

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147431.1 + phase: 0 /pseudo
(493 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
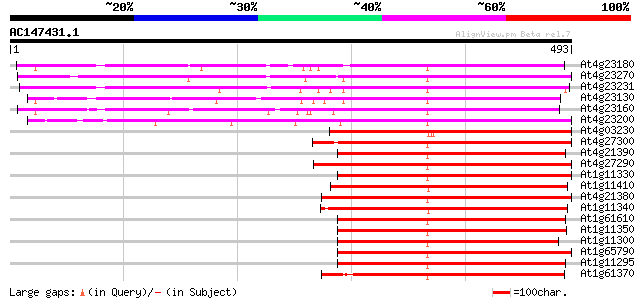
Score E
Sequences producing significant alignments: (bits) Value
At4g23180 receptor-like protein kinase 4 (RLK4) 342 2e-94
At4g23270 serine/threonine kinase 328 3e-90
At4g23231 unknown protein 326 2e-89
At4g23130 receptor-like protein kinase 6 (RLK6) 324 6e-89
At4g23160 putative protein 313 2e-85
At4g23200 serine /threonine kinase - like protein 277 1e-74
At4g03230 putative receptor kinase 261 6e-70
At4g27300 putative receptor protein kinase 261 8e-70
At4g21390 serine/threonine kinase - like protein 260 1e-69
At4g27290 putative receptor like kinase 259 2e-69
At1g11330 receptor-like protein kinase, putative 254 1e-67
At1g11410 putative brassinosteroid insensitive protein; similar ... 253 2e-67
At4g21380 receptor-like serine/threonine protein kinase ARK3 249 3e-66
At1g11340 receptor kinase, putative 249 3e-66
At1g61610 248 5e-66
At1g11350 putative receptor-like protein kinase gi|4008010; simi... 247 1e-65
At1g11300 putative s-locus protein kinase (PPC:1.7.2) 245 4e-65
At1g65790 receptor kinase, putative 244 6e-65
At1g11295 putative s-locus protein kinase (PPC:1.7.2) 243 1e-64
At1g61370 S-like receptor protein kinase; member of gene cluster... 243 2e-64
>At4g23180 receptor-like protein kinase 4 (RLK4)
Length = 669
Score = 342 bits (878), Expect = 2e-94
Identities = 213/561 (37%), Positives = 296/561 (51%), Gaps = 99/561 (17%)
Query: 7 FLSFILLHIFLFTLT---NAQSPYYRLNFCQNSTEKTNNTSYRSNVRNLLLWINTDSATG 63
+ SF L +F F + +AQ P Y + CQN+ T+N++Y +N++ LL +++ +A+
Sbjct: 15 YSSFFFLFLFSFLTSFRVSAQDPTYVYHTCQNTANYTSNSTYNNNLKTLLASLSSRNASY 74
Query: 64 SVSNHIAINSNKTNSNDNDGDVVYGFYDCRGDVTGSFCQFCINAAVRAIAQCCPNGVSAM 123
S A D V G ++CRGDV+ C+ C++ AV CPN A
Sbjct: 75 STGFQNATVGQAP-------DRVTGLFNCRGDVSTEVCRRCVSFAVNDTLTRCPNQKEAT 127
Query: 124 IWYDICVIGYSDHNTSGKISVTPSWNLTGTKNTKDSTELEKAVD---DMRNLIGRVTAEA 180
++YD CV+ YS+ N + T L T+N S +L+ D N V +
Sbjct: 128 LYYDECVLRYSNQNILSTLITTGGVILVNTRNVT-SNQLDLLSDLVLPTLNQAATVALNS 186
Query: 181 NSNWAVGEFDWSDTEKRYGWVQCNSDLTKDGCRYCLETMLDKVPQCCGTKVKWAVVSPSC 240
+ + + +++ + YG VQC DLT+ C CL+ +++++P ++ +++PSC
Sbjct: 187 SKKFGTRKNNFTALQSFYGLVQCTPDLTRQDCSRCLQLVINQIPT---DRIGARIINPSC 243
