

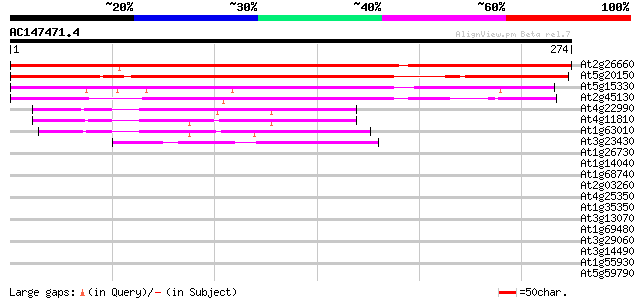
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147471.4 + phase: 0
(274 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g26660 unknown protein 298 2e-81
At5g20150 ids4-like protein 268 3e-72
At5g15330 ids-4 protein - like 183 8e-47
At2g45130 unknown protein 176 2e-44
At4g22990 unknown protein 52 4e-07
At4g11810 putative protein 50 1e-06
At1g63010 tetracycline resistance efflux protein like protein 47 8e-06
At3g23430 PHO1 protein (PHO1) 44 1e-04
At1g26730 unknown protein 39 0.002
At1g14040 unknown protein 39 0.003
At1g68740 putative protein (At1g68740) 36 0.025
At2g03260 unknown protein 34 0.097
At4g25350 putative protein 33 0.16
At1g35350 hypothetical protein 33 0.22
At3g13070 hemolysin-like protein 32 0.28
At1g69480 hypothetical protein 32 0.28
At3g29060 unknown protein 32 0.48
At3g14490 terpene synthase-related protein, putative 31 0.82
At1g55930 unknown protein 31 0.82
At5g59790 unknown protein 30 1.1
>At2g26660 unknown protein
Length = 287
Score = 298 bits (763), Expect = 2e-81
Identities = 158/277 (57%), Positives = 200/277 (72%), Gaps = 7/277 (2%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDD---DGGAE 57
MKF K L+NQIE+TLP+WRDKFLSYK+LKK+LKL+ P+ +++ +KR R D D
Sbjct: 1 MKFGKSLSNQIEETLPEWRDKFLSYKELKKKLKLMEPRSVENRPNKRSRSDSNSVDTDPT 60
Query: 58 GEVTKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSDIELMTVGREI 117
+TKE DF+ LLE E+EKFN FFVE+EEEY+I+ KEL+D+VA AK+S+ E++ + +EI
Sbjct: 61 VGMTKEELDFISLLEDELEKFNSFFVEQEEEYIIRLKELKDQVAKAKNSNEEMINIKKEI 120
Query: 118 VDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVK 177
VDFHGEMVLL NYSALNYTGL KI+KKYDKRTGAL+RLPFIQ VL +PFF D+LN VK
Sbjct: 121 VDFHGEMVLLMNYSALNYTGLAKILKKYDKRTGALIRLPFIQKVLQEPFFTTDLLNTFVK 180
Query: 178 ECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNMENIF 237
ECE ML +FP + +S + E E L VPKE SEI+ ME+++
Sbjct: 181 ECEAMLDRLFPSN----KSRNLDEEGEPTTSGMVKTGTDDSELLRVPKELSEIEYMESLY 236
Query: 238 IKLTTSALDTLKEIRGGSSTVSIYSLPPLHSETLVED 274
+K T SAL LKEIR GSSTVS++SLPPL + L +D
Sbjct: 237 MKSTVSALKVLKEIRSGSSTVSVFSLPPLPASGLEDD 273
>At5g20150 ids4-like protein
Length = 256
Score = 268 bits (684), Expect = 3e-72
Identities = 153/273 (56%), Positives = 189/273 (69%), Gaps = 30/273 (10%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEV 60
MKF K L+NQIEQTLP+W+DKFLSYK+LKK+LKLI K D KR RLD+ +
Sbjct: 1 MKFGKSLSNQIEQTLPEWQDKFLSYKELKKRLKLIGSKTADRPV-KRLRLDEFSVG---I 56
Query: 61 TKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSDIELMTVGREIVDF 120
+KE +F++LLE E+EKFN FFVEKEEEY+I+ KE +D++A AK S +++ + +EIVDF
Sbjct: 57 SKEEINFIQLLEDELEKFNNFFVEKEEEYIIRLKEFRDRIAKAKDSMEKMIKIRKEIVDF 116
