

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147435.10 + phase: 0 /pseudo
(612 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
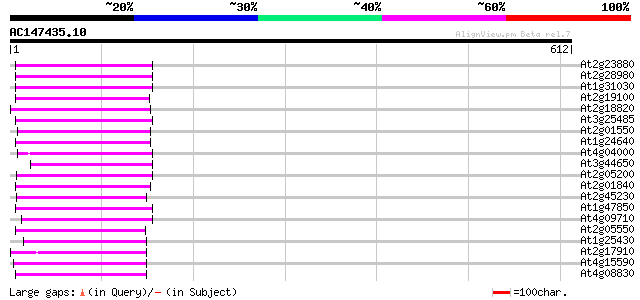
Score E
Sequences producing significant alignments: (bits) Value
At2g23880 putative non-LTR retroelement reverse transcriptase 102 6e-22
At2g28980 putative non-LTR retroelement reverse transcriptase 99 5e-21
At1g31030 F17F8.5 96 6e-20
At2g19100 putative non-LTR retroelement reverse transcriptase 94 3e-19
At2g18820 putative non-LTR retroelement reverse transcriptase 92 1e-18
At3g25485 unknown protein 91 1e-18
At2g01550 putative non-LTR retroelement reverse transcriptase 91 2e-18
At1g24640 hypothetical protein 91 2e-18
At4g04000 putative reverse transcriptase 90 4e-18
At3g44650 putative protein 90 4e-18
At2g05200 putative non-LTR retroelement reverse transcriptase 89 9e-18
At2g01840 putative non-LTR retroelement reverse transcriptase 89 9e-18
At2g45230 putative non-LTR retroelement reverse transcriptase 88 2e-17
At1g47850 hypothetical protein 88 2e-17
At4g09710 RNA-directed DNA polymerase -like protein 86 8e-17
At2g05550 putative non-LTR retroelement reverse transcriptase 86 8e-17
At1g25430 hypothetical protein 84 3e-16
At2g17910 putative non-LTR retroelement reverse transcriptase 83 5e-16
At4g15590 reverse transcriptase like protein 82 9e-16
At4g08830 putative protein 81 1e-15
>At2g23880 putative non-LTR retroelement reverse transcriptase
Length = 1216
Score = 102 bits (254), Expect = 6e-22
Identities = 63/150 (42%), Positives = 82/150 (54%), Gaps = 1/150 (0%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
R SE D+ LL EEI+ V+ +KSPG D Y F K W ++ +EV I
Sbjct: 161 RCSEDDHRLLTRVVTGEEIKKVIFSMPKDKSPGPDGYTSEFYKASWEIIGDEVIIAIQSF 220
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
A LPK + + +ALIPK E+KDYR IS LYK ++K+LA RL +L I
Sbjct: 221 FAKGFLPKGVNSTILALIPKKKEAREIKDYRPISCCNVLYKAISKILANRLKRILPKFIV 280
Query: 127 TTQSAFLKGTNLVDGVLVVNELV-DYAKKS 155
QSAF+K L++ VL+ ELV DY K S
Sbjct: 281 GNQSAFVKDRLLIENVLLATELVKDYHKDS 310
>At2g28980 putative non-LTR retroelement reverse transcriptase
Length = 1529
Score = 99.4 bits (246), Expect = 5e-21
Identities = 61/150 (40%), Positives = 80/150 (52%), Gaps = 1/150 (0%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
R S D ++L EEI+ V+ NKSPG D Y F K W L +
Sbjct: 737 RCSVTDQNILTREVTGEEIQKVLFAMPNNKSPGPDGYTSEFFKATWSLTGPDFIAAIQSF 796
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
LPK + A +ALIPK +E+KDYR IS LYK+++K+LA RL +L S I
Sbjct: 797 FVKGFLPKGLNATILALIPKKDEAIEMKDYRPISCCNVLYKVISKILANRLKLLLPSFIL 856
Query: 127 TTQSAFLKGTNLVDGVLVVNELV-DYAKKS 155
QSAF+K L++ VL+ ELV DY K+S
Sbjct: 857 QNQSAFVKERLLMENVLLATELVKDYHKES 886
>At1g31030 F17F8.5
Length = 872
Score = 95.9 bits (237), Expect = 6e-20
Identities = 55/150 (36%), Positives = 82/150 (54%), Gaps = 1/150 (0%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
R + DN++L EEI+ V+ +KSPG D Y F K W ++ E +
