

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147471.11 - phase: 0 /pseudo
(1453 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
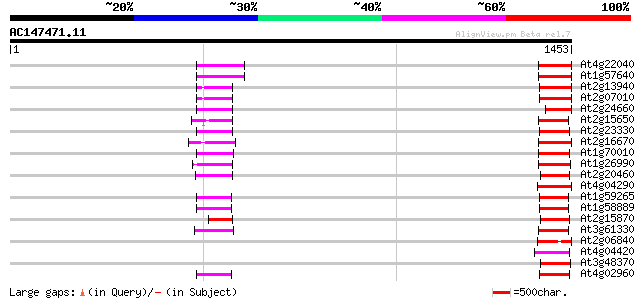
Score E
Sequences producing significant alignments: (bits) Value
At4g22040 LTR retrotransposon like protein 88 3e-17
At1g57640 88 3e-17
At2g13940 putative retroelement pol polyprotein 86 2e-16
At2g07010 putative retroelement pol polyprotein 84 5e-16
At2g24660 putative retroelement pol polyprotein 80 9e-15
At2g15650 putative retroelement pol polyprotein 80 1e-14
At2g23330 putative retroelement pol polyprotein 78 4e-14
At2g16670 putative retroelement pol polyprotein 77 7e-14
At1g70010 hypothetical protein 76 2e-13
At1g26990 polyprotein, putative 76 2e-13
At2g20460 putative retroelement pol polyprotein 75 3e-13
At4g04290 putative reverse transcriptase 74 6e-13
At1g59265 polyprotein, putative 72 2e-12
At1g58889 polyprotein, putative 72 2e-12
At2g15870 putative retroelement pol polyprotein 71 5e-12
At3g61330 copia-type polyprotein 70 7e-12
At2g06840 putative retroelement pol polyprotein 70 7e-12
At4g04420 putative transposon protein 70 9e-12
At3g48370 putative protein 70 9e-12
At4g02960 putative polyprotein of LTR transposon 70 1e-11
>At4g22040 LTR retrotransposon like protein
Length = 1109
Score = 88.2 bits (217), Expect = 3e-17
Identities = 39/83 (46%), Positives = 59/83 (70%)
Query: 1371 EPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLT 1430
+ MR++ D+K AI ++ NPVQH+RTKH+EV+ HFI++ + G+I T +V S LAD+LT
Sbjct: 1026 QAMRIFSDSKSAIALSVNPVQHERTKHVEVDCHFIRDAILDGIIATSFVPSHKQLADILT 1085
Query: 1431 KRLNNNNFEKFVSKLGMIDIHSP 1453
K L F+ KLG++D+H+P
Sbjct: 1086 KALGEKEVRYFLRKLGILDVHAP 1108
Score = 72.0 bits (175), Expect = 2e-12
Identities = 43/126 (34%), Positives = 67/126 (53%), Gaps = 2/126 (1%)
Query: 483 LYHYRVGDPSFRVIKQL-FPLFFKTLDVESFQCELCELAKHKRVTFSVSNKMSTFPFYLV 541
L+H R+G S +++ L L ++ C+ C AK R TF + + S F L+
Sbjct: 291 LWHRRLGHASDKIVNLLPRELLSSGKEILENVCDTCMRAKQTRDTFPLRDNRSMDSFQLI 350
Query: 542 HTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFLHD*KSV*CEH*TD*IR*CQRLF*SQ 601
H +VWGP P+ SGAR+F+T +DD + WVY + D KS +H D + +R F ++
Sbjct: 351 HCDVWGPYRTPSYSGARYFLTIVDDYSRGVWVYLMTD-KSETQKHLKDFMALVERQFDTE 409
Query: 602 FKFILS 607
K + S
Sbjct: 410 IKTVRS 415
>At1g57640
Length = 1444
Score = 88.2 bits (217), Expect = 3e-17
Identities = 39/83 (46%), Positives = 59/83 (70%)
Query: 1371 EPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLT 1430
+ MR++ D+K AI ++ NPVQH+RTKH+EV+ HFI++ + G+I T +V S LAD+LT
Sbjct: 1361 QAMRIFSDSKSAIALSVNPVQHERTKHVEVDCHFIRDAILDGIIATSFVPSHKQLADILT 1420
Query: 1431 KRLNNNNFEKFVSKLGMIDIHSP 1453
K L F+ KLG++D+H+P
