

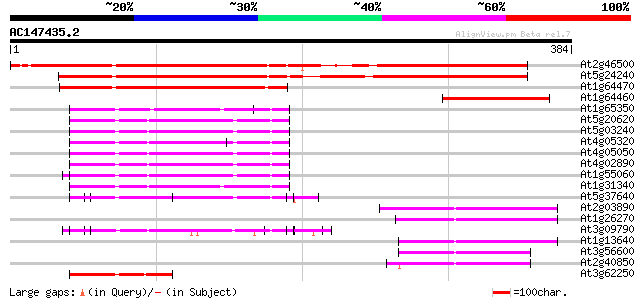
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147435.2 + phase: 0
(384 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g46500 putative ubiquitin 312 2e-85
At5g24240 ubiquitin 288 3e-78
At1g64470 ubiquitin, putative 154 6e-38
At1g64460 unknown protein 90 2e-18
At1g65350 75 8e-14
At5g20620 polyubiquitin 4 UBQ4 74 1e-13
At5g03240 polyubiquitin (ubq3) 74 1e-13
At4g05320 polyubiquitin (ubq10) 74 1e-13
At4g05050 74 1e-13
At4g02890 polyubiquitin 74 1e-13
At1g55060 ubiquitin-like (UBQ12) 74 2e-13
At1g31340 putative ubiquitin (AtRUB1) 74 2e-13
At5g37640 polyubiquitin 4 71 1e-12
At2g03890 unknown protein 69 6e-12
At1g26270 unknown protein 67 2e-11
At3g09790 polyubiquitin 65 5e-11
At1g13640 unknown protein 64 1e-10
At3g56600 putative protein 61 1e-09
At2g40850 putative protein 58 8e-09
At3g62250 ubiquitin extension protein (UBQ5) 46 4e-05
>At2g46500 putative ubiquitin
Length = 566
Score = 312 bits (800), Expect = 2e-85
Identities = 182/358 (50%), Positives = 246/358 (67%), Gaps = 34/358 (9%)
Query: 1 MSSAGVSTLSPVDPTEPLLSPNAFHLPSHLSFEDKSISIFLSVSGSLTPFRVMEWDTIES 60
MSSAGV+ LSPV +EPL+ P S+ D +I I+L++ GS+ P RV+E D+IES
Sbjct: 1 MSSAGVA-LSPVR-SEPLIMPLVRANSCLDSYPDDTIMIYLTLPGSVIPMRVLESDSIES 58
Query: 61 VKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTDGNVLHLIIKLSDLQLINVK 120
VK +IQ F+ QKLV+ GRELARS++ ++DYGV++GN+LHL++KLSDLQ+++VK
Sbjct: 59 VKLRIQSYRG--FVVRNQKLVFGGRELARSNSNMRDYGVSEGNILHLVLKLSDLQVLDVK 116
Query: 121 TSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVCNGERLEDQKLIDDICCKHN 180
T+ GK F VERGR++ Y+K++I+KK F D +EQE++ GE+LEDQ LI+DIC +++
Sbjct: 117 TTCGKHCRFHVERGRNIGYVKKQISKKRGDFVDPDEQEILYEGEKLEDQSLINDIC-RND 175
Query: 181 DAVVHLFVRKRNAKVHRRP----FELSIVTTDLADTKTDDVFENSCGRKFEVGGVEDTDA 236
D+V+HL VR R+AKV +P FELSIV D K GR +A
Sbjct: 176 DSVLHLLVR-RSAKVRVKPVEKNFELSIVAPQAKDKK---------GR----------EA 215
Query: 237 IQRAVPRKLPDRDFLLEPIIVNPKIELASVIRNMVNSAYDGLASEYYPIGSAEGTGGAYF 296
P+KL LEP++VN K ++ V+++M+ SA DGL S P+ S+EGTGGAYF
Sbjct: 216 KSIVPPKKLS-----LEPVVVNSKAKVPLVVKDMIQSASDGLKSGNSPVRSSEGTGGAYF 270
Query: 297 MLDSTGQKYVSVFKPIDEEPMAVNNPRCLPLSLDGEGLKKGTIVGQGAFREVAAYVLE 354
M +G K+V VFKPIDEEPMA NNP+ LPLS +GEGLKKGT VG+GA REVAAY+L+
Sbjct: 271 MQGPSGNKFVGVFKPIDEEPMAENNPQGLPLSPNGEGLKKGTKVGEGALREVAAYILD 328
>At5g24240 ubiquitin
Length = 574
Score = 288 bits (737), Expect = 3e-78
Identities = 155/321 (48%), Positives = 215/321 (66%), Gaps = 20/321 (6%)
Query: 34 DKSISIFLSVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTL 93
D I +FL+++GS+ P RVME D+I SVK +IQ + F KQKL+Y GRE++R+D+
Sbjct: 30 DDPILVFLTIAGSVIPKRVMESDSIASVKLRIQSIKG--FFVKKQKLLYDGREVSRNDSQ 87
Query: 94 LKDYGVTDGNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDD 153
++DYG+ DG +LHL+I+LSDLQ I+V+T GKEF VER R+V Y+K++IA KEK+
Sbjct: 88 IRDYGLADGKLLHLVIRLSDLQAISVRTVDGKEFELVVERSRNVGYVKQQIASKEKELGI 147
Query: 154 LEEQELVCNGERLEDQKLIDDICCKHNDAVVHLFVRKRNAKVHRRPFELSIVTTDLADTK 213
+ EL +GE L+DQ+LI D+C ++ D V+HL + K +AKV +P K
Sbjct: 148 PRDHELTLDGEELDDQRLITDLC-QNGDNVIHLLISK-SAKVRAKPVG-----------K 194
Query: 214 TDDVFENSCGRKFEVGGVEDTDAIQRAVPRKLPDRDFLLEPIIVNPKIELASVIRNMVNS 273
+VF K V G + A P+ +F +EP IVNP+I+L +++ +++S
Sbjct: 195 DFEVFIEDVNHKHNVDGRRGKNISSEAKPK-----EFFVEPFIVNPEIKLPILLKELISS 249
Query: 274 AYDGLASEYYPIGSAEGTGGAYFMLDSTGQKYVSVFKPIDEEPMAVNNPRCLPLSLDGEG 333
+GL PI S++G+GGAYFM D +G KYVSVFKPIDEEPMAVNNP P+S+DGEG
Sbjct: 250 TLEGLEKGNGPIRSSDGSGGAYFMQDPSGHKYVSVFKPIDEEPMAVNNPHGQPVSVDGEG 309
Query: 334 LKKGTIVGQGAFREVAAYVLE 354
LKKGT VG+GA REVAAY+L+
Sbjct: 310 LKKGTQVGEGAIREVAAYILD 330
>At1g64470 ubiquitin, putative