Query: 121 HGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVKECE 180
HGEMVLLENYSALNYTGLVKI+KKYDKRTG L+RLPFIQ VL QPF+ D+L KLVKE E
Sbjct: 117 HGEMVLLENYSALNYTGLVKILKKYDKRTGDLMRLPFIQKVLQQPFYTTDLLFKLVKESE 176
Query: 181 VMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNMENIFIKL 240
ML IFP ANET+ + + E SE + ME++ +K
Sbjct: 177 AMLDQIFP------------------------ANETESEI--IQAELSEHKFMESLHMKS 210
Query: 241 TTSALDTLKEIRGGSSTVSIYSLPPLHSETLVE 273
T +AL LKEIR GSSTVS++SLPPL L E
Sbjct: 211 TIAALRVLKEIRSGSSTVSVFSLPPLQLNGLDE 243
>At5g15330 ids-4 protein - like
Length = 318
Score = 183 bits (465), Expect = 8e-47
Identities = 116/295 (39%), Positives = 163/295 (54%), Gaps = 38/295 (12%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIV-------PKEIDSSCSKRRRLD-- 51
MKF K +E+TLP+WRDKFL YK LKK LK P D + S+ D
Sbjct: 1 MKFGKEFRTHLEETLPEWRDKFLCYKPLKKLLKYYPYYSADFGPANSDHNDSRPVFADTT 60
Query: 52 ------DDGGAEGEVTKEVK---DFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAW 102
DDGG V F+R+L E+EKFN F+V+KEE++VI+ +EL++++
Sbjct: 61 NISSAADDGGVVPGVRPSEDLQGSFVRILNDELEKFNDFYVDKEEDFVIRLQELKERIEQ 120
Query: 103 AKSSD----------IELMTVGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGAL 152
K + E+M + R++V HGEMVLL+NYS+LN+ GLVKI+KKYDKRTG L
Sbjct: 121 VKEKNGEFASESEFSEEMMDIRRDLVTIHGEMVLLKNYSSLNFAGLVKILKKYDKRTGGL 180
Query: 153 LRLPFIQDVLNQPFFKIDVLNKLVKECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETTT 212
LRLPF Q VL+QPFF + L +LV+ECE L ++FP S +EV E +
Sbjct: 181 LRLPFTQLVLHQPFFTTEPLTRLVRECEANLELLFP---------SEAEVVESSSAVQAH 231
Query: 213 ANETKETLDHVPKEFSEIQNMENIFI-KLTTSALDTLKEIRGGSSTVSIYSLPPL 266
++ + + E S EN+ I K T +A+ ++ ++ SST + S L
Sbjct: 232 SSSHQHNSPRISAETSSTLGNENLDIYKSTLAAMRAIRGLQKASSTYNPLSFSSL 286
>At2g45130 unknown protein
Length = 245
Score = 176 bits (445), Expect = 2e-44
Identities = 111/275 (40%), Positives = 153/275 (55%), Gaps = 58/275 (21%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEV 60
MKF K + QI+++LP+WRDKFL YK+LK + P E
Sbjct: 1 MKFGKRIKEQIQESLPEWRDKFLRYKELKNLISSPAPVE--------------------- 39
Query: 61 TKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWA--------KSSDIELMT 112
F+ LL EI+KFN FFVE+EE+++I KELQ ++ + S +
Sbjct: 40 ----SIFVGLLNAEIDKFNAFFVEQEEDFIIHHKELQYRIQRLVEKCGHNDEMSRENISE 95
Query: 113 VGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVL 172
+ ++IV+FHGEMVLL NYS +NYTGL KI+KKYDKRT LR PFIQ VL+QPFFK D++
Sbjct: 96 IRKDIVNFHGEMVLLVNYSNINYTGLAKILKKYDKRTRGGLRSPFIQKVLHQPFFKTDLV 155
Query: 173 NKLVKECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQN 232
++LV+E E + + P ++ +E +E A T+ A
Sbjct: 156 SRLVREWETTMDAVDP------VKVAEAEGYERCAAVTSAAAG----------------- 192