Sbjct: 1421 KALGEKEVRYFLRKLGILDVHAP 1443
Score = 75.9 bits (185), Expect = 2e-13
Identities = 45/126 (35%), Positives = 68/126 (53%), Gaps = 2/126 (1%)
Query: 483 LYHYRVGDPSFRVIKQL-FPLFFKTLDVESFQCELCELAKHKRVTFSVSNKMSTFPFYLV 541
L+H R+G S +++ L L ++ C+ C AK R TF +S+ S F L+
Sbjct: 491 LWHRRLGHASDKIVNLLPRELLSSGKEILENVCDTCMRAKQTRDTFPLSDNRSMDSFQLI 550
Query: 542 HTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFLHD*KSV*CEH*TD*IR*CQRLF*SQ 601
H +VWGP P+ SGAR+F+T +DD + WVY + D KS +H D I +R F ++
Sbjct: 551 HCDVWGPYRAPSYSGARYFLTIVDDYSRGVWVYLMTD-KSETQKHLKDFIALVERQFDTE 609
Query: 602 FKFILS 607
K + S
Sbjct: 610 IKIVRS 615
>At2g13940 putative retroelement pol polyprotein
Length = 1501
Score = 85.9 bits (211), Expect = 2e-16
Identities = 39/82 (47%), Positives = 58/82 (70%)
Query: 1372 PMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTK 1431
P RLYCD+K AI+IA NPV H+RTKH+E + H +++ + G+I T +V + + LAD+ TK
Sbjct: 1419 PARLYCDSKAAIHIAANPVFHERTKHIESDCHSVRDAVRDGIITTQHVRTTEQLADVFTK 1478
Query: 1432 RLNNNNFEKFVSKLGMIDIHSP 1453
L N F +SKLG+ ++H+P
Sbjct: 1479 ALGRNQFLYLMSKLGVQNLHTP 1500
Score = 75.5 bits (184), Expect = 2e-13
Identities = 41/95 (43%), Positives = 53/95 (55%), Gaps = 2/95 (2%)
Query: 483 LYHYRVGDPSFRVIKQLFPLFFKTLD-VESFQCELCELAKHKRVTFSVSNKMSTFPFYLV 541
L+H R+G PSF V+ L PLF KT V S C++C AK R F S + F L+
Sbjct: 529 LWHQRLGHPSFSVLSSL-PLFSKTSSTVTSHSCDVCFRAKQTREVFPESINKTEECFSLI 587
Query: 542 HTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFL 576
H +VWGP VP GA +F+T +DD + W Y L
Sbjct: 588 HCDVWGPYRVPASCGAVYFLTIVDDYSRAVWTYLL 622
>At2g07010 putative retroelement pol polyprotein
Length = 1413
Score = 84.3 bits (207), Expect = 5e-16
Identities = 38/82 (46%), Positives = 58/82 (70%)
Query: 1372 PMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTK 1431
P RL+CD+K AI+IA NPV H+RTKH+E + H +++ + G+I T +V + + LAD+ TK
Sbjct: 1331 PTRLFCDSKAAISIAANPVFHERTKHIERDCHSVRDAVRDGIITTHHVRTSEQLADIFTK 1390
Query: 1432 RLNNNNFEKFVSKLGMIDIHSP 1453
L N F +SKLG+ ++H+P
Sbjct: 1391 ALGRNQFIYLMSKLGIQNLHTP 1412
Score = 77.0 bits (188), Expect = 7e-14
Identities = 41/95 (43%), Positives = 55/95 (57%), Gaps = 2/95 (2%)
Query: 483 LYHYRVGDPSFRVIKQLFPLFF-KTLDVESFQCELCELAKHKRVTFSVSNKMSTFPFYLV 541
L+H R+G PSF V+ L PLF + V S C++C AK R F S+ ST F L+
Sbjct: 512 LWHQRLGHPSFSVLSSL-PLFSGSSCSVSSRSCDVCFRAKQTREVFPDSSNKSTDCFSLI 570
Query: 542 HTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFL 576
H +VWGP VP+ GA +F+T +DD + W Y L
Sbjct: 571 HCDVWGPYRVPSSCGAVYFLTIVDDFSRSVWTYLL 605
>At2g24660 putative retroelement pol polyprotein
Length = 1156
Score = 80.1 bits (196), Expect = 9e-15
Identities = 40/94 (42%), Positives = 53/94 (55%)
