

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147431.9 - phase: 0
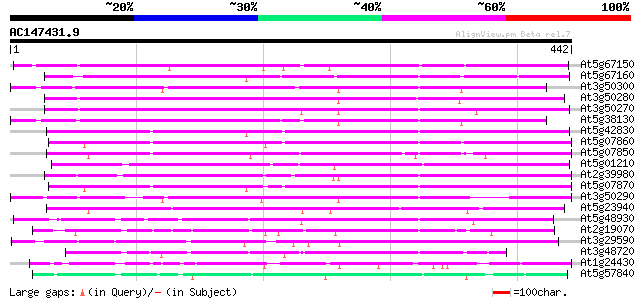
(442 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g67150 anthranilate N-hydroxycinnamoyl/benzoyltransferase-lik... 300 8e-82
At5g67160 anthranilate N-hydroxycinnamoyl/benzoyltransferase-lik... 293 1e-79
At3g50300 anthranilate N-hydroxycinnamoyl/benzoyltransferase - l... 291 5e-79
At3g50280 anthranilate N-hydroxycinnamoyl/benzoyltransferase - l... 287 7e-78
At3g50270 anthranilate N-hydroxycinnamoyl/benzoyltransferase - l... 280 9e-76
At5g38130 anthranilate N-benzoyltransferase -like protein 278 6e-75
At5g42830 N-hydroxycinnamoyl/benzoyltransferase-like protein 260 9e-70
At5g07860 N-hydroxycinnamoyl/benzoyltransferase - like protein 247 8e-66
At5g07850 proanthranilate N-benzoyltransferase -like protein 241 4e-64
At5g01210 anthranilate N-benzoyltransferase - like protein 239 3e-63
At2g39980 putative anthocyanin 5-aromatic acyltransferase 233 2e-61
At5g07870 N-hydroxycinnamoyl/benzoyltransferase - like protein 229 2e-60
At3g50290 putative protein 221 8e-58
At5g23940 acyltransferase 200 1e-51
At5g48930 anthranilate N-benzoyltransferase 117 1e-26
At2g19070 putative anthranilate N-hydroxycinnamoyl/benzoyltransf... 116 3e-26
At3g29590 Anthocyanin 5-aromatic acyltransferase, putative 104 1e-22
At3g48720 unknown protein 100 2e-21
At1g24430 similar to acetyl-CoA:benzylalcoholo acetyltransferase 99 4e-21
At5g57840 N-hydroxycinnamoyl/benzoyltransferase 97 1e-20
>At5g67150 anthranilate N-hydroxycinnamoyl/benzoyltransferase-like
protein
Length = 448
Score = 300 bits (769), Expect = 8e-82
Identities = 176/449 (39%), Positives = 263/449 (58%), Gaps = 20/449 (4%)
Query: 4 IQVLSTTPVHAPNHNPCDPNYHHTIDLTPWDLQFLPFGVNQKGLIYHHPSNLDTSNQIEH 63
+ V+S + V ++N + + I LTPWDL FL Q+GL++ P ++ I
Sbjct: 5 VVVISKSIVRPESYN--EESDRVKIHLTPWDLFFLRSEYPQRGLLFPQPD--PETHIISQ 60
Query: 64 LKHSLSSTLEFFPPFTGRLKIKEHED-NTISCSITCNNVGALFVHAAAENTSVDDIIGAT 122
LK SLS L+ F PF GRL E+ED T S + C+ G F+HA+A++ SV D++
Sbjct: 61 LKSSLSVALKIFYPFAGRLVKVENEDAGTASFYVDCDGSGVKFIHASAKSVSVSDVLEPV 120
Query: 123 YH--PKILHSFFPFNGVKNYEGTSMPLLAVQVTELVNGIFIACTVNHVVADGTSIWHFIN 180
P+ L+ FFP NGV++ EG S L+A QVTEL +G+FI NH+VADG+S W F N
Sbjct: 121 DSNVPEFLNRFFPANGVRSCEGISESLIAFQVTELKDGVFIGFGYNHIVADGSSFWSFFN 180
Query: 181 SWAKISNGSLEISKIPSF-----ERWFSDDIQPPIQFPF--TIEPQNIHHKEEKLNLYER 233
+W++I + F WF D I+ PI+ P T P + L E+
Sbjct: 181 TWSEICFNGFDADHRRKFPPLLLRGWFLDGIEYPIRIPLPETETPNRVVVTSSLLQ--EK 238
Query: 234 MFHFTKENIAKLKFKAN--LEAGTKNISSLQALFTHIWRSIIRSKNLDPEEEVHYVVVIG 291
+F T NI++LK KAN +++ + ISSLQA+ ++WRSIIR+ L+PEE +H +++
Sbjct: 239 IFRVTSRNISELKAKANGEVDSDDRKISSLQAVSAYMWRSIIRNSGLNPEEVIHCKLLVD 298
Query: 292 VRPRLIPLLKKDYFGNALIDPAVSMKVGDLLQDNGLGKGALEMNKMIALQSDEMLKNEYE 351
+R RL P L+K+ FGN + V+ V ++L NGLG AL++NK + Q++E + E
Sbjct: 299 MRGRLNPPLEKECFGNVVGFATVTTTVAEMLH-NGLGWAALQINKTVGSQTNEEFREFAE 357
Query: 352 SWLKTPSFITDAKFNCKTLVGSSSPWFDVYGNDFGWGKPVGVRTGGANKENGNICVFQGV 411
+W+K PS I +AK ++ +SSP F+VYGNDFGWGKP+ VR G N +G + + G+
Sbjct: 358 NWVKKPS-ILNAKAFSNSITIASSPRFNVYGNDFGWGKPIAVRAGPGNTTDGKLIAYPGI 416
Query: 412 EKDSMDLEVCLSHETLEAIGNDPEFMNVV 440
E+ +++ + CLS LE + D EF+ V
Sbjct: 417 EEGNIEFQTCLSSSVLEKLSTDEEFLKHV 445
>At5g67160 anthranilate N-hydroxycinnamoyl/benzoyltransferase-like
protein
Length = 434
Score = 293 bits (750), Expect = 1e-79
Identities = 173/419 (41%), Positives = 243/419 (57%), Gaps = 18/419 (4%)
Query: 28 IDLTPWDLQFLPFGVNQKGLIYHHPSNLDTSNQIEHLKHSLSSTLEFFPPFTGRLKIKEH 87
I LTPWDL L FG Q+GL++H +++ L+ SLS L+ F P GRL ++
Sbjct: 27 IQLTPWDLSRLRFGYLQRGLLFH-------KIEVKQLQASLSVALDRFYPLAGRLVKLKN 79
Query: 88 EDNTISCSITCNNVGALFVHAAAENTSVDDIIGATYH-PKILHSFFPFNGVKNYEGTSMP 146
+D+T+S I+C+ G FVHA A+N + D++ + P SFFP G+KNY G S
Sbjct: 80 DDDTVSFFISCDGSGVEFVHAVAKNIELSDVLELSGSVPGFFASFFPATGIKNYHGVSRS 139
Query: 147 LLAVQVTELVNGIFIACTVNHVVADGTSIWHFINSWAKI----SNGSLEISKIPSFERWF 202
LL VQVTE+ +G+FI N VAD TSIW FIN+W++I S+GS + + WF
Sbjct: 140 LLMVQVTEMKDGVFIGFGYNSTVADATSIWKFINAWSEICSKDSSGSQTFQRRLHLKGWF 199
Query: 203 SDDIQPPIQFPFTIEPQNIHHKEEKLNLYERMFHFTKENIAKLKFKANLEAGTKNISSLQ 262
D+I PI P E + + NL E+MFH TKEN+ KL KAN EA K ISS+Q
Sbjct: 200 FDEIDYPIHIP-DPETKPTSYVTTPTNLQEKMFHVTKENVLKLDAKANDEADQK-ISSIQ 257
Query: 263 ALFTHIWRSIIRSKNLDPEEEVHYVVVIGVRPRLIPLLKKDYFGNALIDPAVSMKVGDLL 322
A+ +IWRS+++ + EEE H + I +R RL P L+++ FGN ++ VG+LL