Query: 233 MENIFIKLTTSALDTLKEIRGGSSTVSIYSLPPLH 267
E IF + T +AL T+KE+R GSST S +SLPPL+
Sbjct: 193 -EGIF-RNTVAALLTMKEMRRGSSTYSAFSLPPLN 225
>At4g22990 unknown protein
Length = 699
Score = 51.6 bits (122), Expect = 4e-07
Identities = 40/160 (25%), Positives = 83/160 (51%), Gaps = 16/160 (10%)
Query: 12 EQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEVTKEVKDFLRLL 71
E+++ +W+ +++YK +KK++K ++++ +RR + +KDF R+L
Sbjct: 10 ERSIQEWQGYYINYKLMKKKVKQY-SRQLEGGNLERRHV-------------LKDFSRML 55
Query: 72 EVEIEKFNGFFVEKEEEYVIKWKELQDKV-AWAKSSDIELMTVGREIVDFHGEMVL-LEN 129
+ +IEK F +E++ + + L+ A + DI M+ +E G+ +L L
Sbjct: 56 DNQIEKIALFMLEQQGLLASRLQTLRGSHDALQEQPDISHMSYLKEEYRAVGQDLLKLLF 115
Query: 130 YSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKI 169
+ +N G+ KI+KK+DKR G +++ N P+ ++
Sbjct: 116 FVEMNAIGIRKILKKFDKRFGYRFTNYYVKTRANHPYSEL 155
>At4g11810 putative protein
Length = 707
Score = 50.1 bits (118), Expect = 1e-06
Identities = 41/162 (25%), Positives = 82/162 (50%), Gaps = 20/162 (12%)
Query: 12 EQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEVTKEVKDFLRLL 71
E+++ +W++ +++YK +KK++K P +I+ RR + +KDF R+L
Sbjct: 10 ERSIEEWQEYYINYKLMKKKVKQYGP-QIEVGSLDRRHV-------------LKDFSRML 55
Query: 72 EVEIEKFNGFFVEKE---EEYVIKWKELQDKVAWAKSSDIELMTVGREIVDFHGEMVL-L 127
+ +IEK F +E++ + K +E D + D+ + RE G+ +L L
Sbjct: 56 DHQIEKIALFMLEQQGLLSSRLQKLREWHDTL--QDEPDLSQIAKLREAYRAVGQDLLKL 113
Query: 128 ENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKI 169
+ +N G+ KI+KK+DKR G +++ + P+ ++
Sbjct: 114 LFFIDMNAIGIRKILKKFDKRFGYRFTNYYVKTRADHPYSQL 155
>At1g63010 tetracycline resistance efflux protein like protein
Length = 699
Score = 47.4 bits (111), Expect = 8e-06
Identities = 42/166 (25%), Positives = 78/166 (46%), Gaps = 20/166 (12%)
Query: 15 LPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEVTKEVKDFLRLLEVE 74
+ +W +++YK +KK++K ++I R + +KDF R+L+ +
Sbjct: 13 IEEWSGYYINYKLMKKKVKQYA-EQIQGGSQHPRHV-------------LKDFSRMLDTQ 58
Query: 75 IEKFNGFFVEKE---EEYVIKWKELQDKVAWAKSSDIELMTVGREIV-DFHGEMVLLENY 130
IE F +E++ + K +E D + + DI + RE D +++ L +
Sbjct: 59 IETTVLFMLEQQGLLSGRLAKLRESHDAIL--EQPDISRIFELREAYRDVGRDLLQLLKF 116
Query: 131 SALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLV 176
LN GL KI+KK+DKR G +++ N P+ ++ + K V
Sbjct: 117 VELNAIGLRKILKKFDKRFGYRFADYYVKTRANHPYSQLQQVFKHV 162
>At3g23430 PHO1 protein (PHO1)
Length = 782
Score = 43.5 bits (101), Expect = 1e-04
Identities = 33/130 (25%), Positives = 55/130 (41%), Gaps = 17/130 (13%)
Query: 51 DDDGGAEGEVTKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSDIEL 110
D GAEG + + + + +L E+ V WK +Q +S+ +EL
Sbjct: 243 DAVAGAEGGIARSIATAMSVLWEEL-------VNNPRSDFTNWKNIQSAEKKIRSAFVEL 295