Query: 241 GMEIDDNKFYNFQTES-------PPSLPNP------------------------------ 263
+ Y F TES PPS+ P
Sbjct: 244 TSRYE---IYAFYTESAVPPPPPPPSISTPPVSAPPRSGKDGNSKVLVIAIVVPIIVAVL 300
Query: 264 ---GGLLLRT-------ITPKSFR-DHVPREDSFNGDLPTIPLTVIQQSTDNFSESFKLG 312
G T TP +F D + DS D TI Q +TD+F ES K+G
Sbjct: 301 LFIAGYCFLTRRARKSYYTPSAFAGDDITTADSLQLDYRTI-----QTATDDFVESNKIG 355
Query: 313 EGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKNEVIFIAKLQHRNLVKLL------- 365
+GGFG VYKGTL DGTE+A KRLS++SGQG EFKNEV+ +AKLQHRNLV+LL
Sbjct: 356 QGGFGEVYKGTLSDGTEVAVKRLSKSSGQGEVEFKNEVVLVAKLQHRNLVRLLGFCLDGE 415
Query: 366 -------------------DEEKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLK 406
D K LDW R II G+ARG+LYLH+DS L +IHRDLK
Sbjct: 416 ERVLVYEYVPNKSLDYFLFDPAKKGQLDWTRRYKIIGGVARGILYLHQDSRLTIIHRDLK 475
Query: 407 ASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFS 466
ASN+LLD +MNPKI+DFG+AR F DQ T R++GTYGYM+PEYAM G +S+KSDV+S
Sbjct: 476 ASNILLDADMNPKIADFGMARIFGLDQTEENTSRIVGTYGYMSPEYAMHGQYSMKSDVYS 535
Query: 467 FGVLVLEIIYGKRNGDFFLSE 487
FGVLVLEII GK+N F+ ++
Sbjct: 536 FGVLVLEIISGKKNSSFYQTD 556
>At4g23270 serine/threonine kinase
Length = 581
Score = 328 bits (842), Expect = 3e-90
Identities = 194/521 (37%), Positives = 278/521 (53%), Gaps = 45/521 (8%)
Query: 8 LSFILLHIFLFTLTNAQSPYYRLNFCQNSTEKTNNTSYRSNVRNLLLWINTDSATGSVSN 67
+SFI L +F +AQ+ +Y + C +T ++N++Y +N++ LL S+ S++
Sbjct: 5 ISFIFLFLFSSITASAQNTFYLYHNCSVTTTFSSNSTYSTNLKTLL------SSLSSLNA 58
Query: 68 HIAINSNKTNSNDNDGDVVYGFYDCRGDVTGSFCQFCINAAVRAIAQCCPNGVSAMIWYD 127
+T + D V G + CR DV+ C+ C+ AV CP + +Y+
Sbjct: 59 SSYSTGFQTATAGQAPDRVTGLFLCRVDVSSEVCRSCVTFAVNETLTRCPKDKEGVFYYE 118
Query: 128 ICVIGYSDHNTSGKISVTPSWNLTGTKN--TKDSTELEKAVDDMRNLIGRVTAEANSNWA 185
C++ YS+ N ++ + +N + + V NL A + WA
Sbjct: 119 QCLLRYSNRNIVATLNTDGGMFMQSARNPLSVKQDQFRDLVLTPMNLAAVEAARSFKKWA 178
Query: 186 VGEFDWSDTEKRYGWVQCNSDLTKDGCRYCLETMLDKVPQCCGTKVKWAVVSPSCGMEID 245
V + D + ++ YG V+C DL + C CL+ +++V K+ ++ PSC D
Sbjct: 179 VRKIDLNASQSLYGMVRCTPDLREQDCLDCLKIGINQVTY---DKIGGRILLPSCASRYD 235
Query: 246 DNKFYNFQTESPP--SLPNPGGLLLRTITPKSFRDHVPREDSFNGDLPT-----IPLTVI 298
+ FYN P S P P +R ++ + P + N D+ T I
Sbjct: 236 NYAFYNESNVGTPQDSSPRPAVFSVRAKNKRTLNEKEPVAEDGN-DITTAGSLQFDFKAI 294
Query: 299 QQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKNEVIFIAKLQH 358
+ +T+ F KLG+GGFG VYKGTL G ++A KRLS+TSGQG +EF+NEV+ +AKLQH