Sbjct: 258 AVLAYIWRSMVKHSGMSREEETHCRLPINMRQRLNPPLEEECFGNVSQTGIATVTVGELL 317
Query: 323 QDNGLGKGALEMNKMIALQSDEMLKNEYESWLKTPSFITDAKFNCKTLVGSSSPWFDVYG 382
D+GLG A+++N M Q+DE K E+W+K + K LV ++S FDVY
Sbjct: 318 -DHGLGWAAMQINNMELSQTDEKAKAFAENWVK--NIKIPVSVGSKDLVVTNSHRFDVYC 374
Query: 383 NDFGWGKPVGVRTGGANKENGNICVFQGVEKDSMDLEVCLSHETLEAIGNDPEFMNVVS 441
NDFGWGKP+ R G NG + VF+G+ + S+D + CL + +E + D EF VS
Sbjct: 375 NDFGWGKPIAARAGPPYL-NGRLVVFKGIGEASLDFQACLLPQVVEKLVKDAEFNEYVS 432
>At3g50300 anthranilate N-hydroxycinnamoyl/benzoyltransferase - like
protein
Length = 448
Score = 291 bits (745), Expect = 5e-79
Identities = 182/435 (41%), Positives = 252/435 (57%), Gaps = 22/435 (5%)
Query: 1 MSSIQVLSTTPVHAPNHNPCDPNYHHTIDLTPWDLQFLPFGVNQKGLIYHHPSNLDTSNQ 60
M + V+S++ V N I LTP DL L Q+ L++ P +
Sbjct: 1 MDDVTVISSSIVQPGNARHSG---REKIHLTPSDLSLLYLNYPQRSLLF--PKPYPETRF 55
Query: 61 IEHLKHSLSSTLEFFPPFTGRL-KIKEHEDNTISCSITCNNV-GALFVHAAAENTSVDDI 118
I LK SLS++LE + P GRL K+ HEDNT+S I C++ G FVHA AE+ SV DI
Sbjct: 56 ISRLKSSLSTSLEIYFPLAGRLVKVNNHEDNTVSFYIDCDDGRGVKFVHAIAESVSVSDI 115
Query: 119 I---GATYHPKILHSFFPFNGVKNYEGTSMPLLAVQVTELVNGIFIACTVNHVVADGTSI 175
+ G+ P FFP NGV++ +G S PLLA+QVTE+ +GI I+ NH+VADG+S+
Sbjct: 116 LQPHGSV--PDFFRLFFPMNGVRSIDGLSEPLLALQVTEIKDGIVISFGYNHLVADGSSM 173
Query: 176 WHFINSWAKIS-NGSLEISKIPSFERWFSDDIQPPIQFPFT-IEPQNIHHKEEKLNLYER 233
W+FI+ W+K+ NG EI + WF D+I PI P + IE + ++E ++ ER
Sbjct: 174 WNFIHVWSKLCLNGQWEIHQPLVLRGWFLDNIDFPIHIPASEIETERARNRE--VSTKER 231
Query: 234 MFHFTKENIAKLKFKANLEAGTKN--ISSLQALFTHIWRSIIRSKNLDPEEEVHYVVVIG 291
+FHFTKE ++ LK KAN E G+ + ISSLQA+ H+WRSI+R L+ EEE V
Sbjct: 232 VFHFTKEKLSDLKAKANDEIGSSDIKISSLQAVLAHLWRSIVRHSGLNQEEESRCGVAAD 291
Query: 292 VRPRLIPLLKKDYFGNALIDPAVSMKVGDLLQDNGLGKGALEMNKMIALQSDEMLKNEYE 351
R RL P L KD FGN + VGDLL D GLG AL++NK + L ++E + E
Sbjct: 292 FRQRLNPPLDKDCFGNVANLGLTTATVGDLL-DRGLGWAALQINKTVRLHTNENFRTFSE 350
Query: 352 SWL---KTPSFITDAKFNCKTLVGSSSPWFDVYGNDFGWGKPVGVRTGGANKENGNICVF 408
+W+ K P ++ + S+SPWF VY NDFG GKP+ VR G AN G + VF
Sbjct: 351 NWVRNGKIPRIDVRSRMGDHGFMVSNSPWFQVYDNDFGLGKPMAVRAGPANGIGGKLVVF 410
Query: 409 QGVEKDSMDLEVCLS 423
+G+E+ S+D+ L+
Sbjct: 411 RGIEEGSIDVHAILT 425
>At3g50280 anthranilate N-hydroxycinnamoyl/benzoyltransferase - like
protein
Length = 443
Score = 287 bits (735), Expect = 7e-78
Identities = 166/418 (39%), Positives = 245/418 (57%), Gaps = 11/418 (2%)
Query: 28 IDLTPWDLQFLPFGVNQKGLIYHHPSNLDTSNQIEHLKHSLSSTLEFFPPFTGRL-KIKE 86
I LTP+DL L Q+GL++ P ++ I L+ SLSS L+ + PF GRL K++
Sbjct: 25 IHLTPFDLNLLYVDYTQRGLLFPKPD--PETHFISRLRTSLSSALDIYFPFAGRLNKVEN 82
Query: 87 HEDNTISCSITCNNVGALFVHAAAENTSVDDIIGATYH-PKILHSFFPFNGVKNYEGTSM 145
HED T+S I C+ GA F+HA +++ SV D++ P F+P NGVK+ +G S
Sbjct: 83 HEDETVSFYINCDGSGAKFIHAVSDSVSVSDLLRPDGSVPDFFRIFYPMNGVKSIDGLSE 142
Query: 146 PLLAVQVTELVNGIFIACTVNHVVADGTSIWHFINSWAKI-SNGSLEISKIPSFERWFSD 204
PLLA+QVTE+ +G+FI NH+VADG SIW+F +W+KI SNG E + + + F D
Sbjct: 143 PLLALQVTEMRDGVFIGFGYNHMVADGASIWNFFRTWSKICSNGQRENLQPLALKGLFVD 202
Query: 205 DIQPPIQFPFTIEPQNIHHKEEKLNLYERMFHFTKENIAKLKFKANLEAGTKN--ISSLQ 262
+ PI P + + +E ER+FHFTK NI+ LK K N E G ++ +SSLQ
Sbjct: 203 GMDFPIHIPVSDTETSPPSRELSPTFKERVFHFTKRNISDLKAKVNGEIGLRDHKVSSLQ 262
Query: 263 ALFTHIWRSIIRSKNLDPEEEVHYVVVIGVRPRLIPLLKKDYFGNALIDPAVSMKVGDLL 322
A+ H+WRSIIR L+ EE+ V + +R RL P L K+ FG+ + + V+ VG+ L
Sbjct: 263 AVSAHMWRSIIRHSGLNQEEKTRCFVAVDLRQRLNPPLDKECFGHVIYNSVVTTTVGE-L 321
Query: 323 QDNGLGKGALEMNKMIALQSDEMLKNEYESW---LKTPSFITDAKFNCKTLVGSSSPWFD 379
D GLG L++N M+ ++E + E+W +K +K +++ SSSP F+
Sbjct: 322 HDQGLGWAFLQINNMLRSLTNEDYRIYAENWVRNMKIQKSGLGSKMTRDSVIVSSSPRFE 381
Query: 380 VYGNDFGWGKPVGVRTGGANKENGNICVFQGVEKDSMDLEVCLSHETLEAIGNDPEFM 437
VY NDFGWGKP+ VR G +N +G + F+G+E+ +D+ L + L + D EF+
Sbjct: 382 VYDNDFGWGKPIAVRAGPSNSISGKLVFFRGIEEGCIDVHAFLLPDVLVKLLADVEFL 439
>At3g50270 anthranilate N-hydroxycinnamoyl/benzoyltransferase - like
protein
Length = 450
Score = 280 bits (717), Expect = 9e-76
Identities = 167/424 (39%), Positives = 250/424 (58%), Gaps = 12/424 (2%)
Query: 28 IDLTPWDLQFLPFGVNQKGLIYHHPSNLDTSNQIEHLKHSLSSTLEFFPPFTGRL-KIKE 86
I LTP+DL L F Q+GL++ P + D I LK SLS LE + PF GRL K +
Sbjct: 27 IHLTPFDLSLLQFDYPQRGLLFSKPDS-DFHLIISRLKDSLSLALEIYFPFAGRLVKTEN 85
Query: 87 HEDNTISCSITCNNVGALFVHAAAENTSVDDIIGATYH-PKILHSFFPFNGVKNYEGTSM 145
ED+T+S I C+ GA F+H A++ SV DI+ P L FFP + K+ +G S+