Query: 111 MTVGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKID 170
+ + LL+ YS+LN KI+KK+DK G +++ V F D
Sbjct: 296 ----------YRGLGLLKTYSSLNMIAFTKIMKKFDKVAGQNASSTYLKVVKRSQFISSD 345
Query: 171 VLNKLVKECE 180
+ +L+ E E
Sbjct: 346 KVVRLMDEVE 355
Score = 30.4 bits (67), Expect = 1.1
Identities = 15/35 (42%), Positives = 24/35 (67%), Gaps = 3/35 (8%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLI 35
+KF K L Q+ +P+W++ F++Y LKKQ+K I
Sbjct: 2 VKFSKELEAQL---IPEWKEAFVNYCLLKKQIKKI 33
>At1g26730 unknown protein
Length = 750
Score = 39.3 bits (90), Expect = 0.002
Identities = 21/63 (33%), Positives = 34/63 (53%)
Query: 118 VDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVK 177
++F+ ++ L+NYS LN + KI+KKYDK P+++ V D +NKL+
Sbjct: 257 IEFYRKLRHLKNYSFLNTLAISKIMKKYDKIASRSAAKPYMEMVDKSYLTSSDEINKLML 316
Query: 178 ECE 180
E
Sbjct: 317 RVE 319
>At1g14040 unknown protein
Length = 798
Score = 38.9 bits (89), Expect = 0.003
Identities = 21/66 (31%), Positives = 37/66 (55%)
Query: 115 REIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNK 174
R ++F+ ++ LL++YS LN KI+KKYDK T P+++ V + D + +
Sbjct: 315 RAFIEFYQKLRLLKSYSFLNVLAFSKILKKYDKITSRDATKPYMKVVDSSYLGSSDEVMR 374
Query: 175 LVKECE 180
L++ E
Sbjct: 375 LMERVE 380
>At1g68740 putative protein (At1g68740)
Length = 784
Score = 35.8 bits (81), Expect = 0.025
Identities = 30/133 (22%), Positives = 50/133 (37%), Gaps = 46/133 (34%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLI---------------VPKEIDSSCS 45
+KF K Q+ +P+W+D F+ Y LKK LK I + K + SS
Sbjct: 2 VKFTKQFEGQL---VPEWKDAFVDYSQLKKDLKKIHLFTNGVEKKHTETSLIKTVKSSLG 58
Query: 46 KRRRLDDDGGAEGEV----------------------------TKEVKDFLRLLEVEIEK 77
+ + G + V T K+F L++++ K
Sbjct: 59 RLSIFGNKGREQSRVIQVHKKLASSGSNNDVYETELLEKIADDTDAAKEFFACLDMQLNK 118
Query: 78 FNGFFVEKEEEYV 90
N F+ KE+E++
Sbjct: 119 VNQFYKTKEKEFL 131
Score = 31.6 bits (70), Expect = 0.48
Identities = 23/71 (32%), Positives = 34/71 (47%), Gaps = 2/71 (2%)
Query: 99 KVAWAKSSDIELMTVGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFI 158
+++ K S E M G F G + L+ Y LN + I+KK+DK TG + LP
Sbjct: 276 QISKKKLSHAEKMIKGALTELFKG-LNYLKTYRNLNILAFMNILKKFDKVTGKQI-LPIY 333
Query: 159 QDVLNQPFFKI 169
V+ +F I
Sbjct: 334 LKVVESSYFNI 344
>At2g03260 unknown protein
Length = 783
Score = 33.9 bits (76), Expect = 0.097
Identities = 15/30 (50%), Positives = 22/30 (73%)
Query: 118 VDFHGEMVLLENYSALNYTGLVKIIKKYDK 147
V+F+ ++ LL++YS LN KI+KKYDK
Sbjct: 307 VEFYQKLRLLKSYSFLNVLAFSKILKKYDK 336
>At4g25350 putative protein
Length = 710
Score = 33.1 bits (74), Expect = 0.16
Identities = 14/30 (46%), Positives = 23/30 (76%)
Query: 118 VDFHGEMVLLENYSALNYTGLVKIIKKYDK 147
++F+ ++ L+NYS LN + + KI+KKYDK