Length = 213
Score = 154 bits (390), Expect = 6e-38
Identities = 77/156 (49%), Positives = 114/156 (72%), Gaps = 3/156 (1%)
Query: 35 KSISIFLSVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLL 94
KS+ +FLSVSGS P ++E D+I VK +IQ C F +QKLV++GRELAR+ + +
Sbjct: 33 KSVLVFLSVSGSTMPMLILESDSIAEVKLRIQTCNG--FRVRRQKLVFSGRELARNASRV 90
Query: 95 KDYGVTDGNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDL 154
KDYGVT G+VLHL++KL D L+ V T+ GK F F V+R R+V Y+K+RI+K+ K F ++
Sbjct: 91 KDYGVTGGSVLHLVLKLYDPLLVTVITTCGKVFQFHVDRRRNVGYLKKRISKEGKGFPEV 150
Query: 155 EEQELVCNGERLEDQKLIDDICCKHNDAVVHLFVRK 190
++QE++ GE+L+D ++ID I CK ++V+HL V+K
Sbjct: 151 DDQEILFKGEKLDDNRIIDGI-CKDGNSVIHLLVKK 185
>At1g64460 unknown protein
Length = 301
Score = 89.7 bits (221), Expect = 2e-18
Identities = 46/74 (62%), Positives = 59/74 (79%), Gaps = 1/74 (1%)
Query: 297 MLDSTGQKYVSVFKPIDEEPMAVNNPRCLPLSLDGEGLKKGTIVGQGAFREVAAYVLETQ 356
M DS+G YVSVFKP+DEEPMAVNNP+ LP+S DG+GLK+GT VG+GA REVAAY+L+
Sbjct: 1 MQDSSGLNYVSVFKPMDEEPMAVNNPQQLPVSSDGQGLKRGTRVGEGATREVAAYLLDHP 60
Query: 357 RV-VTSLSISVLSF 369
+ + S+S V+ F
Sbjct: 61 KSGLRSVSKEVMGF 74
>At1g65350
Length = 294
Score = 74.7 bits (182), Expect = 8e-14
Identities = 50/150 (33%), Positives = 84/150 (55%), Gaps = 7/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 7 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 63
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 64 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIF 121
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++LED + + D K ++ +HL +R R
Sbjct: 122 AGKQLEDGRTLADNIQK--ESTLHLVLRLR 149
Score = 69.7 bits (169), Expect = 3e-12
Identities = 45/150 (30%), Positives = 86/150 (57%), Gaps = 7/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL + +
Sbjct: 83 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLADN--IQK 138
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I ++K++ ++Q L+
Sbjct: 139 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKI--QDKEWIPPDQQRLIF 196
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++LED + + D + ++ +HL +R R
Sbjct: 197 AGKQLEDGRTLADYNIQ-KESTLHLVLRLR 225
Score = 65.9 bits (159), Expect = 4e-11
Identities = 43/126 (34%), Positives = 71/126 (56%), Gaps = 5/126 (3%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 158 TLTGKTITLEVESSDTIDNVKAKIQDKEWIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 214
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 215 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSGTIDNVKAKIQDKEGIPPD--QQRLIF 272
Query: 162 NGERLE 167
G++LE
Sbjct: 273 AGKQLE 278
Score = 36.2 bits (82), Expect = 0.031
Identities = 24/75 (32%), Positives = 41/75 (54%), Gaps = 3/75 (4%)
Query: 117 INVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVCNGERLEDQKLIDDIC 176
I VKT +GK + +VE + +K +I KE D +Q L+ G++LED + + D
Sbjct: 3 IFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIFAGKQLEDGRTLADYN 60
Query: 177 CKHNDAVVHLFVRKR 191
+ ++ +HL +R R
Sbjct: 61 IQ-KESTLHLVLRLR 74
>At5g20620 polyubiquitin 4 UBQ4
Length = 382
Score = 74.3 bits (181), Expect = 1e-13
Identities = 49/150 (32%), Positives = 84/150 (55%), Gaps = 6/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 7 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 63
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 64 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIF 121
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++LED + + D + ++ +HL +R R
Sbjct: 122 AGKQLEDGRTLADYNIQ-KESTLHLVLRLR 150
Score = 74.3 bits (181), Expect = 1e-13