Sbjct: 87 RCTNSDNEMLTREVSSEEIKTVLFSMPKDKSPGPDGYTSEFYKATWDIIGQEFTLPVQSF 146
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
LPK + + +ALIPK + E++DYR IS LYK+++K++A RL +L I+
Sbjct: 147 FQKGFLPKGINSIILALIPKKLAAKEMRDYRPISCCNVLYKVISKIIANRLKLLLPRFIA 206
Query: 127 TTQSAFLKGTNLVDGVLVVNELV-DYAKKS 155
QSAF+K L++ +L+ ELV DY K S
Sbjct: 207 ENQSAFVKDRLLIENLLLATELVKDYHKDS 236
>At2g19100 putative non-LTR retroelement reverse transcriptase
Length = 1447
Score = 93.6 bits (231), Expect = 3e-19
Identities = 51/146 (34%), Positives = 78/146 (52%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
R SE +++LL EEI+ V+ +KSPG D + F K W ++ NE +
Sbjct: 848 RCSETEHELLTRVVTAEEIKKVLFSMPNDKSPGPDGFTSEFFKATWEILGNEFILAIQSF 907
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
A LPK + +ALIPK E+KDYR IS +YK+++K++A RL V I+
Sbjct: 908 FAKGFLPKGINTTILALIPKKKEAKEMKDYRPISCCNVIYKVISKIIANRLKLVPPKFIA 967
Query: 127 TTQSAFLKGTNLVDGVLVVNELVDYA 152
QSAF+K L++ VL+ + ++
Sbjct: 968 GNQSAFVKDRLLIENVLLATDTASFS 993
>At2g18820 putative non-LTR retroelement reverse transcriptase
Length = 1412
Score = 91.7 bits (226), Expect = 1e-18
Identities = 58/153 (37%), Positives = 82/153 (52%), Gaps = 1/153 (0%)
Query: 2 GVNFNRLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRI 61
G+ R S + +LLVA E+ V NKSPG D Y F +E W ++ EV +
Sbjct: 701 GILQYRYSLHEQNLLVAEITEAEVMKVFFSIPLNKSPGPDGYTVEFFRETWSVIGQEVTM 760
Query: 62 MFDQIHANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVL 121
LPK + + +ALIPK + E+KDYR IS LYK ++K+LA RL +L
Sbjct: 761 AIKSFFTYGFLPKGLNSTILALIPKRTYAKEMKDYRPISCCNVLYKAISKLLANRLKCLL 820
Query: 122 SSVISTTQSAFLKGTNLVDGVLVVNELV-DYAK 153
I+ QSAF+ L++ +L+ +ELV DY K
Sbjct: 821 PEFIAPNQSAFISDRLLMENLLLASELVKDYHK 853
>At3g25485 unknown protein
Length = 979
Score = 91.3 bits (225), Expect = 1e-18
Identities = 53/150 (35%), Positives = 81/150 (53%), Gaps = 1/150 (0%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
R + D L+ P EEI+ V+ +KSPG D + F K W ++ E +
Sbjct: 356 RCTAEDRSRLMEPVTGEEIKTVLFFMSSDKSPGPDGFTTEFYKATWEIIGAEFIVAVKSF 415
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
LPK + + +ALIPK E+KDYR IS +YK+++K++A+RL +L I+
Sbjct: 416 FEKGFLPKGVNSTILALIPKKFETKEMKDYRPISCCNVIYKVISKLIAKRLKEILPQFIA 475
Query: 127 TTQSAFLKGTNLVDGVLVVNELV-DYAKKS 155
QSAF+K L+ +L+ E+V DY K+S
Sbjct: 476 GNQSAFVKDRLLIQNLLLATEIVKDYHKES 505
>At2g01550 putative non-LTR retroelement reverse transcriptase
Length = 1449
Score = 90.9 bits (224), Expect = 2e-18
Identities = 53/146 (36%), Positives = 77/146 (52%), Gaps = 1/146 (0%)
Query: 9 SEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIHA 68
S + D+L A +EI + +KSPG D Y F K W ++ E +
Sbjct: 857 SPAEKDMLTASVSAKEIRGALFSMPNDKSPGPDGYTSEFYKRAWDIIGAEFVLAVKSFFE 916
Query: 69 NELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVISTT 128
LPK + +ALIPK E+KDYR IS +YK+++K++A RL VL + I+
Sbjct: 917 KGFLPKGVNTTILALIPKKLEAKEMKDYRPISCCNVIYKVISKIIANRLKHVLPNFIAGN 976
Query: 129 QSAFLKGTNLVDGVLVVNELV-DYAK 153
QSAF+K L++ +L+ ELV DY K