Query: 483 LYHYRVGDPSFRVIKQLFPLFFKTLDVESFQCELCELAKHKRVTFSVSNKMSTFPFYLVH 542
L+H R+G PSF V+ L L +L V S C++C AK R F VS S F L+H
Sbjct: 90 LWHQRLGHPSFSVLSSLPVLTSSSLSVGSRSCDVCFRAKQTREVFPVSTNKSIECFSLIH 149
Query: 543 TNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFL 576
+VWGP VP+ GA +F+T +DD + W Y L
Sbjct: 150 CDVWGPYRVPSSCGAVYFLTIVDDFSRAVWTYLL 183
Score = 64.7 bits (156), Expect = 4e-10
Identities = 28/66 (42%), Positives = 45/66 (67%)
Query: 1388 NPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTKRLNNNNFEKFVSKLGM 1447
NPV H+RTKH+E + H +++ + G+I T +V + + LAD+ TK L + F +SKLG+
Sbjct: 1061 NPVFHERTKHIESDCHSVRDAVRDGIITTHHVRTNEQLADIFTKALGRSQFLYLMSKLGI 1120
Query: 1448 IDIHSP 1453
D+H+P
Sbjct: 1121 QDLHTP 1126
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 79.7 bits (195), Expect = 1e-14
Identities = 39/79 (49%), Positives = 50/79 (62%)
Query: 1369 FDEPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADL 1428
F E + + CDNK AI I NPVQH RTKH+E+ HF++E GLI Y +D LAD+
Sbjct: 1264 FKEGIPILCDNKSAIAIGRNPVQHRRTKHIEIKYHFVREAEHKGLIQLEYCKGEDQLADV 1323
Query: 1429 LTKRLNNNNFEKFVSKLGM 1447
LTK L+ + FE KLG+
Sbjct: 1324 LTKALSVSRFEGLRRKLGV 1342
Score = 57.8 bits (138), Expect = 5e-08
Identities = 36/114 (31%), Positives = 58/114 (50%), Gaps = 13/114 (11%)
Query: 472 ESIKTNREKVLLYHYRVGDPSFRVIKQL--------FPLFFKTLDVESFQCELCELAKHK 523
E++ N + +H R+G S + ++Q+ P F T + C+ C L K
Sbjct: 444 EAMTANVQTEETWHKRLGHVSNKRLQQMQDKELVNGLPRFKVTKET----CKACNLGKQS 499
Query: 524 RVTFSVSNKMSTFP-FYLVHTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFL 576
R +F ++ T +VHT+V GP +I G+R++V F+DD T + WVYFL
Sbjct: 500 RKSFPKESQTKTREKLEIVHTDVCGPMQHQSIDGSRYYVLFLDDYTHMCWVYFL 553
>At2g23330 putative retroelement pol polyprotein
Length = 1496
Score = 77.8 bits (190), Expect = 4e-14
Identities = 36/78 (46%), Positives = 56/78 (71%)
Query: 1372 PMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTK 1431
PM+L+CD++ AI+IA NPV H+RTKH+E + H +++ + LI T ++ ++D +ADLLTK
Sbjct: 1414 PMKLHCDSEAAIHIAANPVFHERTKHIESDCHKVRDAVLDKLITTEHIYTEDQVADLLTK 1473
Query: 1432 RLNNNNFEKFVSKLGMID 1449
L FE+ +S LG+ D
Sbjct: 1474 SLPRPTFERLLSTLGVTD 1491
Score = 62.0 bits (149), Expect = 2e-09
Identities = 31/95 (32%), Positives = 49/95 (50%), Gaps = 1/95 (1%)
Query: 483 LYHYRVGDPSFRVIKQLFPLFFKTLDVESFQ-CELCELAKHKRVTFSVSNKMSTFPFYLV 541
L+H R+G PS V+ L + + C+ C +K R F +SN + F L+
Sbjct: 501 LWHRRLGHPSTSVLLSLPECNRSSQGFDKIDSCDTCFRSKQTREVFPISNNKTMECFSLI 560
Query: 542 HTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFL 576
H +VWGP P+ +GA +F+T +DD + W Y +
Sbjct: 561 HGDVWGPYRTPSTTGAVYFLTLVDDYSRSVWTYLM 595
>At2g16670 putative retroelement pol polyprotein
Length = 1333
Score = 77.0 bits (188), Expect = 7e-14