Identities = 49/150 (32%), Positives = 84/150 (55%), Gaps = 6/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 83 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 139
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 140 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIF 197
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++LED + + D + ++ +HL +R R
Sbjct: 198 AGKQLEDGRTLADYNIQ-KESTLHLVLRLR 226
Score = 74.3 bits (181), Expect = 1e-13
Identities = 49/150 (32%), Positives = 84/150 (55%), Gaps = 6/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 159 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 215
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 216 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIF 273
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++LED + + D + ++ +HL +R R
Sbjct: 274 AGKQLEDGRTLADYNIQ-KESTLHLVLRLR 302
Score = 74.3 bits (181), Expect = 1e-13
Identities = 49/150 (32%), Positives = 84/150 (55%), Gaps = 6/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 235 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 291
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 292 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIF 349
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++LED + + D + ++ +HL +R R
Sbjct: 350 AGKQLEDGRTLADYNIQ-KESTLHLVLRLR 378
Score = 36.2 bits (82), Expect = 0.031
Identities = 24/75 (32%), Positives = 41/75 (54%), Gaps = 3/75 (4%)
Query: 117 INVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVCNGERLEDQKLIDDIC 176
I VKT +GK + +VE + +K +I KE D +Q L+ G++LED + + D
Sbjct: 3 IFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIFAGKQLEDGRTLADYN 60
Query: 177 CKHNDAVVHLFVRKR 191
+ ++ +HL +R R
Sbjct: 61 IQ-KESTLHLVLRLR 74
>At5g03240 polyubiquitin (ubq3)
Length = 306
Score = 74.3 bits (181), Expect = 1e-13
Identities = 49/150 (32%), Positives = 84/150 (55%), Gaps = 6/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 83 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 139
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 140 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIF 197
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++LED + + D + ++ +HL +R R
Sbjct: 198 AGKQLEDGRTLADYNIQ-KESTLHLVLRLR 226
Score = 74.3 bits (181), Expect = 1e-13
Identities = 49/150 (32%), Positives = 84/150 (55%), Gaps = 6/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 7 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 63
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 64 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIF 121
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++LED + + D + ++ +HL +R R
Sbjct: 122 AGKQLEDGRTLADYNIQ-KESTLHLVLRLR 150
Score = 74.3 bits (181), Expect = 1e-13
Identities = 49/150 (32%), Positives = 84/150 (55%), Gaps = 6/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 159 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 215
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 216 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIF 273
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++LED + + D + ++ +HL +R R
Sbjct: 274 AGKQLEDGRTLADYNIQ-KESTLHLVLRLR 302
Score = 36.2 bits (82), Expect = 0.031
Identities = 24/75 (32%), Positives = 41/75 (54%), Gaps = 3/75 (4%)
Query: 117 INVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVCNGERLEDQKLIDDIC 176
I VKT +GK + +VE + +K +I KE D +Q L+ G++LED + + D
Sbjct: 3 IFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIFAGKQLEDGRTLADYN 60
Query: 177 CKHNDAVVHLFVRKR 191
+ ++ +HL +R R
Sbjct: 61 IQ-KESTLHLVLRLR 74
>At4g05320 polyubiquitin (ubq10)
Length = 464
