

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147435.9 + phase: 0
(351 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
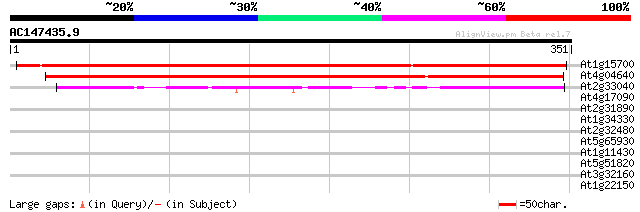
Score E
Sequences producing significant alignments: (bits) Value
At1g15700 unknown protein 456 e-128
At4g04640 ATPase gamma-subunit (AtpC1) 439 e-123
At2g33040 mitochondrial F1-ATPase, gamma subunit (ATP3_ARATH) 121 5e-28
At4g17090 beta-amylase (ct-bmy gene) 30 1.5
At2g31890 unknown protein 30 1.5
At1g34330 peroxidase, putative 30 1.5
At2g32480 unknown protein 29 4.4
At5g65930 kinesin-like calmodulin-binding protein 28 5.8
At1g11430 unknown protein 28 5.8
At5g51820 phosphoglucomutase (emb|CAB64725.1) 28 9.8
At3g32160 hypothetical protein 28 9.8
At1g22150 sulfate tansporter Sultr1;3 (Sultr1;3) 28 9.8
>At1g15700 unknown protein
Length = 386
Score = 456 bits (1172), Expect = e-128
Identities = 243/345 (70%), Positives = 284/345 (81%), Gaps = 3/345 (0%)
Query: 5 TITNGRIPSKPHFPQHFQIRCGIREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVIN 64
++T RI S+ P QIR GIRE+R+RI+SVK TQKITEAM+LVAAARVRRAQ+AVI
Sbjct: 42 SLTPNRISSRSPLPS-IQIRAGIRELRERIDSVKNTQKITEAMRLVAAARVRRAQDAVIK 100
Query: 65 SRPFSEAFAETLHSINQSLQNDDVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKA 124
RPF+E E L+SINQS Q +D+ PL+ VRPVK VAL+V+TGD+GLCGGFNN+V KKA
Sbjct: 101 GRPFTETLVEILYSINQSAQLEDIDFPLSIVRPVKRVALVVVTGDKGLCGGFNNAVTKKA 160
Query: 125 EARVMELKNLGINCVVISVGKKGSSYFNRSGFVEVDRFIDNVG-FPTTKDAQIIADDVFS 183
RV ELK GI+CVVISVGKKG++YF+R +VD+ I+ G FPTTK+AQ+IADDVFS
Sbjct: 161 TLRVQELKQRGIDCVVISVGKKGNAYFSRRDEFDVDKCIEGGGVFPTTKEAQVIADDVFS 220
Query: 184 LFVTEEVDKVELVYTKFVSLVRFNPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSK 243
LFV+EEVDKVELVYTKFVSLV+ +PVI TLLPLS KGE DV G VD +EDE FRLTSK
Sbjct: 221 LFVSEEVDKVELVYTKFVSLVKSDPVIHTLLPLSMKGESCDVKGECVDAIEDEMFRLTSK 280
Query: 244 DGKLALKRDVKKKKMKDGFVPVMEFEQDPAQILDAMMPLYLNSQVLKALQESLASELAAR 303
DGKLA++R K + K P+M+FEQDP QILDAMMPLYLNSQ+L+ALQESLASELA+R
Sbjct: 281 DGKLAVER-TKLEVEKPEISPLMQFEQDPVQILDAMMPLYLNSQILRALQESLASELASR 339
Query: 304 MGAMSNATDNAVELTKELSVAYNRERQAKITGEILEIVAGAEALR 348
M AMSNATDNAVEL K L++AYNR RQAKITGE+LEIVAGAEALR
Sbjct: 340 MNAMSNATDNAVELKKNLTMAYNRARQAKITGELLEIVAGAEALR 384
>At4g04640 ATPase gamma-subunit (AtpC1)
Length = 373
Score = 439 bits (1129), Expect = e-123
Identities = 221/324 (68%), Positives = 269/324 (82%), Gaps = 1/324 (0%)
