 211
effective length of database: 8,673,415
effective search space: 1830090565
effective search space used: 1830090565
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147483.4