Sbjct: 86 LEDDTVSFFIDCDGSGARFLHTEAKSVSVSDILQPDGSVPDFLKYFFPADDFKSCDGVSL 145
Query: 146 PLLAVQVTELVNGIFIACTVNHVVADGTSIWHFINSWAKI-SNGSLEISKIPSFERWFSD 204
PLL +QVTE+ +G+FI+ NH+VADG S+W F ++W+KI S GS K + WF +
Sbjct: 146 PLLVIQVTEMKDGVFISFCYNHMVADGVSMWSFFDTWSKICSTGSCFNYKPLVLKGWFLE 205
Query: 205 DIQPPIQFPFTIEPQNIHHKEEKL--NLYERMFHFTKENIAKLKFKANLEAGTKN---IS 259
I PI P + ++ +E E +FHFTK+NI+ LK KAN E + + +S
Sbjct: 206 GIDYPIHVPVSEAERSPPSREPSSVPITKEWIFHFTKKNISDLKAKANSEIASSSDLEVS 265
Query: 260 SLQALFTHIWRSIIRSKNLDPEEEVHYVVVIGVRPRLIPLLKKDYFGNALIDPAVSMKVG 319
SLQA+ H+WRSIIR + E++ H +V+ +R R+ P L+KD FGN + + V
Sbjct: 266 SLQAVSAHMWRSIIRHSGVSREQKTHCRLVVDLRQRVNPPLEKDCFGNMVYLSSAITTVE 325
Query: 320 DLLQDNGLGKGALEMNKMIALQSDEMLKNEYESWLKTPSFITDAKFN--CKTLVGSSSPW 377
+LL D GLG+ AL++ K+++ Q++E K+ E W++ + + T+V +SSP
Sbjct: 326 ELL-DRGLGEAALQIRKLVSSQTNETCKSFAEDWVRNIKNLNSGIGSKVGNTIVIASSPR 384
Query: 378 FDVYGNDFGWGKPVGVRTGGANKENGNICVFQGVEKDSMDLEVCLSHETLEAIGNDPEFM 437
F+VY DFGWGKP+ +R G +N NG + VFQG+ + S+D++ L + + + D EF+
Sbjct: 385 FEVYNKDFGWGKPIAIRAGPSNSINGKLSVFQGISEGSIDVQAILWGDVIVKLLADLEFL 444
Query: 438 NVVS 441
V+
Sbjct: 445 EHVT 448
>At5g38130 anthranilate N-benzoyltransferase -like protein
Length = 442
Score = 278 bits (710), Expect = 6e-75
Identities = 175/433 (40%), Positives = 252/433 (57%), Gaps = 20/433 (4%)
Query: 1 MSSIQVLSTTPVHAPNHNPCDPNYHHTIDLTPWDLQFLPFGVNQKGLIYHHPSNLDTSNQ 60
M + V+S+T V N N I LTP DL L Q+GL++H P D N
Sbjct: 1 MVEVTVISSTMVRPENINQTG---RQKIHLTPHDLDLLYLFYPQRGLLFHKP---DPENS 54
Query: 61 I-EHLKHSLSSTLEFFPPFTGRL-KIKEHEDNTISCSITCNNVGALFVHAAAENTSVDDI 118
I L SLS+ LE + PF GRL K+ HED+T+S I C+ +GA FVHA AE+ +V+D+
Sbjct: 55 IIPRLMASLSTALEIYFPFAGRLVKVNNHEDDTVSFYIDCDGLGAKFVHAKAESITVNDV 114
Query: 119 IGATYH-PKILHSFFPFNGVKNYEG-TSMPLLAVQVTELVNGIFIACTVNHVVADGTSIW 176
+ + P + FFP N V++ + S PLLA+QVTE+ +G+FI+ NH+VADGT W
Sbjct: 115 LQSHGSVPYFISKFFPANNVQSRDALVSEPLLALQVTEMKDGVFISFGYNHMVADGTCFW 174
Query: 177 HFINSWAKIS-NGSLEISKIPSFERWFSDDIQPPIQFPFTIEPQNIHHKEEKLNLYERMF 235
F ++W+KI NGS + + WF D I P+ P +E + + E ER+F
Sbjct: 175 KFFHTWSKICLNGSDPSIQSIVLKDWFCDGIDYPVHVP-VLEMETLPRWEPSTK--ERVF 231
Query: 236 HFTKENIAKLKFKANLEAGTKN--ISSLQALFTHIWRSIIRSKNLDPEEEVHYVVVIGVR 293
H +K+NI LK KAN E T + ISSLQA+ ++W SIIR L+ EEE V +R
Sbjct: 232 HLSKKNILDLKAKANNEIDTNDLKISSLQAVVAYLWLSIIRHSGLNREEETQCNVAADMR 291
Query: 294 PRLIPLLKKDYFGNALIDPAVSMKVGDLLQDNGLGKGALEMNKMIALQSDEMLKNEYESW 353
PRL PLLKK+ FGN + VG+LL D+GLG AL+++K + +++E + ++W
Sbjct: 292 PRLNPLLKKECFGNVTNLATATTTVGELL-DHGLGWTALQISKSVRSETNESYEVFAKNW 350
Query: 354 L---KTPSFITDAKFNCKTLVGSSSPWFDVYGNDFGWGKPVGVRTGGANKENGNICVFQG 410
+ K P ++ +L+ SSSP F+VY +DFGWGKP+ R G A+ G + +F+G
Sbjct: 351 VRNVKRPKTSFGSRLANNSLIISSSPRFEVYEHDFGWGKPIAARAGPADGAGGMLVMFRG 410
Query: 411 VEKDSMDLEVCLS 423
VE+ S+D+ L+
Sbjct: 411 VEEGSIDVHATLN 423
>At5g42830 N-hydroxycinnamoyl/benzoyltransferase-like protein
Length = 450
Score = 260 bits (665), Expect = 9e-70
Identities = 161/423 (38%), Positives = 235/423 (55%), Gaps = 14/423 (3%)
Query: 30 LTPWDLQFLPFGVNQKGLIYHHPSNLDTSNQIEHLKHSLSSTLEFFPPFTGRLK-IKEHE 88
L+PWD L QKGL++H P +E LK SL+ TL F P GRL + +
Sbjct: 30 LSPWDYAMLSVQYIQKGLLFHKPPLDSIDTLLEKLKDSLAVTLVHFYPLAGRLSSLTTEK 89
Query: 89 DNTISCSITCNNV-GALFVHAAAENTSVDDIIGATYHPKILHSFFPFNGVKNYEGTSMPL 147
+ S + CN+ GA F++A ++ + DI+GA Y P I+ SFF + N++G +M L
Sbjct: 90 PKSYSVFVDCNDSPGAGFIYATSD-LCIKDIVGAKYVPSIVQSFFDHHKAVNHDGHTMSL 148
Query: 148 LAVQVTELVNGIFIACTVNHVVADGTSIWHFINSWAKI-------SNGSLEISKIPSFER 200
L+VQVTELV+GIFI ++NH + DGT+ W F +W++I N L + P +R
Sbjct: 149 LSVQVTELVDGIFIGLSMNHAMGDGTAFWKFFTAWSEIFQGQESNQNDDLCLKNPPVLKR 208
Query: 201 WFSDDIQPPIQFPFTIEPQNIHHKEEKLNLYERMFHFTKENIAKLKFKANLEAGTKNISS 260
+ + P P++ P E L ERMF F+ E I LK + N GT +ISS
Sbjct: 209 YIPEGYGPLFSLPYS-HPDEFIRTYESPILKERMFCFSSETIRMLKTRVNQICGTTSISS 267
Query: 261 LQALFTHIWRSIIRSKNLDPEEEVHYVVVIGVRPRLIPLLKKDYFGNALIDPAVSMKVGD 320
Q+L IWR I R++ L + E V R R+ P L KDYFGN L + K G+
Sbjct: 268 FQSLTAVIWRCITRARRLPLDRETSCRVAADNRGRMYPPLHKDYFGNCLSALRTAAKAGE 327
Query: 321 LLQDNGLGKGALEMNKMIALQSDEMLKNEYESWLKTP-SFITDAKFNCKTLVGSSSPWFD 379
LL +N LG AL++++ +A + E + + WLK+P + D F +++ SSP F+
Sbjct: 328 LL-ENDLGFAALKVHQAVAEHTSEKVSQMIDQWLKSPYIYHIDRLFEPMSVMMGSSPRFN 386
Query: 380 VYGNDFGWGKPVGVRTGGANKENGNICVFQGVE-KDSMDLEVCLSHETLEAIGNDPEFMN 438
YG +FG GK V +R+G A+K +G + + G E S+DLEVCL E +EA+ +D EFM+
Sbjct: 387 KYGCEFGLGKGVTLRSGYAHKFDGKVSAYPGREGGGSIDLEVCLVPEFMEALESDEEFMS 446