Sbjct: 977 QSAFVKDRLLIENLLLATELVKDYHK 1002
>At1g24640 hypothetical protein
Length = 1270
Score = 90.5 bits (223), Expect = 2e-18
Identities = 51/147 (34%), Positives = 79/147 (53%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
R+SEV N+ LV +EI+ V +PG D + F + +W + N+V +
Sbjct: 378 RVSEVMNESLVGEVSAQEIKEAVFSIKPASAPGPDGMSALFFQHYWSTVGNQVTSEVKKF 437
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
A+ ++P + LIPK P E+ D R ISL LYK+++K++A+RL L ++S
Sbjct: 438 FADGIMPAEWNYTHLCLIPKTQHPTEMVDLRPISLCSVLYKIISKIMAKRLQPWLPEIVS 497
Query: 127 TTQSAFLKGTNLVDGVLVVNELVDYAK 153
TQSAF+ + D +LV +ELV K
Sbjct: 498 DTQSAFVSERLITDNILVAHELVHSLK 524
>At4g04000 putative reverse transcriptase
Length = 1077
Score = 89.7 bits (221), Expect = 4e-18
Identities = 49/147 (33%), Positives = 83/147 (56%), Gaps = 1/147 (0%)
Query: 9 SEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIHA 68
++++N L+ P + EI+ +K+PG D ++ +F + W + E+
Sbjct: 394 TDMNNTLISLPT-VAEIKQACLSIHPDKAPGRDGFSASFFQSNWSTVSEEITAEVQAFFI 452
Query: 69 NELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVISTT 128
+E+LP+ + V LIPK+ +P ++ DYR I+L YK++AK+LA+RL +L S IS
Sbjct: 453 SEILPQKINHTHVRLIPKIPAPQKVSDYRPIALCSVYYKIIAKLLAKRLQPILHSCISEN 512
Query: 129 QSAFLKGTNLVDGVLVVNELVDYAKKS 155
QSAF+ + D VL+ +E + Y K S
Sbjct: 513 QSAFVPQRAISDNVLITHEALHYLKNS 539
>At3g44650 putative protein
Length = 762
Score = 89.7 bits (221), Expect = 4e-18
Identities = 52/134 (38%), Positives = 76/134 (55%), Gaps = 1/134 (0%)
Query: 23 EEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIHANELLPKSMLAYFVA 82
EEI+ V+ +KSPG D + F KE W ++ E + A LPK + + +A
Sbjct: 9 EEIKKVLFSMPNDKSPGPDGFTSEFFKESWEILGPEFILAIQSFFALGFLPKGVNSTILA 68
Query: 83 LIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVISTTQSAFLKGTNLVDGV 142
LIPK E+KDYR IS +YK+++K+LA RL +L I+ QS+F+K L++ V
Sbjct: 69 LIPKKLESKEMKDYRPISCCNVMYKVISKILANRLKLLLPQFIAGNQSSFVKDRLLIENV 128
Query: 143 LVVNELV-DYAKKS 155
L+ +LV DY K S
Sbjct: 129 LLATDLVKDYHKDS 142
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 88.6 bits (218), Expect = 9e-18
Identities = 45/148 (30%), Positives = 82/148 (55%)
Query: 8 LSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIH 67
+S+ DND L + EE++ V + +K+PG D + F +W+++ +V
Sbjct: 311 VSQGDNDFLTRIPNDEEVKDAVFSINASKAPGPDGFTAGFYHSYWHIISTDVGREIRLFF 370
Query: 68 ANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIST 127
++ P+ M + LIPK P ++ DYR I+L YK++AK++ +R+ +L +IS
Sbjct: 371 TSKNFPRRMNETHIRLIPKDLGPRKVADYRPIALCNIFYKIVAKIMTKRMQLILPKLISE 430
Query: 128 TQSAFLKGTNLVDGVLVVNELVDYAKKS 155
QSAF+ G + D VL+ +E++ + + S
Sbjct: 431 NQSAFVPGRVISDNVLITHEVLHFLRTS 458
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 88.6 bits (218), Expect = 9e-18
Identities = 48/147 (32%), Positives = 81/147 (54%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
+++E N L+ +E+ V +++PG D + AF W L+ N+V +M
Sbjct: 777 KVTEQMNHELLQSVTDQEVRDAVFAIGADRAPGFDGFTAAFYHHLWDLIGNDVCLMVRHF 836
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