Identities = 35/83 (42%), Positives = 56/83 (67%)
Query: 1371 EPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLT 1430
+PM + CD+K AI IA NPV H+RTKH+E++ HF++++ G+I +V + LAD+ T
Sbjct: 1250 QPMIMCCDSKSAIYIATNPVFHERTKHIEIDYHFVRDEFVKGVITPRHVGTTSQLADIFT 1309
Query: 1431 KRLNNNNFEKFVSKLGMIDIHSP 1453
K L + F F KLG+ ++++P
Sbjct: 1310 KPLGRDCFSAFRIKLGIRNLYAP 1332
Score = 66.2 bits (160), Expect = 1e-10
Identities = 41/126 (32%), Positives = 66/126 (51%), Gaps = 8/126 (6%)
Query: 464 YLATTFFLESIKTNREKVL-LYHYRVGDPSFRVIKQLFPLFFKTLDVESFQ----CELCE 518
+ +T S+ EK L+H R+G P+ RV+ L P ++ V S C++C
Sbjct: 371 HFRSTEIAASVTVKEEKNYELWHSRMGHPAARVVS-LIPE--SSVSVSSTHLNKACDVCH 427
Query: 519 LAKHKRVTFSVSNKMSTFPFYLVHTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFLHD 578
AK R +F +S + F L++ ++WGP P+ +GAR+F+T IDD + W+Y L+D
Sbjct: 428 RAKQTRNSFPLSINKTLRIFELIYCDLWGPYRTPSHTGARYFLTIIDDYSRGVWLYLLND 487
Query: 579 *KSV*C 584
C
Sbjct: 488 KSEAPC 493
>At1g70010 hypothetical protein
Length = 1315
Score = 75.9 bits (185), Expect = 2e-13
Identities = 36/98 (36%), Positives = 56/98 (56%), Gaps = 2/98 (2%)
Query: 483 LYHYRVGDPSFRVIKQLFPL--FFKTLDVESFQCELCELAKHKRVTFSVSNKMSTFPFYL 540
L+H R+G PS + ++ + L F K + F C +C ++K K + F N S+ PF L
Sbjct: 406 LWHKRLGHPSVQKLQPMSSLLSFPKQKNNTDFHCRVCHISKQKHLPFVSHNNKSSRPFDL 465
Query: 541 VHTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFLHD 578
+H + WGP +V G R+F+T +DD + TWVY L +
Sbjct: 466 IHIDTWGPFSVQTHDGYRYFLTIVDDYSRATWVYLLRN 503
Score = 70.9 bits (172), Expect = 5e-12
Identities = 32/80 (40%), Positives = 57/80 (71%)
Query: 1371 EPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLT 1430
+P L+CDN+ AI+IA+N V H+RTKH+E + H ++E+L GL +++++ +AD T
Sbjct: 1232 KPTLLFCDNEAAIHIANNHVFHERTKHIESDCHSVRERLLKGLFELYHINTELQIADPFT 1291
Query: 1431 KRLNNNNFEKFVSKLGMIDI 1450
K L ++F + +SK+G+++I
Sbjct: 1292 KPLYPSHFHRLISKMGLLNI 1311
>At1g26990 polyprotein, putative
Length = 1436
Score = 75.9 bits (185), Expect = 2e-13
Identities = 36/83 (43%), Positives = 58/83 (69%)
Query: 1369 FDEPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADL 1428
F P LYCDN+ A++IA+N V H+RTKH+E + H ++E +++G++ T +V + + LAD
Sbjct: 1350 FTHPAYLYCDNEAALHIANNSVFHERTKHIENDCHKVRECIEAGILKTIFVRTDNQLADT 1409
Query: 1429 LTKRLNNNNFEKFVSKLGMIDIH 1451
LTK L F + SKLG+++I+
Sbjct: 1410 LTKPLYPKPFRENNSKLGLLNIY 1432
Score = 75.1 bits (183), Expect = 3e-13
Identities = 37/107 (34%), Positives = 58/107 (53%), Gaps = 6/107 (5%)
Query: 473 SIKTNREKVLLYHYRVGDPSFRVIKQLFPLFF---KTLDVESFQCELCELAKHKRVTFSV 529
S+K E ++H R+G PSF I L + + ++ +S C +C L+K K + F
Sbjct: 550 SVKNESE---MWHKRLGHPSFAKIDTLSDVLMLPKQKINKDSSHCHVCHLSKQKHLPFKS 606
Query: 530 SNKMSTFPFYLVHTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFL 576