Query: 439 VVS 441
+VS
Sbjct: 447 LVS 449
>At5g07860 N-hydroxycinnamoyl/benzoyltransferase - like protein
Length = 454
Score = 247 bits (631), Expect = 8e-66
Identities = 156/423 (36%), Positives = 236/423 (54%), Gaps = 15/423 (3%)
Query: 31 TPWDLQFLPFGVNQKGLIYHHPSNLDT-------SNQIEHLKHSLSSTLEFFPPFTGRLK 83
+P D L QKGL++ PS ++ ++ LK SL+ L F P GR+
Sbjct: 32 SPMDHVILSIHYIQKGLLFLKPSFSESVTPKEFMETLLQKLKDSLAIALVHFYPLAGRIS 91
Query: 84 -IKEHEDNTISCSITCNNVGALFVHAAAENTSVDDIIGATYHPKILHSFFPFNGVKNYEG 142
+K ++ + S + CNN A F+HA ++ SV DI+G+ Y P ++ SFF + + +G
Sbjct: 92 TLKTNDSRSHSVFVDCNNSPAGFIHAESD-LSVSDILGSKYVPLVVQSFFDHHKALSRDG 150
Query: 143 TSMPLLAVQVTELVNGIFIACTVNHVVADGTSIWHFINSWAKISNGSLEISKIPSFER-- 200
+M LL+V+VTELV+G+FI ++NH + DG+S WHF NS ++I N + +K +
Sbjct: 151 DTMTLLSVKVTELVDGVFIGLSMNHSLGDGSSFWHFFNSLSEIFNSQEDNNKFLCLKNPP 210
Query: 201 WFSDDIQPPIQFPFTIEPQNIHHKEEKLNLYERMFHFTKENIAKLKFKANLEAGTKNISS 260
F + P PF+ EP + E+ L ERMFHF+ E + LK KAN E GT ISS
Sbjct: 211 IFREVSGPMYSLPFS-EPDESISQSERPVLKERMFHFSSETVRSLKSKANEECGTTKISS 269
Query: 261 LQALFTHIWRSIIRSKNLDPEEEVHYVVVIGVRPRLIPLLKKDYFGNALIDPAVSMKVGD 320
LQ+L IWRSI R++ L ++E + G R R+ P L ++FGN + + GD
Sbjct: 270 LQSLTALIWRSITRARKLPNDQETTCRLAAGNRSRMNPPLPMNHFGNYISLVIATTTTGD 329
Query: 321 LLQDNGLGKGALEMNKMIALQSDEMLKNEYESWLKTPSFITDAKFNCKTLVGSSSPWFDV 380
LL +N G AL++++ + + E + + + WLK D F+ + SSP F+
Sbjct: 330 LL-ENEFGCAALKLHQAVTEHTGEKISADMDRWLKA-HLKLDGFFSPNIVHMGSSPRFNK 387
Query: 381 YGNDFGWGKPVGVRTGGANKENGNICVFQGVEKD-SMDLEVCLSHETLEAIGNDPEFMNV 439
YG++FG GK V VR+G K +G + + G E S+DLEVCL E +EA+ D EFM++
Sbjct: 388 YGSEFGMGKAVAVRSGYGGKYDGKVSAYPGREGGASIDLEVCLPPECMEALELDQEFMSL 447
Query: 440 VSN 442
VS+
Sbjct: 448 VSS 450
>At5g07850 proanthranilate N-benzoyltransferase -like protein
Length = 456
Score = 241 bits (616), Expect = 4e-64
Identities = 162/434 (37%), Positives = 240/434 (54%), Gaps = 31/434 (7%)
Query: 30 LTPWDLQFLPFGVNQKGLIYHHPSNLDTSNQ-----IEHLKHSLSSTLEFFPPFTGRLK- 83
L+P DL L Q GL++ + + ++ L+ SL+ TL F P GRL
Sbjct: 31 LSPLDLVMLSMHYLQNGLLFLKSDDATKTKDFMETWLQKLRDSLAETLVHFYPLAGRLST 90
Query: 84 IKEHEDNTISCSITCNNV-GALFVHAAAENTSVDDIIGATYHPKILHSFFPFNGVKNYEG 142
+K + S + CN+ GA F+HA ++ SV DI+G+ Y P ++ SFF + +++G
Sbjct: 91 LKTDNPRSYSVFVDCNDSPGAGFIHAKSD-LSVRDIVGSNYVPLVVQSFFDHHKAVSHDG 149
Query: 143 TSMPLLAVQVTELVNGIFIACTVNHVVADGTSIWHFINSWAKISNG----SLEISKIPSF 198
+M L +V+VTELV+G+FI ++NH V DG S+WHF NS ++I N +L + P
Sbjct: 150 HTMSLFSVKVTELVDGVFIGFSLNHAVGDGGSLWHFFNSLSEIFNAQETDNLLLKNPPVL 209
Query: 199 ERWFSDDIQPPIQFPFTIEPQNIHHKEEKLNLYERMFHFTKENIAKLKFKANLEAGTKNI 258
RWF P PFT + I E + L ER+FHF+ E I LK KAN E+ T I
Sbjct: 210 SRWFPKGYGPVYNLPFTHSDEFISRFESPV-LKERIFHFSSETITSLKSKANEESRTTTI 268
Query: 259 SSLQALFTHIWRSIIRSKNLDPEEEVHYVVVIGVRPRLIPLLKKDYFGNALIDPAVSMKV 318
SS QAL +WR I R++NL + E+ + +L P L Y GN L AV K
Sbjct: 269 SSFQALAAFMWRCITRARNLPYDHEIRCSLAANNGTKLDPPLSLSYLGNCL--SAVKSKT 326
Query: 319 ---GDLLQDNGLGKGALEMNKMIALQSDEMLKNEYESWLKTPSFITDAKFNCKTLVGS-- 373
G+LL +N LG AL+M++ + + E++ ++WLK+ S++ F+ + L+G+
Sbjct: 327 VTSGELL-ENDLGWAALKMHEAVIGNTSEVVSETIKNWLKS-SYV----FHLEKLLGAMV 380
Query: 374 ----SSPWFDVYGNDFGWGKPVGVRTGGANKENGNICVFQGVE-KDSMDLEVCLSHETLE 428
SSP F +Y +FG GK V VR+G K +G I + G E ++DLEVCL E +E
Sbjct: 381 VHIGSSPRFKMYECEFGMGKAVAVRSGYGGKFDGKISAYAGREGGGTIDLEVCLLPEFME 440
Query: 429 AIGNDPEFMNVVSN 442
A+ +D EFM+VVS+
Sbjct: 441 ALESDQEFMSVVSS 454
>At5g01210 anthranilate N-benzoyltransferase - like protein
Length = 475
Score = 239 bits (609), Expect = 3e-63
Identities = 155/447 (34%), Positives = 218/447 (48%), Gaps = 47/447 (10%)
Query: 34 DLQFLPFGVNQKG-LIYHHPSNLDTSNQIEHLKHSLSSTLEFFPPFTGRLKIKEHEDNTI 92
DL L QKG L+ P + + + L+ SLSSTL FP GR
Sbjct: 31 DLPMLSCHYIQKGVLLTSPPPSFSFDDLVSSLRRSLSSTLSLFPALAGRFSTTPAGH--- 87
Query: 93 SCSITCNNVGALFVHAAAENTSVDDII-GATYHPKILHSFFPFNGVKNYEGTSMPLLAVQ 151
SI CN+ G FV A+A++ + D++ P + FF F + +Y G PL AVQ
Sbjct: 88 -ISIVCNDAGVDFVAASAKHVKLSDVLLPGEDVPLLFREFFVFERLVSYNGHHKPLAAVQ 146
Query: 152 VTELVNGIFIACTVNHVVADGTSIWHFINSWAKISNGSLEISKIPSFERWFSDDIQPPIQ 211
VTEL +G+FI CTVNH V DGTS WHF N++A +++G+ +I +P F R D P+
Sbjct: 147 VTELHDGVFIGCTVNHSVTDGTSFWHFFNTFADVTSGACKIKHLPDFSRHTVFD--SPVV 204
Query: 212 FPFTIEPQNIHHKEEKLNLYERMFHFTKENIAKLKFKANLEAG----------------- 254
P + ++ L ER+FHF++E I KLK + N
Sbjct: 205 LPVPPGGPRVTFDADQ-PLRERIFHFSREAITKLKQRTNNRVNGIETAVNDGRKCNGEIN 263