Query: 23 IRCGIREIRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQS 82
++ +RE+RDRI+SVK TQKITEAMKLVAAA+VRRAQEAV+N RPFSE E L++IN+
Sbjct: 49 LQASLRELRDRIDSVKNTQKITEAMKLVAAAKVRRAQEAVVNGRPFSETLVEVLYNINEQ 108
Query: 83 LQNDDVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVVIS 142
LQ DDV VPLT VRPVK VAL+V+TGDRGLCGGFNN + KKAEAR+ ELK LG+ VIS
Sbjct: 109 LQTDDVDVPLTKVRPVKKVALVVVTGDRGLCGGFNNFIIKKAEARIKELKGLGLEYTVIS 168
Query: 143 VGKKGSSYFNRSGFVEVDRFIDNVGFPTTKDAQIIADDVFSLFVTEEVDKVELVYTKFVS 202
VGKKG+SYF R ++ VD++++ PT K+AQ +ADDVFSLF++EEVDKVEL+YTKFVS
Sbjct: 169 VGKKGNSYFLRRPYIPVDKYLEAGTLPTAKEAQAVADDVFSLFISEEVDKVELLYTKFVS 228
Query: 203 LVRFNPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGF 262
LV+ PVI TLLPLS KGE+ D+NG VD EDEFFRLT+K+GKL ++R+ + D F
Sbjct: 229 LVKSEPVIHTLLPLSPKGEICDINGTCVDAAEDEFFRLTTKEGKLTVERETFRTPTAD-F 287
Query: 263 VPVMEFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKELS 322
P+++FEQDP QILDA++PLYLNSQ+L+ALQESLASELAARM AMS+A+DNA +L K LS
Sbjct: 288 SPILQFEQDPVQILDALLPLYLNSQILRALQESLASELAARMSAMSSASDNASDLKKSLS 347
Query: 323 VAYNRERQAKITGEILEIVAGAEA 346
+ YNR+RQAKITGEILEIVAGA A
Sbjct: 348 MVYNRKRQAKITGEILEIVAGANA 371
>At2g33040 mitochondrial F1-ATPase, gamma subunit (ATP3_ARATH)
Length = 325
Score = 121 bits (304), Expect = 5e-28
Identities = 97/322 (30%), Positives = 158/322 (48%), Gaps = 52/322 (16%)
Query: 30 IRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSINQSLQNDDVV 89
+R+R+ SVK QKIT+AMK+VAA+++R Q NSR + F L N S+
Sbjct: 48 VRNRMKSVKNIQKITKAMKMVAASKLRAVQGRAENSRGLWQPFTALLGD-NPSID----- 101
Query: 90 VPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKNLGINCVV--ISVGKKG 147
VK ++ ++ D+GLCGG N++V K + R + N G V + VG+K
Sbjct: 102 --------VKKSVVVTLSSDKGLCGGINSTVVKVS--RALYKLNAGPEKEVQFVIVGEKA 151
Query: 148 SSYFNRSGFVEVDRFIDNVGFPTTKDAQI--IADDVFSLFVTEEVDKVELVYTKFVSLVR 205
+ R ++ + + AQ+ +ADD+ E D + +VY KF S+V
Sbjct: 152 KAIMFRDSKNDIVLSVTELNKNPLNYAQVSVLADDILK---NVEFDALRIVYNKFHSVVA 208
Query: 206 FNPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGFVPV 265
F P + T+L S +++E E + GKL ++ +++ G
Sbjct: 209 FLPTVSTVL--------------SPEIIEKE----SEIGGKLG---ELDSYEIEGG---- 243
Query: 266 MEFEQDPAQILDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKELSVAY 325
+ +IL + + + A+ E+ SE+ ARM AM +++ NA E+ L++ Y
Sbjct: 244 ----ETKGEILQNLAEFQFSCVMFNAVLENACSEMGARMSAMDSSSRNAGEMLDRLTLTY 299