N + F LVH + WGP +VP + R+F+T +DD + TW+Y L
Sbjct: 607 VNHIREKAFELVHIDTWGPFSVPTVDSYRYFLTIVDDFSRATWIYLL 653
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 75.1 bits (183), Expect = 3e-13
Identities = 35/98 (35%), Positives = 58/98 (58%), Gaps = 2/98 (2%)
Query: 481 VLLYHYRVGDPSFRVIKQLFPLFFKTL--DVESFQCELCELAKHKRVTFSVSNKMSTFPF 538
V ++H R+G PSF + L + T + +S C +C LAK K+++F +N + F
Sbjct: 566 VSVWHKRLGHPSFSRLDSLSEVLGTTRHKNKKSAYCHVCHLAKQKKLSFPSANNICNSTF 625
Query: 539 YLVHTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFL 576
L+H +VWGP +V + G ++F+T +DD + TW+Y L
Sbjct: 626 ELLHIDVWGPFSVETVEGYKYFLTIVDDHSRATWIYLL 663
Score = 68.6 bits (166), Expect = 3e-11
Identities = 33/76 (43%), Positives = 52/76 (68%)
Query: 1375 LYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTKRLN 1434
LY D+ A+ IA NPV H+RTKH+E++ H ++EKLD+G + +V ++D +AD+LTK L
Sbjct: 1381 LYSDSTAAVYIATNPVFHERTKHIEIDCHTVREKLDNGQLKLLHVKTKDQVADILTKPLF 1440
Query: 1435 NNNFEKFVSKLGMIDI 1450
F +SK+ + +I
Sbjct: 1441 PYQFAHLLSKMSIQNI 1456
>At4g04290 putative reverse transcriptase
Length = 403
Score = 73.9 bits (180), Expect = 6e-13
Identities = 34/86 (39%), Positives = 55/86 (63%)
Query: 1368 DFDEPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLAD 1427
D P+ ++CDN+ AI+IA N V H+RTKH+EV+ H ++E++ G+I Y S++ L D
Sbjct: 318 DTPNPIPMHCDNQAAIHIASNSVFHERTKHIEVDCHKVREQVQLGVILPHYTESEEQLVD 377
Query: 1428 LLTKRLNNNNFEKFVSKLGMIDIHSP 1453
+ TK + E +KLG++D+ P
Sbjct: 378 VFTKGASTKVCEYIHNKLGVVDLTRP 403
>At1g59265 polyprotein, putative
Length = 1466
Score = 72.0 bits (175), Expect = 2e-12
Identities = 33/76 (43%), Positives = 48/76 (62%)
Query: 1372 PMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTK 1431
P +YCDN A + NPV H R KH+ ++ HFI+ ++ SG + +VS+ D LAD LTK
Sbjct: 1385 PPVIYCDNVGATYLCANPVFHSRMKHIAIDYHFIRNQVQSGALRVVHVSTHDQLADTLTK 1444
Query: 1432 RLNNNNFEKFVSKLGM 1447
L+ F+ F SK+G+
Sbjct: 1445 PLSRTAFQNFASKIGV 1460
Score = 44.3 bits (103), Expect = 5e-04
Identities = 26/94 (27%), Positives = 46/94 (48%), Gaps = 4/94 (4%)
Query: 484 YHYRVGDPSFRVIKQL---FPLFFKTLDVESFQCELCELAKHKRVTFSVSNKMSTFPFYL 540
+H R+G P+ ++ + + L + C C + K +V FS S ST P
Sbjct: 467 WHARLGHPAPSILNSVISNYSLSVLNPSHKFLSCSDCLINKSNKVPFSQSTINSTRPLEY 526
Query: 541 VHTNVWGPSNVPNISGARWFVTFIDDCTWVTWVY 574
++++VW S + + R++V F+D T TW+Y
Sbjct: 527 IYSDVWS-SPILSHDNYRYYVIFVDHFTRYTWLY 559
>At1g58889 polyprotein, putative
Length = 1466
Score = 72.0 bits (175), Expect = 2e-12
Identities = 33/76 (43%), Positives = 48/76 (62%)
Query: 1372 PMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTK 1431
P +YCDN A + NPV H R KH+ ++ HFI+ ++ SG + +VS+ D LAD LTK