Sbjct: 249 IEFYQKLRHLKNYSFLNASAVSKIMKKYDK 278
>At1g35350 hypothetical protein
Length = 747
Score = 32.7 bits (73), Expect = 0.22
Identities = 14/30 (46%), Positives = 22/30 (72%)
Query: 118 VDFHGEMVLLENYSALNYTGLVKIIKKYDK 147
++F+ ++ L+NYS LN + KI+KKYDK
Sbjct: 257 IEFYRKLRHLKNYSFLNTLAISKIMKKYDK 286
Score = 27.3 bits (59), Expect = 9.0
Identities = 30/124 (24%), Positives = 54/124 (43%), Gaps = 12/124 (9%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLI--VPKEIDSSCSKRRRLDDDGGAEG 58
MKF K + Q +P+W+ ++ Y LK L+ I K +S +R+L G
Sbjct: 1 MKFGK---EYVAQMIPEWQQAYMDYTCLKTILREIKTSQKRSESQGVLKRKLSGRRNFSG 57
Query: 59 EVTKEVKDF----LRLLEVEIEKFNG-FFVEKEEEYVIKWKEL--QDKVAWAKSSDIELM 111
+ + F L ++ + G EK E ++K E+ + ++ + K+ D+E
Sbjct: 58 LTKRYSRTFSSRDLENHDIMVHATTGDDGFEKYETTILKVSEVGRESELVFFKTLDLEFD 117
Query: 112 TVGR 115
V R
Sbjct: 118 KVNR 121
>At3g13070 hemolysin-like protein
Length = 661
Score = 32.3 bits (72), Expect = 0.28
Identities = 49/216 (22%), Positives = 96/216 (43%), Gaps = 27/216 (12%)
Query: 54 GGAEGEVTK-EVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQD---KVAWAKSSDIE 109
G +E VT+ E+K LR E+ +G E+E++ + E++D + D+
Sbjct: 312 GRSEPYVTEDELKLMLRGAEL-----SGAIEEEEQDMIENVLEIKDTHVREVMTPLVDVV 366
Query: 110 LMTVGREIVDFHGEMVLLENYSAL--------NYTGLVKIIKKYDK-RTGALLRLPFIQD 160
+ +VDFH M + YS + N G+ + D + G LL + D
Sbjct: 367 AIDASASLVDFHS-MWVTHQYSRVPVFEQRIDNIVGIAYAMDLLDYVQKGDLLESTSVGD 425
Query: 161 VLNQP-FFKIDVLN--KLVKEC---EVMLSIIFPKSGPLGQSLSTSEVFEEVARETTTAN 214
+ ++P +F D ++ L++E +V ++++ + G ++ +V EE+ E N
Sbjct: 426 MAHKPAYFVPDSMSVWNLLREFRIRKVHMAVVLNEYGGTIGIVTLEDVVEEIVGEIFDEN 485
Query: 215 ETKETLDHVPKEFSEIQNMENIFIKLTTSALDTLKE 250
++KE + K + E I+ +++D L E
Sbjct: 486 DSKEEIQ--KKTGYIVMRDEGIYDVDANTSIDQLSE 519
>At1g69480 hypothetical protein
Length = 777
Score = 32.3 bits (72), Expect = 0.28
Identities = 18/63 (28%), Positives = 33/63 (51%)
Query: 119 DFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVKE 178
+F+ ++ L+ YS +N KI+KKY+K +++ V N D +N+L++
Sbjct: 282 EFYQKLRRLKEYSFMNLLAFSKIMKKYEKIASRNASRNYMKIVDNSLIGSSDEVNRLLER 341
Query: 179 CEV 181
EV
Sbjct: 342 VEV 344
Score = 28.9 bits (63), Expect = 3.1
Identities = 14/35 (40%), Positives = 21/35 (60%), Gaps = 3/35 (8%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLI 35
MKF KI Q+ +P+W + ++ Y LK+ LK I
Sbjct: 1 MKFGKIFKKQM---VPEWVEAYVDYNGLKRVLKEI 32
>At3g29060 unknown protein
Length = 794
Score = 31.6 bits (70), Expect = 0.48
Identities = 19/68 (27%), Positives = 32/68 (46%), Gaps = 6/68 (8%)
Query: 113 VGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDVL 172
+ R V+F+ ++ S LN KI+KKYDK T P++ V + D +
Sbjct: 300 MNRAFVEFYQKL------SFLNQLAFAKILKKYDKTTSRNASKPYLNTVDHSYLGSCDEV 353