Score = 74.3 bits (181), Expect = 1e-13
Identities = 49/150 (32%), Positives = 84/150 (55%), Gaps = 6/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 159 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 215
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 216 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIF 273
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++LED + + D + ++ +HL +R R
Sbjct: 274 AGKQLEDGRTLADYNIQ-KESTLHLVLRLR 302
Score = 74.3 bits (181), Expect = 1e-13
Identities = 49/150 (32%), Positives = 84/150 (55%), Gaps = 6/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 235 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 291
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 292 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIF 349
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++LED + + D + ++ +HL +R R
Sbjct: 350 AGKQLEDGRTLADYNIQ-KESTLHLVLRLR 378
Score = 74.3 bits (181), Expect = 1e-13
Identities = 49/150 (32%), Positives = 84/150 (55%), Gaps = 6/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 83 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 139
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 140 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIF 197
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++LED + + D + ++ +HL +R R
Sbjct: 198 AGKQLEDGRTLADYNIQ-KESTLHLVLRLR 226
Score = 74.3 bits (181), Expect = 1e-13
Identities = 49/150 (32%), Positives = 84/150 (55%), Gaps = 6/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 7 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 63
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 64 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIF 121
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++LED + + D + ++ +HL +R R
Sbjct: 122 AGKQLEDGRTLADYNIQ-KESTLHLVLRLR 150
Score = 63.2 bits (152), Expect = 2e-10
Identities = 37/107 (34%), Positives = 61/107 (56%), Gaps = 3/107 (2%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 311 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 367
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKE 148
+ LHL+++L I VKT +GK + +VE + +K +I KE
Sbjct: 368 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKE 414
Score = 36.2 bits (82), Expect = 0.031
Identities = 24/75 (32%), Positives = 41/75 (54%), Gaps = 3/75 (4%)
Query: 117 INVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVCNGERLEDQKLIDDIC 176
I VKT +GK + +VE + +K +I KE D +Q L+ G++LED + + D
Sbjct: 3 IFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIFAGKQLEDGRTLADYN 60
Query: 177 CKHNDAVVHLFVRKR 191
+ ++ +HL +R R
Sbjct: 61 IQ-KESTLHLVLRLR 74
>At4g05050
Length = 229
Score = 74.3 bits (181), Expect = 1e-13
Identities = 49/150 (32%), Positives = 84/150 (55%), Gaps = 6/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 7 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 63
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 64 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIF 121
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++LED + + D + ++ +HL +R R
Sbjct: 122 AGKQLEDGRTLADYNIQ-KESTLHLVLRLR 150
Score = 74.3 bits (181), Expect = 1e-13
Identities = 49/150 (32%), Positives = 84/150 (55%), Gaps = 6/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 83 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 139
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 140 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIF 197
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++LED + + D + ++ +HL +R R
Sbjct: 198 AGKQLEDGRTLADYNIQ-KESTLHLVLRLR 226
