

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147482.11 - phase: 0
(109 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
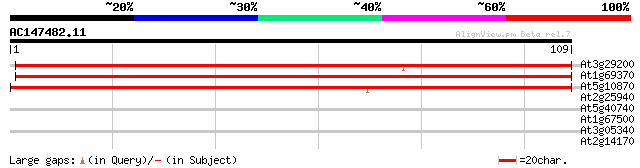
At3g29200 chorismate mutase 152 2e-38
At1g69370 chorismate mutase like protein 134 7e-33
At5g10870 chorismate mutase CM2 122 2e-29
At2g25940 putative vacuolar processing enzyme 27 2.2
At5g40740 unknown protein 26 3.7
At1g67500 putative DNA polymerase zeta catalytic subunit 26 3.7
At3g05340 hypothetical protein 25 4.9
At2g14170 F15N24.6 25 6.4
>At3g29200 chorismate mutase
Length = 340
Score = 152 bits (385), Expect = 2e-38
Identities = 75/115 (65%), Positives = 95/115 (82%), Gaps = 7/115 (6%)
Query: 2 LSKRIHYGKFVAEAKFQAAPDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTYG 61
LSKRIHYGKFVAEAKFQA+P++Y++AI AQDKD LM++LT+P VE++IK+RV +K +TYG
Sbjct: 226 LSKRIHYGKFVAEAKFQASPEAYESAIKAQDKDALMDMLTFPTVEDAIKKRVEMKTRTYG 285
Query: 62 QEVAVNLKDQKTEP-------VYKINPSLVADLYSDWIMPLTKEVQVAYLLRKLD 109
QEV V +++++ E VYKI+P LV DLY DWIMPLTKEVQV YLLR+LD
Sbjct: 286 QEVKVGMEEKEEEEEEGNESHVYKISPILVGDLYGDWIMPLTKEVQVEYLLRRLD 340
>At1g69370 chorismate mutase like protein
Length = 316
Score = 134 bits (337), Expect = 7e-33
Identities = 64/108 (59%), Positives = 85/108 (78%)
Query: 2 LSKRIHYGKFVAEAKFQAAPDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTYG 61
LSKRIH+GKFVAEAKF+ P +Y+ AI QD+ +LM+LLTY VEE +K+RV +KA+ +G
Sbjct: 209 LSKRIHFGKFVAEAKFRENPAAYETAIKEQDRTQLMQLLTYETVEEVVKKRVEIKARIFG 268
Query: 62 QEVAVNLKDQKTEPVYKINPSLVADLYSDWIMPLTKEVQVAYLLRKLD 109
Q++ +N + + +P YKI PSLVA LY + IMPLTKEVQ+ YLLR+LD
Sbjct: 269 QDITINDPETEADPSYKIQPSLVAKLYGERIMPLTKEVQIEYLLRRLD 316
>At5g10870 chorismate mutase CM2
Length = 265
Score = 122 bits (307), Expect = 2e-29
Identities = 60/110 (54%), Positives = 84/110 (75%), Gaps = 1/110 (0%)
Query: 1 ALSKRIHYGKFVAEAKFQAAPDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTY 60
ALS+RIHYGKFVAE KF+ AP Y+ AI AQD++ LM+LLT+ +VEE +K+RV KA+T+
Sbjct: 156 ALSRRIHYGKFVAEVKFRDAPQDYEPAIRAQDREALMKLLTFEKVEEMVKKRVQKKAETF 215
Query: 61 GQEVAVNL-KDQKTEPVYKINPSLVADLYSDWIMPLTKEVQVAYLLRKLD 109
GQEV N +++ YK++P L + +Y +W++PLTK V+V YLLR+LD
Sbjct: 216 GQEVKFNSGYGDESKKKYKVDPLLASRIYGEWLIPLTKLVEVEYLLRRLD 265
>At2g25940 putative vacuolar processing enzyme
Length = 478
Score = 26.6 bits (57), Expect = 2.2
Identities = 14/43 (32%), Positives = 20/43 (45%)
Query: 22 DSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTYGQEV 64
D Y A I + ++ T E E +K+R A K+YG V
Sbjct: 254 DLYSVAWIEDSEKHNLQTETLHEQYELVKKRTAGSGKSYGSHV 296
>At5g40740 unknown protein
Length = 741
Score = 25.8 bits (55), Expect = 3.7
Identities = 19/85 (22%), Positives = 35/85 (40%), Gaps = 9/85 (10%)
Query: 13 AEAKFQAAPDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTYGQEVAVNLKDQK 72
A +K+ PD+Y D L Y + + + Q+ ++ DQ
Sbjct: 658 APSKYSDIPDTY---------DDLDSFKDYDNGNGFLSVAGSNSVASDAQQSFYDIDDQV 708
Query: 73 TEPVYKINPSLVADLYSDWIMPLTK 97
P ++ SL++D Y D + PL++
Sbjct: 709 FSPPLLMDSSLLSDAYEDLLAPLSE 733
>At1g67500 putative DNA polymerase zeta catalytic subunit
Length = 1871
Score = 25.8 bits (55), Expect = 3.7
Identities = 17/60 (28%), Positives = 26/60 (43%), Gaps = 9/60 (15%)
Query: 17 FQAAPDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTYGQEVAVNLKDQKTEPV 76
F A D++ A ++ D D + LL K R +A GQE+A + + PV
Sbjct: 1512 FVNANDNWNARVVYGDTDSMFVLL---------KGRTVKEAFVVGQEIASAITEMNPHPV 1562
>At3g05340 hypothetical protein
Length = 770
Score = 25.4 bits (54), Expect = 4.9
Identities = 11/21 (52%), Positives = 14/21 (66%)
Query: 9 GKFVAEAKFQAAPDSYKAAII 29
G++ AE FQ APDS A I+
Sbjct: 545 GEYAAEQLFQTAPDSSSAHIL 565
>At2g14170 F15N24.6
Length = 607
Score = 25.0 bits (53), Expect = 6.4
Identities = 16/73 (21%), Positives = 31/73 (41%), Gaps = 4/73 (5%)
Query: 20 APDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTYGQEVAVNLKDQKTEPVYKI 79
APD ++ +I + K + Y E +++ K R + T + + + P +
Sbjct: 30 APDQHRVSIHSSLKSKTKRRRLYKEADDNTKLRSSSSTTTTTTTMLLRISGNNLRP---L 86
Query: 80 NPSLVADLYSDWI 92
P +A L S W+
Sbjct: 87 RPQFLA-LRSSWL 98
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.132 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,159,822
Number of Sequences: 26719
Number of extensions: 73581
Number of successful extensions: 213
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 205
Number of HSP's gapped (non-prelim): 8
length of query: 109
length of database: 11,318,596
effective HSP length: 85
effective length of query: 24
effective length of database: 9,047,481
effective search space: 217139544
effective search space used: 217139544
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 52 (24.6 bits)
Medicago: description of AC147482.11