Query: 173 NKLVKECE 180
++L+ E
Sbjct: 354 SRLMSRVE 361
>At3g14490 terpene synthase-related protein, putative
Length = 601
Score = 30.8 bits (68), Expect = 0.82
Identities = 38/175 (21%), Positives = 73/175 (41%), Gaps = 31/175 (17%)
Query: 94 KELQDKVAWAKSSDIELMTVGREIVDF------HGEMVLLE-----NYSALNYTGLVKII 142
K +Q+ + A+ ++E++ V RE + F H E +L NY L+Y +K +
Sbjct: 242 KHIQNALYRARYHNLEIL-VAREYISFYEQEEDHDETLLKFAKLNFNYCQLHYIQELKDL 300
Query: 143 KKYDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVKECEVMLSIIFPKSGPLGQSLSTS-- 200
K+ K +LP+I+D + + +F L++ F LG+ + T
Sbjct: 301 TKWWKELDLASKLPYIRDRIVEVYFG-------------ALALYFEPRYSLGRIIVTKIT 347
Query: 201 ---EVFEEVARETTTANETKETLDHVPK-EFSEIQNMENIFIKLTTSALDTLKEI 251
VF + T E +D + + +I+ + + + +TL+EI
Sbjct: 348 MIVTVFNDTCDAYGTLPEVTSLVDSFQRWDLGDIEKLPSYVKIVFRGVFETLEEI 402
>At1g55930 unknown protein
Length = 846
Score = 30.8 bits (68), Expect = 0.82
Identities = 50/216 (23%), Positives = 94/216 (43%), Gaps = 27/216 (12%)
Query: 54 GGAEGEVTK-EVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQD---KVAWAKSSDIE 109
G +E VT+ E+K LR E+ +G E+E++ + E++D + D+
Sbjct: 500 GRSEPYVTEDELKLMLRGAEL-----SGAIEEEEQDMIENVLEIKDTHVREVMTPLVDVV 554
Query: 110 LMTVGREIVDFHGEMVLLENYSAL--------NYTGLVKIIKKYDK-RTGALLRLPFIQD 160
+ +VDFH V YS + N G+ + D G LL + D
Sbjct: 555 AIDGSGSLVDFHNFWVT-HQYSRVPVFEQRIDNIVGIAYAMDLLDYVPKGKLLESTTVVD 613
Query: 161 VLNQP-FFKIDVLN--KLVKEC---EVMLSIIFPKSGPLGQSLSTSEVFEEVARETTTAN 214
+ ++P FF D ++ L++E +V ++++ + G ++ +V EE+ E N
Sbjct: 614 MAHKPAFFVPDSMSVWNLLREFRIRKVHMAVVLNEYGGTIGIVTLEDVVEEIVGEIFDEN 673
Query: 215 ETKETLDHVPKEFSEIQNMENIFIKLTTSALDTLKE 250
++KE + K + E I+ +++D L E
Sbjct: 674 DSKEEIQ--KKTGYIVMRAEGIYDVDANTSIDQLSE 707
>At5g59790 unknown protein
Length = 423
Score = 30.4 bits (67), Expect = 1.1
Identities = 18/49 (36%), Positives = 28/49 (56%), Gaps = 2/49 (4%)
Query: 42 SSCSKRRRLDDDGGAEGEVTKEVKDFLRLLEVEIEKFNGFFVEKEEEYV 90
S C+ R +D+G E V KE+K F R+ + E F+G +E ++E V
Sbjct: 327 SGCTITRGAEDNG--EERVDKELKSFGRVQLEDKEYFSGSLIETKKELV 373
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.136 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,036,778
Number of Sequences: 26719
Number of extensions: 255762
Number of successful extensions: 1006
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 951
Number of HSP's gapped (non-prelim): 69
length of query: 274
length of database: 11,318,596
effective HSP length: 98
effective length of query: 176
effective length of database: 8,700,134
effective search space: 1531223584
effective search space used: 1531223584
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC147471.4