++++ + + LIPK+ P + DYR ISL + YK+++K+L +RL L VIS
Sbjct: 837 FESDVMDNQINQTQICLIPKIIDPKHMSDYRPISLCTASYKIISKILIKRLKQCLGDVIS 896
Query: 127 TTQSAFLKGTNLVDGVLVVNELVDYAK 153
+Q+AF+ G N+ D VLV +EL+ K
Sbjct: 897 DSQAAFVPGQNISDNVLVAHELLHSLK 923
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 87.8 bits (216), Expect = 2e-17
Identities = 46/142 (32%), Positives = 81/142 (56%)
Query: 8 LSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIH 67
+S+ N+ L+AP EE++ + +K PG D N ++FW M +++ M
Sbjct: 416 VSDQMNNNLLAPITKEEVQRATFSINPHKCPGPDGMNGFLYQQFWETMGDQITEMVQAFF 475
Query: 68 ANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIST 127
+ + + M + LIPK+ ++ D+R ISL +YK++ K++A RL +L S+IS
Sbjct: 476 RSGSIEEGMNKTNICLIPKILKAEKMTDFRPISLCNVIYKVIGKLMANRLKKILPSLISE 535
Query: 128 TQSAFLKGTNLVDGVLVVNELV 149
TQ+AF+KG + D +L+ +EL+
Sbjct: 536 TQAAFVKGRLISDNILIAHELL 557
>At1g47850 hypothetical protein
Length = 799
Score = 87.8 bits (216), Expect = 2e-17
Identities = 53/150 (35%), Positives = 75/150 (49%), Gaps = 1/150 (0%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
R S D D+L EE + V+ NK PG D Y F K W + +
Sbjct: 13 RCSATDQDMLTREVTSEENQKVLFAMPSNKFPGPDGYTSEFFKATWSITGQDFIAAIKSF 72
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
LPK + A +ALIPK ++DYR IS +YK+++K++A RL +L + I
Sbjct: 73 FIKGFLPKGLNATILALIPKKDEATLMRDYRPISCCNVIYKVISKIIANRLKVMLPTFIL 132
Query: 127 TTQSAFLKGTNLVDGVLVVNELV-DYAKKS 155
QSAF++ L++ VL+ ELV DY K S
Sbjct: 133 QNQSAFVRERLLIENVLLATELVKDYHKDS 162
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 85.5 bits (210), Expect = 8e-17
Identities = 46/143 (32%), Positives = 80/143 (55%)
Query: 13 NDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIHANELL 72
N+ L+ + EI+ + +K+PG D ++ +F +W +++ +V + L
Sbjct: 362 NEELIKISSLLEIKEALFSISADKAPGPDGFSASFFHAYWDIIEADVSRDIRSFFVDSCL 421
Query: 73 PKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVISTTQSAF 132
+ V LIPK+S+P ++ DYR I+L YK++AK+L RRL LS +IS QSAF
Sbjct: 422 SPRLNETHVTLIPKISAPRKVSDYRPIALCNVQYKIVAKILTRRLQPWLSELISLHQSAF 481
Query: 133 LKGTNLVDGVLVVNELVDYAKKS 155
+ G + D VL+ +E++ + + S
Sbjct: 482 VPGRAIADNVLITHEILHFLRVS 504
>At2g05550 putative non-LTR retroelement reverse transcriptase
Length = 977
Score = 85.5 bits (210), Expect = 8e-17
Identities = 48/142 (33%), Positives = 77/142 (53%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
+++E D +L EEI+ VV +KS G D Y F K W ++ E +
Sbjct: 555 KVAEEDKGMLTTEVTKEEIKRVVFSMPKDKSLGPDGYRTEFYKATWDIIGEEFVLAIKSF 614
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
LPK + + +ALIPK +E+KDYR IS +YK+++K++A RL +L + I+
Sbjct: 615 FEKGFLPKGVNSTILALIPKKLEEIEMKDYRPISCCNVIYKVISKIIANRLKRLLPNFIA 674
Query: 127 TTQSAFLKGTNLVDGVLVVNEL 148
QSAF++ L++ +L+ EL
Sbjct: 675 GNQSAFVQDRLLLENLLLTTEL 696
>At1g25430 hypothetical protein
Length = 1213
Score = 83.6 bits (205), Expect = 3e-16
Identities = 49/134 (36%), Positives = 74/134 (54%)
Query: 16 LVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIHANELLPKS 75
L + F E+I + NKS G D + F + W ++ EV + ++ L K