Score = 36.2 bits (82), Expect = 0.031
Identities = 24/75 (32%), Positives = 41/75 (54%), Gaps = 3/75 (4%)
Query: 117 INVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVCNGERLEDQKLIDDIC 176
I VKT +GK + +VE + +K +I KE D +Q L+ G++LED + + D
Sbjct: 3 IFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIFAGKQLEDGRTLADYN 60
Query: 177 CKHNDAVVHLFVRKR 191
+ ++ +HL +R R
Sbjct: 61 IQ-KESTLHLVLRLR 74
>At4g02890 polyubiquitin
Length = 229
Score = 74.3 bits (181), Expect = 1e-13
Identities = 49/150 (32%), Positives = 84/150 (55%), Gaps = 6/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 7 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 63
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 64 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIF 121
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++LED + + D + ++ +HL +R R
Sbjct: 122 AGKQLEDGRTLADYNIQ-KESTLHLVLRLR 150
Score = 74.3 bits (181), Expect = 1e-13
Identities = 49/150 (32%), Positives = 84/150 (55%), Gaps = 6/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 83 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 139
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 140 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIF 197
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++LED + + D + ++ +HL +R R
Sbjct: 198 AGKQLEDGRTLADYNIQ-KESTLHLVLRLR 226
Score = 36.2 bits (82), Expect = 0.031
Identities = 24/75 (32%), Positives = 41/75 (54%), Gaps = 3/75 (4%)
Query: 117 INVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVCNGERLEDQKLIDDIC 176
I VKT +GK + +VE + +K +I KE D +Q L+ G++LED + + D
Sbjct: 3 IFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIFAGKQLEDGRTLADYN 60
Query: 177 CKHNDAVVHLFVRKR 191
+ ++ +HL +R R
Sbjct: 61 IQ-KESTLHLVLRLR 74
>At1g55060 ubiquitin-like (UBQ12)
Length = 230
Score = 73.6 bits (179), Expect = 2e-13
Identities = 51/156 (32%), Positives = 87/156 (55%), Gaps = 7/156 (4%)
Query: 37 ISIFL-SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLK 95
+ IFL +++G V DTI++VK KIQ E +P ++ +L++AG++L TL
Sbjct: 1 MQIFLKTLTGKTKVLEVESSDTIDNVKAKIQDIEGIP--PDQHRLIFAGKQLEDGRTLA- 57
Query: 96 DYGVTDGNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLE 155
DY V + + LHL+++ I VKT +GK + +VE + +K +I KE D
Sbjct: 58 DYNVQEDSTLHLLLRFRGGMQIFVKTLTGKTITLEVESSDTIDNLKAKIQDKEGIPPD-- 115
Query: 156 EQELVCNGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
+Q L+ G++LED + + D + ++ +HL +R R
Sbjct: 116 QQRLIFAGKQLEDGRTLADYNIQ-KESTLHLVLRLR 150
Score = 71.2 bits (173), Expect = 9e-13
Identities = 47/150 (31%), Positives = 83/150 (55%), Gaps = 6/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI+++K KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 83 TLTGKTITLEVESSDTIDNLKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 139
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I VKT +GK + +VE + +K +I KE D +Q L+
Sbjct: 140 ESTLHLVLRLRGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGISPD--QQRLIF 197
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++ ED + + D + ++ +HL +R R
Sbjct: 198 AGKQHEDGRTLADYNIQ-KESTLHLVLRLR 226
>At1g31340 putative ubiquitin (AtRUB1)
Length = 156
Score = 73.6 bits (179), Expect = 2e-13
Identities = 49/150 (32%), Positives = 82/150 (54%), Gaps = 6/150 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 7 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQK 63
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L +I VKT +GKE +E + IK R+ +EK+ +Q L+
Sbjct: 64 ESTLHLVLRLRGGTMIKVKTLTGKEIEIDIEPTDTIDRIKERV--EEKEGIPPVQQRLIY 121
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKR 191