Query: 326 NRERQAKITGEILEIVAGAEAL 347
NR RQA IT E++EI++GA AL
Sbjct: 300 NRTRQASITTELIEIISGASAL 321
>At4g17090 beta-amylase (ct-bmy gene)
Length = 548
Score = 30.4 bits (67), Expect = 1.5
Identities = 25/99 (25%), Positives = 41/99 (41%), Gaps = 16/99 (16%)
Query: 74 ETLHSINQSLQNDDVVVPLTAVRPVKTVALIVITGDRGLCGGFNNSVAKKAEARVMELKN 133
E LH ++ +D VP+ + P+ TV + G N A A +M LK
Sbjct: 70 EKLHVLSYPHSKNDASVPVFVMLPLDTVT---------MSGHLNKPRAMNAS--LMALKG 118
Query: 134 LGINCVVIS-----VGKKGSSYFNRSGFVEVDRFIDNVG 167
G+ V++ V K G +N G+ E+ + + G
Sbjct: 119 AGVEGVMVDAWWGLVEKDGPMNYNWEGYAELIQMVQKHG 157
>At2g31890 unknown protein
Length = 671
Score = 30.4 bits (67), Expect = 1.5
Identities = 33/128 (25%), Positives = 53/128 (40%), Gaps = 16/128 (12%)
Query: 236 EFFRLTSKDGKLALKRDVKKKKMKDGFVPVMEFEQDPAQ-------ILDAMMPL-----Y 283
E L K K + K VKK K+K +P +F+ + + D M L
Sbjct: 157 EVMPLKKKKKKKSKKVIVKKDKVKSKSIPEDDFDTEDEDLDFEDGFVEDKMGDLRKRVSS 216
Query: 284 LNSQVLKALQESLASELAARMGAMSNATD--NAVELTKELSVAYNRERQAKITGEILEIV 341
L + + +E + +LA R+ S +D + L K + A E ++T E I+
Sbjct: 217 LAGGMFEEKKEKMKEQLAQRLSQFSGPSDRMKEINLNKAIIEAQTAEEVLEVTAE--TIM 274
Query: 342 AGAEALRP 349
A A+ L P
Sbjct: 275 AVAKGLSP 282
>At1g34330 peroxidase, putative
Length = 271
Score = 30.4 bits (67), Expect = 1.5
Identities = 20/67 (29%), Positives = 32/67 (46%), Gaps = 7/67 (10%)
Query: 227 GNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGFVPVMEFEQDPAQILDAMMPLYLNS 286
G+SV V F+ KD K+ K K + G DP+ +LD M PL +++
Sbjct: 147 GHSVGVAHCSLFQDRLKDPKMDSKLKAKLQNTCRG-------PNDPSVVLDQMTPLEVDN 199
Query: 287 QVLKALQ 293
Q+ K ++
Sbjct: 200 QIYKQIK 206
>At2g32480 unknown protein
Length = 447
Score = 28.9 bits (63), Expect = 4.4
Identities = 28/121 (23%), Positives = 52/121 (42%), Gaps = 12/121 (9%)
Query: 171 TKDAQIIADDVFSLFVTEEVDKVELVYTKFVSLVRFNPVIQTLLPLSKKGEVFDVNGNSV 230
++D + D + ++ TE +K V +V+ NP + + + GE FD+
Sbjct: 225 SRDGLLSGDVILAVDGTELSKTGPDAVSKIVDIVKRNPKSNVVFRIERGGEDFDIR---- 280
Query: 231 DVLEDEFFRLTSKDGKLALKRDVKKKKMKDGFVPV------MEFEQDPAQILDAMMPLYL 284
V D+ F T K G + L +V+ K++ +P EF + +LD + +
Sbjct: 281 -VTPDKNFDGTGKIG-VQLSPNVRITKVRPRNIPETFRFVGREFMGLSSNVLDGLKQTFF 338
Query: 285 N 285
N
Sbjct: 339 N 339
>At5g65930 kinesin-like calmodulin-binding protein
Length = 1260
Score = 28.5 bits (62), Expect = 5.8
Identities = 27/125 (21%), Positives = 56/125 (44%)