Query: 255 -------------------TKNISSLQALFTHIWRSIIRSKNLDPEEEVHYVVVIGVRPR 295
T ISS Q+L +WRS+ R++NLDP + + + + R R
Sbjct: 264 GKITTVLDSFLNNKKSYDRTAEISSFQSLSAQLWRSVTRARNLDPSKTTTFRMAVNCRHR 323
Query: 296 LIPLLKKDYFGNALIDPAVSMKVGDLLQDNGLGKGALEMNKMIALQSDEMLKNEYESWLK 355
L P + YFGNA+ GDLL + L A ++++ + D ++ +W
Sbjct: 324 LEPKMDPYYFGNAIQSIPTLASAGDLLSKD-LRWSAEQLHRNVVAHDDATVRRGIAAWES 382
Query: 356 TPSFITDAKFNCKTLVGSSSPWFDVYGNDFGWGKPVGVRTGGANKENGNICVFQGVE-KD 414
P + ++ SSP F +Y NDFGWGKP+ VR+GGANK +G I F G E
Sbjct: 383 DPRLFPLGNPDGASITMGSSPRFPMYDNDFGWGKPLAVRSGGANKFDGKISAFPGREGNG 442
Query: 415 SMDLEVCLSHETLEAIGNDPEFMNVVS 441
S+DLEV L+ ET+ I ND EFM VS
Sbjct: 443 SVDLEVVLAPETMTGIENDAEFMQYVS 469
>At2g39980 putative anthocyanin 5-aromatic acyltransferase
Length = 482
Score = 233 bits (593), Expect = 2e-61
Identities = 158/464 (34%), Positives = 232/464 (49%), Gaps = 58/464 (12%)
Query: 28 IDLTPWDLQFLPFGVNQKGLIYHHPSNLDTSNQIEHLKHSLSSTLEFFPPFTGRLKIKEH 87
+ L+ DL L QKG ++ P NL I HLKHSLS TL FPP GRL
Sbjct: 27 LKLSVSDLPMLSCHYIQKGCLFTCP-NLPLPALISHLKHSLSITLTHFPPLAGRLSTS-- 83
Query: 88 EDNTISCSITCNNVGALFVHAAAENTSVDDIIGATYHPKILHSFFPFNGVKNYEGTSMPL 147
++ +TCN+ GA FV A A++ V D+I P ++ FF ++ +YEG + P+
Sbjct: 84 --SSGHVFLTCNDAGADFVFAQAKSIHVSDVIAGIDVPDVVKEFFTYDRAVSYEGHNRPI 141
Query: 148 LAVQVTELVNGIFIACTVNHVVADGTSIWHFINSWAKISNGSLEISKIPSFERWFSDDIQ 207
LAVQVTEL +G+FI C+VNH V DGTS+W+FIN++A++S G+ +++ P F R S I
Sbjct: 142 LAVQVTELNDGVFIGCSVNHAVTDGTSLWNFINTFAEVSRGAKNVTRQPDFTR-ESVLIS 200
Query: 208 PPIQFPFTIEPQNIHHKEEKLNLYERMFHFTKENIAKLKFKANLEA-------------- 253
P + P+ +E L ER+F F++E+I +LK N +
Sbjct: 201 PAVLKVPQGGPKVTF--DENAPLRERIFSFSRESIQELKAVVNKKKWLTVDNGEIDGVEL 258
Query: 254 ----------GTKN------------------------ISSLQALFTHIWRSIIRSKNLD 279
G +N ISS Q+L +WR+I R++ L
Sbjct: 259 LGKQSNDKLNGKENGILTEMLESLFGRNDAVSKPVAVEISSFQSLCALLWRAITRARKLP 318
Query: 280 PEEEVHYVVVIGVRPRLIPLLKKDYFGNALIDPAVSMKVGDLLQDNGLGKGALEMNKMIA 339
+ + + + R RL P L +YFGNA+ ++L L A ++N+ +A
Sbjct: 319 SSKTTTFRMAVNCRHRLSPKLNPEYFGNAIQSVPTFATAAEVL-SRDLKWCADQLNQSVA 377
Query: 340 LQSDEMLKNEYESWLKTPSFITDAKFNCKTLVGSSSPWFDVYGNDFGWGKPVGVRTGGAN 399
D +++ W P + ++ SSP F +Y NDFGWG+PV VR+G +N
Sbjct: 378 AHQDGRIRSVVADWEANPRCFPLGNADGASVTMGSSPRFPMYDNDFGWGRPVAVRSGRSN 437
Query: 400 KENGNICVFQGVE-KDSMDLEVCLSHETLEAIGNDPEFMNVVSN 442
K +G I F G E ++DLEV LS ET+ I +D EFM V+N
Sbjct: 438 KFDGKISAFPGREGNGTVDLEVVLSPETMAGIESDGEFMRYVTN 481
>At5g07870 N-hydroxycinnamoyl/benzoyltransferase - like protein
Length = 464
Score = 229 bits (584), Expect = 2e-60
Identities = 157/437 (35%), Positives = 235/437 (52%), Gaps = 31/437 (7%)
Query: 31 TPWDLQFLPFGVNQKGLIYHHPSNLDT-------SNQIEHLKHSLSSTLEFFPPFTGRLK 83
+P D L QKGL++ PS ++ ++ LK SL+ L F P GRL
Sbjct: 32 SPTDHIMLSIHYIQKGLLFLKPSFSESVKPKEFMETLLQKLKDSLAIALVHFYPLAGRLS 91
Query: 84 -IKEHEDNTISCSITCNNV-GALFVHAAAENTSVDDIIGATYHPKILHSFFPFNGVKNYE 141
+K + S + CNN GA F+HA ++ SV DI+G+TY P ++ S F + N +
Sbjct: 92 TLKTDNSRSHSVFVDCNNSPGARFIHAESD-LSVSDILGSTYVPLVVQSLFDHHKALNRD 150
Query: 142 GTSMPLLAVQVTELVNGIFIACTVNHVVADGTSIWHFINSWAKI--------------SN 187
G +M LL+++VTELV+G+FI ++NH + DG+S W F NS ++I +N
Sbjct: 151 GYTMSLLSIKVTELVDGVFIGLSMNHSLGDGSSFWQFFNSLSEIFNSQEETIGNNNNNNN 210
Query: 188 GSLEISKIPSFERWFSDDIQPPIQFPFTIEPQNIHHKEEKLNLYERMFHFTKENIAKLKF 247
+L K P R + P PF+ EP + E L ERMFHF+ E + LK
Sbjct: 211 NALLCLKNPPIIR---EATGPMYSLPFS-EPNESLSQSEPPVLKERMFHFSSETVRSLKS 266
Query: 248 KANLEAGTKNISSLQALFTHIWRSIIRSKNLDPEEEVHYVVVIGVRPRLIPLLKKDYFGN 307
KAN E GT ISS QAL IWRSI R++ L ++E G R R+ P L + FG
Sbjct: 267 KANQECGTTMISSFQALTALIWRSITRARKLPNDQETTCRFAAGNRSRMNPPLPTNQFGV 326
Query: 308 ALIDPAVSMKVGDLLQDNGLGKGALEMNKMIALQSDEMLKNEYESWLKTPSFITDAKFNC 367
+ + K+G+LL +N G AL++++ + + E + E + LK+P + + +
Sbjct: 327 YISLVKTTTKIGNLL-ENEFGWIALKLHQAVTEHTGEKISYEIDQMLKSPLPLQAYRLSN 385
Query: 368 KTLVG-SSSPWFDVYGNDFGWGKPVGVRTGGANKENGNICVFQGVEKD-SMDLEVCLSHE 425
+V SSP F+ YG++FG GK V VR+G K +G + + G + S+DLEVCL E
Sbjct: 386 LNIVHMGSSPRFNKYGSEFGMGKAVAVRSGYGGKYDGKVSAYPGRQGGASIDLEVCLLPE 445
Query: 426 TLEAIGNDPEFMNVVSN 442
+EA+ +D EFM++VS+
Sbjct: 446 FMEALESDQEFMSLVSS 462
>At3g50290 putative protein
Length = 648
Score = 221 bits (562), Expect = 8e-58
Identities = 158/452 (34%), Positives = 230/452 (49%), Gaps = 61/452 (13%)
Query: 1 MSSIQVLSTTPVHAPNHNPCDPNYHHTIDLTPWDLQFLPFGVNQKGLIYHHPSNLDTSNQ 60
M+ + V+S++ V + N H LTP+DL L Q+GL++ P +