G++L D K D + +V+HL + R
Sbjct: 122 AGKQLADDKTAKDYNIE-GGSVLHLVLALR 150
Score = 38.9 bits (89), Expect = 0.005
Identities = 31/109 (28%), Positives = 53/109 (48%), Gaps = 9/109 (8%)
Query: 117 INVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVCNGERLEDQKLIDDIC 176
I VKT +GK + +VE + +K +I KE D +Q L+ G++LED + + D
Sbjct: 3 IFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIFAGKQLEDGRTLADYN 60
Query: 177 CKHNDAVVHLFVRKRNAKVHR------RPFELSIVTTDLADTKTDDVFE 219
+ ++ +HL +R R + + + E+ I TD D + V E
Sbjct: 61 IQ-KESTLHLVLRLRGGTMIKVKTLTGKEIEIDIEPTDTIDRIKERVEE 108
>At5g37640 polyubiquitin 4
Length = 322
Score = 70.9 bits (172), Expect = 1e-12
Identities = 50/166 (30%), Positives = 88/166 (52%), Gaps = 12/166 (7%)
Query: 52 VMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTDGNVLHLIIKL 111
V+ DTI +VK KIQ E +P ++Q+L+++G+ L TL DY + ++LHL ++L
Sbjct: 19 VVSSDTINNVKAKIQDIEGIPL--DQQRLIFSGKLLDDGRTLA-DYSIQKDSILHLALRL 75
Query: 112 SDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVCNGERLEDQKL 171
I VKT +GK + +VE + +K +I KE D +Q L+ G++L+D +
Sbjct: 76 RGGMQIFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGVPPD--QQRLIFAGKQLDDGRT 133
Query: 172 IDDICCKHNDAVVHLFVRKRNA------KVHRRPFELSIVTTDLAD 211
+ D + ++ +HL +R R + R+ L + ++D D
Sbjct: 134 LADYNIQ-KESTLHLVLRLRGGMQIFVRTLTRKTIALEVESSDTTD 178
Score = 67.8 bits (164), Expect = 1e-11
Identities = 45/139 (32%), Positives = 75/139 (53%), Gaps = 5/139 (3%)
Query: 56 DTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTDGNVLHLIIKLSDLQ 115
DT ++VK KIQ E +P ++Q+L++AG++L TL DY + + LHL+++L
Sbjct: 175 DTTDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRTLA-DYNIQKESTLHLVLRLCGGM 231
Query: 116 LINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVCNGERLEDQKLIDDI 175
I V T +GK + +VE + +K +I KE+ D +Q L+ GE+LED
Sbjct: 232 QIFVNTLTGKTITLEVESSDTIDNVKAKIQDKERIQPD--QQRLIFAGEQLEDGYYTLAD 289
Query: 176 CCKHNDAVVHLFVRKRNAK 194
++ +HL +R R +
Sbjct: 290 YNIQKESTLHLVLRLRGGE 308
Score = 67.0 bits (162), Expect = 2e-11
Identities = 46/148 (31%), Positives = 81/148 (54%), Gaps = 6/148 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L TL DY +
Sbjct: 85 TLTGKTITLEVESSDTIDNVKAKIQDKEGVP--PDQQRLIFAGKQLDDGRTLA-DYNIQK 141
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
+ LHL+++L I V+T + K + +VE +K +I KE D +Q L+
Sbjct: 142 ESTLHLVLRLRGGMQIFVRTLTRKTIALEVESSDTTDNVKAKIQDKEGIPPD--QQRLIF 199
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVR 189
G++LED + + D + ++ +HL +R
Sbjct: 200 AGKQLEDGRTLADYNIQ-KESTLHLVLR 226
Score = 42.7 bits (99), Expect = 3e-04
Identities = 23/70 (32%), Positives = 40/70 (56%), Gaps = 2/70 (2%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E + ++Q+L++AG +L L DY +
Sbjct: 237 TLTGKTITLEVESSDTIDNVKAKIQDKERIQ--PDQQRLIFAGEQLEDGYYTLADYNIQK 294
Query: 102 GNVLHLIIKL 111
+ LHL+++L
Sbjct: 295 ESTLHLVLRL 304
>At2g03890 unknown protein
Length = 650
Score = 68.6 bits (166), Expect = 6e-12
Identities = 40/123 (32%), Positives = 64/123 (51%), Gaps = 2/123 (1%)
Query: 254 PIIVNPKIELASVIRNMVNSAYDGLASEYYPIGSAEGTGGAYFMLDSTGQKYVSVFKPID 313
PI + + S++++MV + P+ G GGAY+ + G+ V++ KP D
Sbjct: 139 PIEILGHSDCFSIVKHMVKDIVKAMKMGVEPLPVHSGLGGAYYFRNKRGES-VAIVKPTD 197
Query: 314 EEPMAVNNPR-CLPLSLDGEGLKKGTIVGQGAFREVAAYVLETQRVVTSLSISVLSFTRP 372
EEP A NNP+ + +L GLK VG+ FREVAAY+L+ R +++ T
Sbjct: 198 EEPFAPNNPKGFVGKALGQPGLKSSVRVGETGFREVAAYLLDYGRFANVPPTALVKITHS 257
Query: 373 IFS 375
+F+
Sbjct: 258 VFN 260
>At1g26270 unknown protein
Length = 630
Score = 67.0 bits (162), Expect = 2e-11
Identities = 38/112 (33%), Positives = 59/112 (51%), Gaps = 2/112 (1%)