Sbjct: 1385 PPVIYCDNVGATYLCANPVFHSRMKHIAIDYHFIRNQVQSGALRVVHVSTHDQLADTLTK 1444
Query: 1432 RLNNNNFEKFVSKLGM 1447
L+ F+ F SK+G+
Sbjct: 1445 PLSRTAFQNFASKIGV 1460
Score = 44.3 bits (103), Expect = 5e-04
Identities = 26/94 (27%), Positives = 46/94 (48%), Gaps = 4/94 (4%)
Query: 484 YHYRVGDPSFRVIKQL---FPLFFKTLDVESFQCELCELAKHKRVTFSVSNKMSTFPFYL 540
+H R+G P+ ++ + + L + C C + K +V FS S ST P
Sbjct: 467 WHARLGHPAPSILNSVISNYSLSVLNPSHKFLSCSDCLINKSNKVPFSQSTINSTRPLEY 526
Query: 541 VHTNVWGPSNVPNISGARWFVTFIDDCTWVTWVY 574
++++VW S + + R++V F+D T TW+Y
Sbjct: 527 IYSDVWS-SPILSHDNYRYYVIFVDHFTRYTWLY 559
>At2g15870 putative retroelement pol polyprotein
Length = 1264
Score = 70.9 bits (172), Expect = 5e-12
Identities = 33/76 (43%), Positives = 53/76 (69%)
Query: 1375 LYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTKRLN 1434
L+ D+ AI IA NPV H+RTKH+E++ H ++EK+D G + +V ++D +AD++TK L
Sbjct: 1184 LFSDSTAAIYIAINPVFHERTKHIEIDCHTVREKIDDGELKLLHVRTEDQVADIMTKPLF 1243
Query: 1435 NNNFEKFVSKLGMIDI 1450
N FE SK+ +++I
Sbjct: 1244 PNQFEHLKSKMSILNI 1259
Score = 64.3 bits (155), Expect = 5e-10
Identities = 23/63 (36%), Positives = 41/63 (64%)
Query: 514 CELCELAKHKRVTFSVSNKMSTFPFYLVHTNVWGPSNVPNISGARWFVTFIDDCTWVTWV 573
C +C LAKH++++F N + F ++H ++WGP +V + G ++F+T +DD + TW+
Sbjct: 449 CHVCHLAKHRKLSFPSQNNVCNEIFEMLHIDIWGPFSVETVDGYQYFLTIVDDHSRATWI 508
Query: 574 YFL 576
Y L
Sbjct: 509 YLL 511
>At3g61330 copia-type polyprotein
Length = 1352
Score = 70.5 bits (171), Expect = 7e-12
Identities = 33/78 (42%), Positives = 48/78 (61%)
Query: 1370 DEPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLL 1429
+EP +++ DNK AI +A NPV HDR+KH++ H+I+E + + YV + D +AD
Sbjct: 1264 EEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYIRECVSKKDVQLEYVKTHDQVADFF 1323
Query: 1430 TKRLNNNNFEKFVSKLGM 1447
TK L NF K S LG+
Sbjct: 1324 TKPLKRENFIKMRSLLGV 1341
Score = 50.8 bits (120), Expect = 6e-06
Identities = 32/106 (30%), Positives = 55/106 (51%), Gaps = 5/106 (4%)
Query: 478 REKVLLYHYRVGDPSFRVIKQL----FPLFFKTLDVESFQCELCELAKHKRVTF-SVSNK 532
+E+ L+H R G +F ++ L ++ + CE C L K +++F S+
Sbjct: 462 KEESWLWHLRFGHLNFGGLELLSRKEMVRGLPCINHPNQVCEGCLLGKQFKMSFPKESSS 521
Query: 533 MSTFPFYLVHTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFLHD 578
+ P L+HT+V GP ++ + +F+ FIDD + TWVYFL +
Sbjct: 522 RAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKE 567
>At2g06840 putative retroelement pol polyprotein
Length = 1102
Score = 70.5 bits (171), Expect = 7e-12
Identities = 35/86 (40%), Positives = 55/86 (63%), Gaps = 4/86 (4%)
Query: 1368 DFDEPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLAD 1427
D +P+ L+CD++ AI+IA NPV H+RTKH+E + H +++ + +I TP++S+ D
Sbjct: 1020 DHTQPILLFCDSQAAIHIAANPVFHERTKHIEKDCHQVRDAVTDKVISTPHIST----TD 1075