Sbjct: 1 MADVTVISSSIVQPRSTNQSGRTKIH---LTPFDLNLLYVDYPQRGLLFRKPE--PETRF 55
Query: 61 IEHLKHSLSSTLEFFPPFTGRL-KIKEHEDNTISCSITCNNVGALFVHAAAENTSVDDII 119
I LK SLS+ LE + PF GRL K++ E++T FV+A +++ SV DI+
Sbjct: 56 ISRLKTSLSTALEIYFPFAGRLVKVENLENDT-------------FVYAESKSLSVSDIL 102
Query: 120 ---GATYHPKILHSFFPFNGVKNYEGTSMPLLAVQVTELVNGIFIACTVNHVVADGTSIW 176
G+ P + FFP VK+ G S PLLA+QVTEL +G+FI+ NH+VADG SIW
Sbjct: 103 QPHGSV--PDFMSHFFPSIDVKSIYGLSEPLLALQVTELKDGVFISFGYNHMVADGASIW 160
Query: 177 HFINSWAKISNGSLEISKIPS---FERWFSDDIQPPIQFPFT-IEPQNIHHKEEKLNLYE 232
+F N+W+KI + + K WF D I I P + + + E
Sbjct: 161 NFFNTWSKICSNEYDHQKNLKPLVLRGWFLDKIDYRIHIPASEADAPTSDEISQMPTSKE 220
Query: 233 RMFHFTKENIAKLKFKANLEAGTKN--ISSLQALFTHIWRSIIRSKNLDPEEEVHYVVVI 290
R+FHFTK+NI++LK KAN E G+ + ISSLQAL H+WRSIIR + EEE H V +
Sbjct: 221 RVFHFTKKNISELKTKANGETGSSDLIISSLQALSAHLWRSIIRHSGMKREEETHCKVAV 280
Query: 291 GVRPRLIPLLKKDYFGNALIDPAVSMKVGDLLQDNGLGKGALEMNKMIALQSDEMLKNEY 350
+R RL P L K+ FGN ++ VG+LL D+ +G AL+++KM+ Q+DE K
Sbjct: 281 DLRQRLNPPLDKECFGNVNNLGVATVTVGELL-DHEIGWAALQISKMVRSQTDEKYKTFA 339
Query: 351 ESWLKTPSFITDAKFNCKTLVGSSSPWFDVYGNDFGWGKPVGVRTGGANKENGNICVFQG 410
E+W+ A R G N +G + +FQG
Sbjct: 340 ENWVNNVKISKTA------------------------------RGGPGNSISGKLVLFQG 369
Query: 411 VEKDSMDLEVCLSHETLEAIGNDPEFMNVVSN 442
+E+ S+D+ L + L + D EFM + +
Sbjct: 370 IEEGSIDVHATLWCDVLVKLLADVEFMENIKS 401
>At5g23940 acyltransferase
Length = 484
Score = 200 bits (509), Expect = 1e-51
Identities = 140/428 (32%), Positives = 210/428 (48%), Gaps = 24/428 (5%)
Query: 30 LTPWDLQFLPFGVNQKGLIYHHPSNLDTSNQ------IEHLKHSLSSTLEFFPPFTGRLK 83
LT +DL +L F NQK L+Y + LD +E+LK L LE F G+L
Sbjct: 25 LTTFDLPYLAFYYNQKFLLYKFQNLLDLEEPTFQNEVVENLKDGLGLVLEDFYQLAGKLA 84
Query: 84 IKEHEDNTISCSITCNNV-GALFVHAAAENTSVDDIIGATYHPKILHSFFPFNGVKNYEG 142
+ + + + G F A A + +VDD+ K P+NG+ N EG
Sbjct: 85 KDDEGVFRVEYDAEDSEINGVEFSVAHAADVTVDDLTAEDGTAKF-KELVPYNGILNLEG 143
Query: 143 TSMPLLAVQVTELVNGIFIACTVNHVVADGTSIWHFINSWAKISNGSLEISKIPSFERWF 202
S PLLAVQVT+L +G+ + NH V DGTS WHF++SWA+I G+ IS P +R
Sbjct: 144 LSRPLLAVQVTKLKDGLAMGLAFNHAVLDGTSTWHFMSSWAEICRGAQSISTQPFLDRSK 203
Query: 203 SDDIQPPIQFPFTIEPQNIHHKEEKLN-------LYERMFHFTKENIAKLKFKANL---E 252
+ D + + +P + E+ N L E++F F+ + +K +AN
Sbjct: 204 ARDTRVKLDLTAPKDPNETSNGEDAANPTVEPPQLVEKIFRFSDFAVHTIKSRANSVIPS 263
Query: 253 AGTKNISSLQALFTHIWRSIIRSKNLDPEEEVHYVVVIGVRPRLIPLLKKDYFGNALIDP 312
+K S+ Q+L +HIWR + ++ L PE+ + V R R+ P + ++YFGN LI
Sbjct: 264 DSSKPFSTFQSLTSHIWRHVTLARGLKPEDITIFTVFADCRRRVDPPMPEEYFGN-LIQA 322
Query: 313 AVSMKVGDLLQDNGLGKGALEMNKMIALQSDEMLKNEYESWLKTPSF--ITDAKFNCKTL 370
+ LL +G GA + K IA ++ + W K+P DA NC +
Sbjct: 323 IFTGTAAGLLAAHGPEFGASVIQKAIAAHDASVIDARNDEWEKSPKIFQFKDAGVNCVAV 382
Query: 371 VGSSSPWFDVYGNDFGWGKPVGVRTGGANKENGNICVFQGVEKD-SMDLEVCLSHETLEA 429
SSP F VY DFG+GKP VR+G N+ NG + ++QG S+D+E+ L +E
Sbjct: 383 --GSSPRFRVYEVDFGFGKPETVRSGSNNRFNGMMYLYQGKAGGISIDVEITLEASVMEK 440
Query: 430 IGNDPEFM 437
+ EF+
Sbjct: 441 LVKSKEFL 448
>At5g48930 anthranilate N-benzoyltransferase
Length = 433
Score = 117 bits (294), Expect = 1e-26
Identities = 111/439 (25%), Positives = 179/439 (40%), Gaps = 31/439 (7%)
Query: 4 IQVLSTTPVHAPNHNPCDPNYHHTIDLTPWDLQFLPFGVNQKGLIYHHPSNLDTSNQIEH 63
I + +T V P ++ +DL +P + + ++ P+ +
Sbjct: 3 INIRDSTMVRPATETPITNLWNSNVDLV------IP-RFHTPSVYFYRPTGASNFFDPQV 55
Query: 64 LKHSLSSTLEFFPPFTGRLKIKEHEDNTISCSITCNNVGALFVHAAAENTSVDDIIGATY 123
+K +LS L F P GRLK D+ I CN G LFV A+ SV D G
Sbjct: 56 MKEALSKALVPFYPMAGRLK----RDDDGRIEIDCNGAGVLFV--VADTPSVIDDFGDFA 109
Query: 124 HPKILHSFFPFNGVKNYEGT-SMPLLAVQVTEL-VNGIFIACTVNHVVADGTSIWHFINS 181
L P V + G S PLL +QVT G + + H ADG S HFIN+
Sbjct: 110 PTLNLRQLIP--EVDHSAGIHSFPLLVLQVTFFKCGGASLGVGMQHHAADGFSGLHFINT 167
Query: 182 WAKISNGSLEISKIPSFERWFSDDIQPPIQFPFTIEPQNIHHKEEKL--------NLYER 233
W+ ++ G L+++ P +R PP +E Q + L N
Sbjct: 168 WSDMARG-LDLTIPPFIDRTLLRARDPPQPAFHHVEYQPAPSMKIPLDPSKSGPENTTVS 226
Query: 234 MFHFTKENIAKLKFKANLEAGTKNISSLQALFTHIWRSIIRSKNLDPEEEVHYVVVIGVR 293
+F T++ + LK K+ + T + SS + L H+WRS+ +++ L ++E + R
Sbjct: 227 IFKLTRDQLVALKAKSKEDGNTVSYSSYEMLAGHVWRSVGKARGLPNDQETKLYIATDGR 286
Query: 294 PRLIPLLKKDYFGNALIDPAVSMKVGDLLQDNGLGKGALEMNKMIALQSDEMLKNEYESW 353
RL P L YFGN + GDLL A +++ + D L++ +
Sbjct: 287 SRLRPQLPPGYFGNVIFTATPLAVAGDLL-SKPTWYAAGQIHDFLVRMDDNYLRSALDYL 345
Query: 354 LKTPSFITDAK----FNCKTLVGSSSPWFDVYGNDFGWGKPVGVRTGGANKENGNICVFQ 409