Query: 265 SVIRNMVNSAYDGLASEYYPIGSAEGTGGAYFMLDSTGQKYVSVFKPIDEEPMAVNNPR- 323
S +R M + P+ G GGAY+ +S G+ V++ KP DEEP A NNP+
Sbjct: 146 SFVRQMAKDITKAVKKGIDPVAVNSGLGGAYYFKNSRGES-VAIVKPTDEEPYAPNNPKG 204
Query: 324 CLPLSLDGEGLKKGTIVGQGAFREVAAYVLETQRVVTSLSISVLSFTRPIFS 375
+ +L GLK+ VG+ +REVAAY+L+ + +++ T IF+
Sbjct: 205 FVGKALGQPGLKRSVRVGETGYREVAAYLLDKEHFANVPPTALVKITHSIFN 256
>At3g09790 polyubiquitin
Length = 631
Score = 65.5 bits (158), Expect = 5e-11
Identities = 42/134 (31%), Positives = 76/134 (56%), Gaps = 6/134 (4%)
Query: 56 DTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTDGNVLHLIIKLSDLQ 115
D+I +VK KIQ E +P ++Q+L++AG++L TL DY + + LHL+++L
Sbjct: 23 DSIHNVKAKIQNKEGIPL--DQQRLIFAGKQLEDGLTLA-DYNIQKESTLHLVLRLRGGM 79
Query: 116 LINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVCNGERLEDQKLIDDI 175
I V+T +GK + +V+ + +K +I KE +Q L+ G++LED + + D
Sbjct: 80 QIFVQTLTGKTITLEVKSSDTIDNVKAKIQDKEGILP--RQQRLIFAGKQLEDGRTLADY 137
Query: 176 CCKHNDAVVHLFVR 189
+ ++ +HL +R
Sbjct: 138 NIQ-KESTLHLVLR 150
Score = 65.1 bits (157), Expect = 6e-11
Identities = 45/139 (32%), Positives = 73/139 (52%), Gaps = 6/139 (4%)
Query: 56 DTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTDGNVLHLIIKLSDLQ 115
DTI++VK KIQ +P ++Q+L++ GR L S TLL DY + G+ +H +
Sbjct: 496 DTIDNVKVKIQHKVGIPL--DRQRLIFGGRVLVGSRTLL-DYNIQKGSTIHQLFLQRGGM 552
Query: 116 LINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVCNGERLEDQKLIDDI 175
I +KT +GK +VE + +K +I KE D +Q L+ G++LED + D
Sbjct: 553 QIFIKTLTGKTIILEVESSDTIANVKEKIQVKEGIKPD--QQMLIFFGQQLEDGVTLGDY 610
Query: 176 CCKHNDAVVHLFVRKRNAK 194
H + ++L +R R +
Sbjct: 611 DI-HKKSTLYLVLRLRQRR 628
Score = 64.7 bits (156), Expect = 8e-11
Identities = 49/174 (28%), Positives = 88/174 (50%), Gaps = 13/174 (7%)
Query: 52 VMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTDGNVLHLIIKL 111
V WDTIE+VK +Q E + N Q+L++ G+EL + L DY + + LHL++ +
Sbjct: 335 VESWDTIENVKAMVQDKEGIQPQPNLQRLIFLGKEL-KDGCTLADYSIQKESTLHLVLGM 393
Query: 112 SDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVCNGERLEDQKL 171
I VK GK + +V +K +K +I ++K ++Q L+ G +L+D +
Sbjct: 394 Q----IFVKLFGGKIITLEVLSSDTIKSVKAKI--QDKVGSPPDQQILLFRGGQLQDGRT 447
Query: 172 IDDICCKHNDAVVHLFVRKRNA-KVHRRPFELSIVT----TDLADTKTDDVFEN 220
+ D + N++ +HLF R+ ++ + F S T T + ++ D +N
Sbjct: 448 LGDYNIR-NESTLHLFFHIRHGMQIFVKTFSFSGETPTCKTITLEVESSDTIDN 500
Score = 63.2 bits (152), Expect = 2e-10
Identities = 46/140 (32%), Positives = 74/140 (52%), Gaps = 12/140 (8%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E + L +Q+L++AG++L TL DY +
Sbjct: 85 TLTGKTITLEVKSSDTIDNVKAKIQDKEGI--LPRQQRLIFAGKQLEDGRTLA-DYNIQK 141
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEF-----SFQVERGRDVKYIKRRIAKKEKQFDDLEE 156
+ LHL+++L I V T SGK F + +VE ++ +K +I +E D
Sbjct: 142 ESTLHLVLRLCGGMQIFVSTFSGKNFTSDTLTLKVESSDTIENVKAKIQDREGLRPD--H 199
Query: 157 QELVCNGERL--EDQKLIDD 174
Q L+ +GE L ED + + D
Sbjct: 200 QRLIFHGEELFTEDNRTLAD 219
Score = 52.4 bits (124), Expect = 4e-07
Identities = 44/173 (25%), Positives = 82/173 (46%), Gaps = 7/173 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
S +G V DTI++VK K+Q E +P + +L++AG+ L TL Y +
Sbjct: 249 SFTGENFILEVESSDTIDNVKAKLQDKERIPM--DLHRLIFAGKPLEGGRTLAH-YNIQK 305
Query: 102 GNVLHLIIKLSDLQLINVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVC 161
G+ L+L+ + I VKT + K + +VE ++ +K + KE Q L+
Sbjct: 306 GSTLYLVTRFRCGMQIFVKTLTRKRINLEVESWDTIENVKAMVQDKEGIQPQPNLQRLIF 365
Query: 162 NGERLEDQKLIDDICCKHNDAVVHLFVRKRNAKVHRRPFELSIVTTDLADTKT 214
G+ L+D + D + ++ +HL + ++ + F I+T ++ + T
Sbjct: 366 LGKELKDGCTLADYSIQ-KESTLHLVL---GMQIFVKLFGGKIITLEVLSSDT 414