Query: 216 LSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGFVPVMEFEQDPAQI 275
L K + +V + V D RL S++ L +++KKK ++ + +EQ+ +
Sbjct: 732 LYKIQKELEVRNKELHVAVDNSKRLLSENKILEQNLNIEKKKKEEVEIHQKRYEQEKKVL 791
Query: 276 LDAMMPLYLNSQVLKALQESLASELAARMGAMSNATDNAVELTKELSVAYNRERQAKITG 335
+ L +VL +S S + ++ M +N EL + + + +R+ + T
Sbjct: 792 KLRVSELENKLEVLAQDLDSAESTIESKNSDMLLLQNNLKELEELREMKEDIDRKNEQTA 851
Query: 336 EILEI 340
IL++
Sbjct: 852 AILKM 856
>At1g11430 unknown protein
Length = 232
Score = 28.5 bits (62), Expect = 5.8
Identities = 15/33 (45%), Positives = 22/33 (66%), Gaps = 2/33 (6%)
Query: 265 VMEFEQDPAQILDAMMPLYLN--SQVLKALQES 295
VMEF +DPA D M+ YLN + VL +++E+
Sbjct: 93 VMEFPKDPAPSRDQMIDTYLNTLATVLGSMEEA 125
>At5g51820 phosphoglucomutase (emb|CAB64725.1)
Length = 623
Score = 27.7 bits (60), Expect = 9.8
Identities = 16/48 (33%), Positives = 24/48 (49%)
Query: 207 NPVIQTLLPLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVK 254
N +I+ L + K + DV GN V D+F DG +A K+ V+
Sbjct: 505 NKMIEYLREILSKSKAGDVYGNYVLQFADDFSYTDPVDGSVASKQGVR 552
>At3g32160 hypothetical protein
Length = 148
Score = 27.7 bits (60), Expect = 9.8
Identities = 18/60 (30%), Positives = 29/60 (48%), Gaps = 4/60 (6%)
Query: 215 PLSKKGEVFDVNGNSVDVLEDEFFRLTSKDGKLALKRDVKKKKMKDGFVPVMEFEQDPAQ 274
PL K E +D +D + D++ RL + +L LKR + + ++ V E DP Q
Sbjct: 40 PLKDKKEAYDF----MDKVVDKYRRLCDERQRLGLKRQMLEARISCVERKVRSMESDPFQ 95
>At1g22150 sulfate tansporter Sultr1;3 (Sultr1;3)
Length = 656
Score = 27.7 bits (60), Expect = 9.8
Identities = 19/71 (26%), Positives = 36/71 (49%), Gaps = 8/71 (11%)
Query: 30 IRDRINSVKTTQKITEAMKLVAAARVRRAQEAVINSRPFSEAFAETLHSIN---QSLQND 86
+R+RI + +T+ ++V AAR+ R Q +I P ++ +H++ +SLQ
Sbjct: 552 VRERIQ-----RWLTDEEEMVEAARLPRIQFLIIEMSPVTDIDTSGIHALEDLYKSLQKR 606
Query: 87 DVVVPLTAVRP 97
D+ + L P
Sbjct: 607 DIQLVLANPGP 617
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,781,646
Number of Sequences: 26719
Number of extensions: 265501
Number of successful extensions: 770
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 759
Number of HSP's gapped (non-prelim): 12
length of query: 351
length of database: 11,318,596
effective HSP length: 100
effective length of query: 251
effective length of database: 8,646,696
effective search space: 2170320696
effective search space used: 2170320696
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147435.9