Query: 1428 LLTKRLNNNNFEKFVSKLGMIDIHSP 1453
LLTK L FE+ +S LG + P
Sbjct: 1076 LLTKALPRPTFERLLSTLGTCNYDLP 1101
>At4g04420 putative transposon protein
Length = 1008
Score = 70.1 bits (170), Expect = 9e-12
Identities = 33/92 (35%), Positives = 54/92 (57%)
Query: 1359 KHLERLEDKDFDEPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPY 1418
KH+ F +P+ + CDN+ AI I+ NPVQH TKH+ + HF++E ++ I +
Sbjct: 917 KHMGLDYGMSFSDPLLVKCDNESAIAISKNPVQHSITKHIAIRHHFVRELVEEKQITVEH 976
Query: 1419 VSSQDNLADLLTKRLNNNNFEKFVSKLGMIDI 1450
V ++ LAD+ TK L+ N F + LG+ ++
Sbjct: 977 VPTEIQLADIFTKPLDLNMFVNLLKSLGIGEV 1008
>At3g48370 putative protein
Length = 96
Score = 70.1 bits (170), Expect = 9e-12
Identities = 34/78 (43%), Positives = 54/78 (68%)
Query: 1375 LYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTKRLN 1434
L+ D+ AI IA N V H+RTKH+E++ H ++EKLD G + +V+S+D +AD+LTK L
Sbjct: 5 LFSDSTAAIYIAKNHVFHERTKHVEIDCHTVREKLDDGSLKMLHVNSEDQIADILTKPLF 64
Query: 1435 NNNFEKFVSKLGMIDIHS 1452
+ F SK+ +I+I++
Sbjct: 65 PHQFAHLQSKMSLINIYA 82
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 69.7 bits (169), Expect = 1e-11
Identities = 33/79 (41%), Positives = 49/79 (61%)
Query: 1372 PMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTK 1431
P +YCDN A + NPV H R KH+ ++ HFI+ ++ SG + +VS+ D LAD LTK
Sbjct: 1368 PPVIYCDNVGATYLCANPVFHSRMKHIALDYHFIRNQVQSGALRVVHVSTHDQLADTLTK 1427
Query: 1432 RLNNNNFEKFVSKLGMIDI 1450
L+ F+ F K+G+I +
Sbjct: 1428 PLSRVAFQNFSRKIGVIKV 1446
Score = 45.8 bits (107), Expect = 2e-04
Identities = 27/94 (28%), Positives = 46/94 (48%), Gaps = 4/94 (4%)
Query: 484 YHYRVGDPSFRVIKQLFP---LFFKTLDVESFQCELCELAKHKRVTFSVSNKMSTFPFYL 540
+H R+G PS ++ + L + C C + K +V FS S S+ P
Sbjct: 446 WHSRLGHPSLAILNSVISNHSLPVLNPSHKLLSCSDCFINKSHKVPFSNSTITSSKPLEY 505
Query: 541 VHTNVWGPSNVPNISGARWFVTFIDDCTWVTWVY 574
++++VW S + +I R++V F+D T TW+Y
Sbjct: 506 IYSDVWS-SPILSIDNYRYYVIFVDHFTRYTWLY 538
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.364 0.163 0.626
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,219,342
Number of Sequences: 26719
Number of extensions: 990513
Number of successful extensions: 5334
Number of sequences better than 10.0: 98
Number of HSP's better than 10.0 without gapping: 93
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 5057
Number of HSP's gapped (non-prelim): 244
length of query: 1453
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1341
effective length of database: 8,326,068
effective search space: 11165257188
effective search space used: 11165257188
T: 11
A: 40
X1: 14 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 36 (21.5 bits)
S2: 67 (30.4 bits)
Medicago: description of AC147471.11