Score = 46.2 bits (108), Expect = 3e-05
Identities = 44/167 (26%), Positives = 76/167 (45%), Gaps = 13/167 (7%)
Query: 37 ISIFLSV-SGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLK 95
+ IF+ + G + V+ DTI+SVK KIQ P ++Q L++ G +L T L
Sbjct: 393 MQIFVKLFGGKIITLEVLSSDTIKSVKAKIQDKVGSP--PDQQILLFRGGQLQDGRT-LG 449
Query: 96 DYGVTDGNVLHLIIKLSDLQLINVKTSS-------GKEFSFQVERGRDVKYIKRRIAKKE 148
DY + + + LHL + I VKT S K + +VE + +K +I +
Sbjct: 450 DYNIRNESTLHLFFHIRHGMQIFVKTFSFSGETPTCKTITLEVESSDTIDNVKVKI--QH 507
Query: 149 KQFDDLEEQELVCNGERLEDQKLIDDICCKHNDAVVHLFVRKRNAKV 195
K L+ Q L+ G L + + D + + LF+++ ++
Sbjct: 508 KVGIPLDRQRLIFGGRVLVGSRTLLDYNIQKGSTIHQLFLQRGGMQI 554
>At1g13640 unknown protein
Length = 622
Score = 64.3 bits (155), Expect = 1e-10
Identities = 38/110 (34%), Positives = 55/110 (49%), Gaps = 2/110 (1%)
Query: 267 IRNMVNSAYDGLASEYYPIGSAEGTGGAYFMLDSTGQKYVSVFKPIDEEPMAVNNPR-CL 325
++ N + PI G GGAY+ D GQ V++ KP DEEP A NNP+ +
Sbjct: 144 LKQTANDIVKAMKMGVEPIPVNGGLGGAYYFRDEKGQS-VAIVKPTDEEPFAPNNPKGFV 202
Query: 326 PLSLDGEGLKKGTIVGQGAFREVAAYVLETQRVVTSLSISVLSFTRPIFS 375
+L GLK VG+ FREVAAY+L+ +++ T +F+
Sbjct: 203 GKALGQPGLKPSVRVGETGFREVAAYLLDYDHFANVPPTALVKITHSVFN 252
>At3g56600 putative protein
Length = 533
Score = 60.8 bits (146), Expect = 1e-09
Identities = 34/91 (37%), Positives = 52/91 (56%), Gaps = 2/91 (2%)
Query: 267 IRNMVNSAYDGLASEYYPIGSAEGTGGAYFMLDSTGQKYVSVFKPIDEEPMAVNNPR-CL 325
+R++V + S P+ G GGAY + G ++V KP+DEEP+A NNP+
Sbjct: 87 VRSLVAEVTIAIVSGAQPLLLPSGLGGAYLLQTEKGNN-IAVAKPVDEEPLAFNNPKGSG 145
Query: 326 PLSLDGEGLKKGTIVGQGAFREVAAYVLETQ 356
L+L G+K+ VG+ RE+AAY+L+ Q
Sbjct: 146 GLTLGQPGMKRSIRVGESGIRELAAYLLDHQ 176
>At2g40850 putative protein
Length = 561
Score = 58.2 bits (139), Expect = 8e-09
Identities = 37/104 (35%), Positives = 54/104 (51%), Gaps = 7/104 (6%)
Query: 259 PKIELAS-----VIRNMVNSAYDGLASEYYPIGSAEGTGGAYFMLDSTGQKYVSVFKPID 313
P+IE+ +R +V + S P+ G GGAY + G ++V KP+D
Sbjct: 94 PRIEILGGQRVPTVRALVAEVTMAMVSGAQPLLLPSGMGGAYLLQTGKGHN-IAVAKPVD 152
Query: 314 EEPMAVNNP-RCLPLSLDGEGLKKGTIVGQGAFREVAAYVLETQ 356
EEP+A NNP + L L G+K VG+ RE+AAY+L+ Q
Sbjct: 153 EEPLAFNNPKKSGNLMLGQPGMKHSIPVGETGIRELAAYLLDYQ 196
>At3g62250 ubiquitin extension protein (UBQ5)
Length = 157
Score = 45.8 bits (107), Expect = 4e-05
Identities = 25/70 (35%), Positives = 43/70 (60%), Gaps = 3/70 (4%)
Query: 42 SVSGSLTPFRVMEWDTIESVKFKIQRCESLPFLTNKQKLVYAGRELARSDTLLKDYGVTD 101
+++G V DTI++VK KIQ E +P ++Q+L++AG++L T L DY +
Sbjct: 7 TLTGKTITLEVESSDTIDNVKAKIQDKEGIP--PDQQRLIFAGKQLEDGRT-LADYNIQK 63
Query: 102 GNVLHLIIKL 111
+ LHL+++L
Sbjct: 64 ESTLHLVLRL 73
Score = 37.7 bits (86), Expect = 0.011
Identities = 27/89 (30%), Positives = 46/89 (51%), Gaps = 9/89 (10%)
Query: 117 INVKTSSGKEFSFQVERGRDVKYIKRRIAKKEKQFDDLEEQELVCNGERLEDQKLIDDIC 176
I VKT +GK + +VE + +K +I KE D +Q L+ G++LED + + D
Sbjct: 3 IFVKTLTGKTITLEVESSDTIDNVKAKIQDKEGIPPD--QQRLIFAGKQLEDGRTLADYN 60
Query: 177 CKHNDAVVHLFVR------KRNAKVHRRP 199
+ ++ +HL +R KR K + +P
Sbjct: 61 IQ-KESTLHLVLRLRGGAKKRKKKTYTKP 88
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.137 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,474,335
Number of Sequences: 26719
Number of extensions: 373090
Number of successful extensions: 1151
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 989
Number of HSP's gapped (non-prelim): 88
length of query: 384
length of database: 11,318,596
effective HSP length: 101
effective length of query: 283
effective length of database: 8,619,977
effective search space: 2439453491
effective search space used: 2439453491
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC147435.2