P + + C L +S +Y DFGWG+P+ + GG E + +
Sbjct: 346 EMQPDLSALVRGAHTYKCPNLGITSWVRLPIYDADFGWGRPIFMGPGGIPYEGLSFVLPS 405
Query: 410 GVEKDSMDLEVCLSHETLE 428
S+ + + L E ++
Sbjct: 406 PTNDGSLSVAIALQSEHMK 424
>At2g19070 putative anthranilate
N-hydroxycinnamoyl/benzoyltransferase
Length = 451
Score = 116 bits (290), Expect = 3e-26
Identities = 119/452 (26%), Positives = 187/452 (41%), Gaps = 64/452 (14%)
Query: 19 PCDPNYHHTIDLTPWDLQFLPFGVNQKGLIYH--------HPSNLDTSNQIEHLKHSLSS 70
P +P + L WD Q G I H PS N +E LK SLS
Sbjct: 14 PAEPTWSGRFPLAEWD---------QVGTITHIPTLYFYDKPSESFQGNVVEILKTSLSR 64
Query: 71 TLEFFPPFTGRLKIKEHEDNTISCSITCNNVGALFVHAAAENTSVDDIIGATYHPKILHS 130
L F P GRL+ + CN G F+ A +E + D + P+ +
Sbjct: 65 VLVHFYPMAGRLRWLPRG----RFELNCNAEGVEFIEAESEG-KLSDFKDFSPTPEF-EN 118
Query: 131 FFPFNGVKNYEGTSMPLLAVQVTEL-VNGIFIACTVNHVVADGTSIWHFINSWAKISNGS 189
P KN T +PL QVT+ GI ++ V+H + DG S H I+ W +++ G
Sbjct: 119 LMPQVNYKNPIET-IPLFLAQVTKFKCGGISLSVNVSHAIVDGQSALHLISEWGRLARGE 177
Query: 190 LEISKIPSFER---WFSDDIQPPI-----------QFPFTI-EPQNIHHKEEKLNLYERM 234
+ +P +R W + + P + Q PF I E N+ +++K + M
Sbjct: 178 -PLETVPFLDRKILWAGEPLPPFVSPPKFDHKEFDQPPFLIGETDNVEERKKKTIVV--M 234
Query: 235 FHFTKENIAKLKFKANLEAGT---KNISSLQALFTHIWRSIIRSKNLDPEEEVHYVVVIG 291
+ + KL+ KAN + K + + + H+WR +++ PE+ + I
Sbjct: 235 LPLSTSQLQKLRSKANGSKHSDPAKGFTRYETVTGHVWRCACKARGHSPEQPTALGICID 294
Query: 292 VRPRLIPLLKKDYFGNALIDPAVSMKVGDLLQDNGLGKGALEMNKMIALQSDEMLKNEYE 351
R R+ P L + YFGNA +D + G+L+ N LG A ++K I ++E + E
Sbjct: 295 TRSRMEPPLPRGYFGNATLDVVAASTSGELI-SNELGFAASLISKAIKNVTNEYVMIGIE 353
Query: 352 SWLKTPSFITDAKFNCKTLVGSSSPWF--------------DVYGNDFGWGKPVGVRTGG 397
+LK + KF +GS+ F +YG DFGWGK G
Sbjct: 354 -YLKNQKDL--KKFQDLHALGSTEGPFYGNPNLGVVSWLTLPMYGLDFGWGKEFYTGPGT 410
Query: 398 ANKENGNICVFQGVEKDSMDLEVCLSHETLEA 429
+ + ++ + E S+ L CL +EA
Sbjct: 411 HDFDGDSLILPDQNEDGSVILATCLQVAHMEA 442
>At3g29590 Anthocyanin 5-aromatic acyltransferase, putative
Length = 449
Score = 104 bits (259), Expect = 1e-22
Identities = 110/455 (24%), Positives = 190/455 (41%), Gaps = 42/455 (9%)
Query: 2 SSIQVLSTTPVHAPNHNPCDPNYHHTIDLTPWDLQFLPFGVNQKGLIYHHPSNLDTSNQI 61
S++ +L V P+ N T+ LT +DL +L + L YH P L S+ I
Sbjct: 6 SAVNILEVVQVSPPSSNSL------TLPLTYFDLGWLKLHPVDRVLFYHVPE-LTRSSLI 58
Query: 62 EHLKHSLSSTLEFFPPFTGRLKIKEHEDNTISCSITCNNVGALFVHAAAENTSVDDIIGA 121
LK SLS+TL + P GRL + + S + ++ A+++ A N + + G
Sbjct: 59 SKLKSSLSATLLHYLPLAGRL-VWDSIKTKPSIVYSPDDKDAVYLTVAESNGDLSHLSGD 117
Query: 122 TYHPKI-LHSFFPFNGVKNYEGTSMPLLAVQVTELVN-GIFIACTVNHVVADGTSIWHFI 179
P HS P V + S +LAVQVT N G + T +H V DG + F+
Sbjct: 118 EPRPATEFHSLVPELPVSD---ESARVLAVQVTFFPNQGFSLGVTAHHAVLDGKTTAMFL 174
Query: 180 NSWA---KISNGSLEISKIPSFERWFSDDIQPPIQFPFTIEPQNIH------HKEEKLNL 230
+WA K +L +PS +R +Q P +E + ++ + + L L
Sbjct: 175 KAWAHNCKQEQEALPHDLVPSLDRII-------VQDPTGLETKLLNRWISASNNKPSLKL 227
Query: 231 YERM----------FHFTKENIAKLKFKANLEAGTKNI--SSLQALFTHIWRSIIRSKNL 278
+ + T+E+I KL+ + E+ K + S+ + ++ +++ +
Sbjct: 228 FPSKIIGSDILRVTYRLTREDIKKLRERVETESHAKQLRLSTFVITYAYVITCMVKMRGG 287
Query: 279 DPEEEVHYVVVIGVRPRLIPLLKKDYFGNALIDPAVSMKVGDLLQDNGLGKGALEMNKMI 338
DP V R RL P L +FGN ++ + + + G GKG + + +
Sbjct: 288 DPTRFVCVGFASDFRSRLNPPLPPTFFGNCIVGSGDFDVKAEPILEEGEGKGFITAVETL 347
Query: 339 ALQSDEMLKNEYESWLKTPSFITDAKFNCKTLVG-SSSPWFDVYGNDFGWGKPVGVRTGG 397
+ + E + P + ++ + S +YG+DFGWGKPV V
Sbjct: 348 TGWVNGLCPENIEKNMLLPFEAFKRMEPGRQMISVAGSTRLGIYGSDFGWGKPVKVEIVT 407
Query: 398 ANKENGNICVFQGVEKDSMDLEVCLSHETLEAIGN 432
+K+ G +++ VCL + +E G+
Sbjct: 408 IDKDASVSLSESGDGSGGVEVGVCLKKDDVERFGS 442
>At3g48720 unknown protein
Length = 430
Score = 100 bits (249), Expect = 2e-21
Identities = 96/359 (26%), Positives = 153/359 (41%), Gaps = 27/359 (7%)
Query: 45 KGLIYHHPSNLDTSNQIEHLKHSLSSTLEFFPPFTGRLKIKEHEDNTISCSITCNNVGAL 104
K L Y+ + +K SLS L + P GRL I ++ C G +
Sbjct: 42 KTLYYYKSESRTNQESYNVIKKSLSEVLVHYYPVAGRLTISPEG----KIAVNCTGEGVV 97
Query: 105 FVHAAAENTSVDDI---IGATYHPKILHSFFPFNGVKNYEGTSMPLLAVQVTELVNGIFI 161
V A A N +D I I + + G +N +P + VQVT G F+
Sbjct: 98 VVEAEA-NCGIDTIKEAISENRMETLEKLVYDVPGARNI--LEIPPVVVQVTNFKCGGFV 154
Query: 162 -ACTVNHVVADGTSIWHFINSWAKISNGSLEISKIPSFERWFSDDIQPP-IQFPFT---- 215
++H + DG + F+NSW +++ G L +S P +R PP I+FP
Sbjct: 155 LGLGMSHNMFDGVAAAEFLNSWCEMAKG-LPLSVPPFLDRTILRSRNPPKIEFPHNEFDE 213
Query: 216 ---IEPQNIHHKEEKLNLYERMFHFTKENIAKLKFKANLEAGTKNISSLQALFTHIWRSI 272
I + EEKL +Y + F F E + KLK A E +S+ QAL +W+S