Sbjct: 443 LESTFSNEDIRAALFSLPRNKSCGPDGFTAEFFIDSWSIVGAEVTDAIKEFFSSGCLLKQ 502
Query: 76 MLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVISTTQSAFLKG 135
A + LIPK+ +P D+R IS L +LYK++A++L RL +LS VIS+ QSAFL G
Sbjct: 503 WNATTIVLIPKIVNPTCTSDFRPISCLNTLYKVIARLLTDRLQRLLSGVISSAQSAFLPG 562
Query: 136 TNLVDGVLVVNELV 149
+L + VL+ +LV
Sbjct: 563 RSLAENVLLATDLV 576
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 82.8 bits (203), Expect = 5e-16
Identities = 45/149 (30%), Positives = 82/149 (54%), Gaps = 1/149 (0%)
Query: 1 DGVNFNRLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVR 60
+G+ SE++++L+ +E V + +PG D + F ++ W L+K+++
Sbjct: 388 EGLQAKVTSEMNHNLIQEVTELEVYNAVF-SINKESAPGPDGFTALFFQQHWDLVKHQIL 446
Query: 61 IMFDQIHANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGV 120
+LP+ + LIPK++SP + D R ISL LYK+++K+L +RL
Sbjct: 447 TEIFGFFETGVLPQDWNHTHICLIPKITSPQRMSDLRPISLCSVLYKIISKILTQRLKKH 506
Query: 121 LSSVISTTQSAFLKGTNLVDGVLVVNELV 149
L +++STTQSAF+ + D +LV +E++
Sbjct: 507 LPAIVSTTQSAFVPQRLISDNILVAHEMI 535
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 82.0 bits (201), Expect = 9e-16
Identities = 45/145 (31%), Positives = 75/145 (51%)
Query: 5 FNRLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFD 64
F RL+ + + L PF +E+ V VR K+PG D Y F ++ W + V
Sbjct: 229 FPRLTREEKNNLNRPFTRDEVVVAVRSMGRFKAPGPDGYQPVFYQQCWETVGESVSKFVM 288
Query: 65 QIHANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSV 124
+ + +LPKS + L+ KV+ P + +R +SL L+K++ K++ RL V+S +
Sbjct: 289 EFFESGVLPKSTNDVLLVLLAKVAKPERITQFRPVSLCNVLFKIITKMMVIRLKNVISKL 348
Query: 125 ISTTQSAFLKGTNLVDGVLVVNELV 149
I Q++F+ G D ++VV E V
Sbjct: 349 IGPAQASFIPGRLSFDNIVVVQEAV 373
>At4g08830 putative protein
Length = 947
Score = 81.3 bits (199), Expect = 1e-15
Identities = 46/143 (32%), Positives = 74/143 (51%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
RL+ + L PF E+EV +R K+PG D Y F + W ++ N V
Sbjct: 173 RLTREEVLSLNKPFLATEVEVAIRSMGKYKAPGPDGYQPIFYQSCWEVVGNSVIRFALDF 232
Query: 67 HANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIS 126
++ +LP V LIPKV P + +R ISL +++K++ K++ RL V+ +I
Sbjct: 233 FSSGILPPQTNDALVVLIPKVPKPERMNQFRPISLCNAIFKMITKMMVLRLKKVIGKLIG 292
Query: 127 TTQSAFLKGTNLVDGVLVVNELV 149
+QS+F+ G +D ++VV E V
Sbjct: 293 PSQSSFIPGRLSLDNIVVVQEAV 315
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.358 0.165 0.605
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,815,446
Number of Sequences: 26719
Number of extensions: 460620
Number of successful extensions: 2588
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 46
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 2461
Number of HSP's gapped (non-prelim): 107
length of query: 612
length of database: 11,318,596
effective HSP length: 105
effective length of query: 507
effective length of database: 8,513,101
effective search space: 4316142207
effective search space used: 4316142207
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC147435.10