Sbjct: 214 IEDISDTGKIYDEEKL-IY-KSFLFEPEKLEKLKIMAIEENNNNKVSTFQALTGFLWKSR 271
Query: 273 IRSKNLDPEEEVHYVVVIGVRPRLIPLLKKDYFGNALIDPAVSMKVGDLLQDNGLGKGAL 332
+ P++ V + R R IP L + Y GN ++ + G+L+ N L
Sbjct: 272 CEALRFKPDQRVKLLFAADGRSRFIPRLPQGYCGNGIVLTGLVTSSGELV-GNPLSHSVG 330
Query: 333 EMNKMIALQSDEMLKNEYESWLKTPSFITDAKFNCKTLVGSSSPWFDVYGNDFGWGKPV 391
+ +++ L +D +++ + + T N L+ S S ++ DFGWG+PV
Sbjct: 331 LVKRLVELVTDGFMRSAMDYFEVNR---TRPSMNATLLITSWSK-LTLHKLDFGWGEPV 385
>At1g24430 similar to acetyl-CoA:benzylalcoholo acetyltransferase
Length = 435
Score = 99.4 bits (246), Expect = 4e-21
Identities = 115/451 (25%), Positives = 188/451 (41%), Gaps = 69/451 (15%)
Query: 16 NHNPCDPNYHHTIDLTPWDLQFLPFGVNQKGLIYHHPSNLDTSNQIEHLKHSLSSTLEFF 75
NH PC + L P F+PF YH+ +NL + +H+K SLS L +
Sbjct: 21 NHPPCHHHLSFLDQLAP--PIFMPFL-----FFYHNKTNLSDKERSDHIKSSLSEILNLY 73
Query: 76 PPFTGRLKIKEHEDNTISCSITCNNVGALFVHAAAENTSVDDIIGATYHPKILHSFFPFN 135
P GR+K + CN+VG FV A A+ ++ I+ +P L+ PF
Sbjct: 74 YPLAGRIK-------NSGDVVVCNDVGVSFVEAKAD-CNMSQIL-ENPNPNELNKLHPF- 123
Query: 136 GVKNYEGTSMPLLAVQVTEL-VNGIFIACTVNHVVADGTSIWHFINSWAKISNGSLEISK 194
+ +E + +P L VQ+T G+ + ++H + D S F+NSWA + G +
Sbjct: 124 --EFHEVSDVP-LTVQLTFFECGGLALGIGLSHKLCDALSGLIFVNSWAAFARGQTDEII 180
Query: 195 IPSFE--RWFSDDIQPPIQFPFTIEPQNIHHKEEKLNLYERMFHFTKENIAKLKFKANLE 252
PSF+ + F P IE N+ K N+ R F F + ++ L+ +
Sbjct: 181 TPSFDLAKMFP---------PCDIENLNMATGITKENIVTRRFVFLRSSVESLRERF--- 228
Query: 253 AGTKNI--SSLQALFTHIWRSIIRSKNLDPEEEVHYVVV--IGVRPRLIPLLKKDYFGNA 308
+G K I + ++ L IW + S N D + Y ++ + +R + P + + FGN
Sbjct: 229 SGNKKIRATRVEVLSVFIWSRFMASTNHDDKTGKIYTLIHPVNLRRQADPDIPDNMFGNI 288
Query: 309 L-IDPAVSMKVGDLLQDNGLGKGAL------EMNKMIA-----LQSD-----EMLKNEYE 351
+ V M + + +N K +L E+ K+ A LQ D E L +
Sbjct: 289 MRFSVTVPMMI---INENDEEKASLVDQMREEIRKIDAVYVKKLQEDNRGHLEFLNKQAS 345
Query: 352 SWLKTPSFITDAKFNCKTLVGSSSPWFDVYGNDFGWGKPVGVRTGGANKENGNICVFQGV 411
++ CK F VY DFGWGKP+ V + + +N +
Sbjct: 346 GFVNGEIVSFSFTSLCK---------FPVYEADFGWGKPLWVASARMSYKN-LVAFIDTK 395
Query: 412 EKDSMDLEVCLSHETLEAIGNDPEFMNVVSN 442
E D ++ + L + D E + VS+
Sbjct: 396 EGDGIEAWINLDQNDMSRFEADEELLRYVSS 426
>At5g57840 N-hydroxycinnamoyl/benzoyltransferase
Length = 443
Score = 97.4 bits (241), Expect = 1e-20
Identities = 112/447 (25%), Positives = 178/447 (39%), Gaps = 45/447 (10%)
Query: 19 PCDPNYHHTIDLTPWDLQFLPFGVNQKGLIYHHPSNLDTSNQIEHLKHSLSSTLEFFPPF 78
P + H + L+ DL + ++ L ++ P + + L +LS L FF P
Sbjct: 13 PAEETPTHNLWLSNLDL--IQVRLHMGTLYFYKPCSSSDRPNTQSLIEALSKVLVFFYPA 70
Query: 79 TGRLKIKEHEDNTISCSITCNNVGALFVHAAAENTSVDDIIGATYHPKILHSFFPFNGVK 138
GRL+ ++ + CN G LFV A +++T D IG L P
Sbjct: 71 AGRLQ----KNTNGRLEVQCNGEGVLFVEAESDSTVQD--IGLLTQSLDLSQLVP---TV 121
Query: 139 NYEG--TSMPLLAVQVTELVNG-IFIACTVNHVVADGTSIWHFINSWAKISNGSLEISKI 195
+Y G +S PLL QVT G I + +++H + TS+ + + +W+ + G L +
Sbjct: 122 DYAGDISSYPLLLFQVTYFKCGTICVGSSIHHTFGEATSLGYIMEAWSLTARGLL-VKLT 180
Query: 196 PSFERWFSDDIQPPIQ-FPFTIEPQNIHHKEEKLNLYERMFHFTKENIAKLKF------- 247
P +R PP FP T H +L R + IA LK
Sbjct: 181 PFLDRTVLHARNPPSSVFPHTEYQSPPFHNHPMKSLAYRSNPESDSAIATLKLTRLQLKA 240
Query: 248 -KANLEAGTKNISSLQALFTHIWR-SIIRSKNLDPEEEVHYVVVIGVRPRLIPLLKKDYF 305
KA E S+ + L H WR + +++L E ++I RPRL P L + Y
Sbjct: 241 LKARAEIADSKFSTYEVLVAHTWRCASFANEDLSEEHSTRLHIIIDGRPRLQPKLPQGYI 300
Query: 306 GNALIDPAVSMKVGDLLQDNGLGKGALEMNKMIALQSDEMLKNEYESWLKTPS------- 358
GN L +G +++ + +++ I +E L++ + + P
Sbjct: 301 GNTLFHARPVSPLGAFHRES-FSETVERVHREIRKMDNEYLRSAIDYLERHPDLDQLVPG 359
Query: 359 -----FITDAKFNCKTLVGSSSPWFDVYGNDFGWGKPVGVRTGGANKENGNICVFQGVEK 413
F A F +L+ ++ Y DFGWGK R N+ G + E+
Sbjct: 360 EGNTIFSCAANFCIVSLIKQTA-----YEMDFGWGKAYYKRASHLNEGKGFV-TGNPDEE 413
Query: 414 DSMDLEVCLSHETLEAIGN-DPEFMNV 439
SM L +CL L EF+NV
Sbjct: 414 GSMMLTMCLKKTQLSKFRKLFYEFLNV 440
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,861,963
Number of Sequences: 26719
Number of extensions: 494099
Number of successful extensions: 1364
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 51
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 1125
Number of HSP's gapped (non-prelim): 85
length of query: 442
length of database: 11,318,596
effective HSP length: 102
effective length of query: 340
effective length of database: 8,593,258
effective search space: 2921707720
effective search space used: 2921707720
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147431.9