Sbjct: 295 EAATNCFLPINKLGQGGFGEVYKGTLSSGLQVAVKRLSKTSGQGEKEFENEVVVVAKLQH 354
Query: 359 RNLVKLL--------------------------DEEKHKHLDWKLRLSIIKGIARGLLYL 392
RNLVKLL D LDW R II GIARG+LYL
Sbjct: 355 RNLVKLLGYCLEGEEKILVYEFVPNKSLDHFLFDSTMKMKLDWTRRYKIIGGIARGILYL 414
Query: 393 HEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGYMAPEY 452
H+DS L +IHRDLKA N+LLDD+MNPKI+DFG+AR F DQ T+RV+GTYGYM+PEY
Sbjct: 415 HQDSRLTIIHRDLKAGNILLDDDMNPKIADFGMARIFGMDQTEAMTRRVVGTYGYMSPEY 474
Query: 453 AMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNLETL 493
AM G FS+KSDV+SFGVLVLEII G +N + + ++ L
Sbjct: 475 AMYGQFSMKSDVYSFGVLVLEIISGMKNSSLYQMDESVGNL 515
>At4g23231 unknown protein
Length = 627
Score = 326 bits (835), Expect = 2e-89
Identities = 203/557 (36%), Positives = 296/557 (52%), Gaps = 83/557 (14%)
Query: 9 SFILLHIFLFTLT-NAQSPYYRLNFCQNSTEKTNNTSYRSNVRNLLLWINTDSATGSVSN 67
SFI L FL + T + ++ +Y + C N+T + N+ Y +N+R LL ++ +A+ S
Sbjct: 6 SFIFLFFFLTSFTASVENVFYIKHICPNTTTYSRNSPYLTNLRTLLSSLSAPNASYSTGF 65
Query: 68 HIAINSNKTNSNDNDGDVVYGFYDCRGDVTGSFCQFCINAAVRAIAQCCPNGVSAMIWYD 127
A D V G + CRGDV+ + C+ C+ ++ CP+ ++ +YD
Sbjct: 66 QSARAGQAP-------DRVTGLFLCRGDVSPAVCRNCVAFSINDTLVQCPSERKSVFYYD 118
Query: 128 ICVIGYSDHNTSGKISVTPSWNLTGTKNTKDSTELEKAVDDMRNLIGRVTAEANSN---W 184
C++ YSD N ++ +W + D ++ + D + + + + +A S+ +
Sbjct: 119 ECMLRYSDQNILSTLAYDGAWIRMNGNISIDQNQMNRFKDFVSSTMNQAAVKAASSPRKF 178
Query: 185 AVGEFDWSDTEKRYGWVQCNSDLTKDGCRYCLETMLDKVPQCCGTKVKWAVVSPSCGMEI 244
+ W+ + YG VQC DLT+ C CLE+ + +P K + SC
Sbjct: 179 YTVKATWTALQTLYGLVQCTPDLTRQDCFSCLESSIKLMPLY---KTGGRTLYSSCNSRY 235
Query: 245 DDNKFYNFQT----ESPPSLPNPGGLLLRT--ITPKSFRDHV------------------ 280
+ FYN T ++PP LP L+ + + KS+ +V
Sbjct: 236 ELFAFYNETTVRTQQAPPPLPPSSTPLVTSPSLPGKSWNSNVLVVAIVLTILVAALLLIA 295
Query: 281 ------------PREDSFNGDLPT-----IPLTVIQQSTDNFSESFKLGEGGFGPVYKGT 323
+F+GD T + +I+ +T+ FSE+ K+G+GGFG VYKGT
Sbjct: 296 GYCFAKRVKNSSDNAPAFDGDDITTESLQLDYRMIRAATNKFSENNKIGQGGFGEVYKGT 355
Query: 324 LPDGTEIAAKRLSETSGQGLEEFKNEVIFIAKLQHRNLVKLL------------------ 365
+GTE+A KRLS++SGQG EFKNEV+ +AKLQHRNLV+LL
Sbjct: 356 FSNGTEVAVKRLSKSSGQGDTEFKNEVVVVAKLQHRNLVRLLGFSIGGGERILVYEYMPN 415
Query: 366 --------DEEKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMN 417
D K LDW R +I GIARG+LYLH+DS L +IHRDLKASN+LLD +MN
Sbjct: 416 KSLDYFLFDPAKQNQLDWTRRYKVIGGIARGILYLHQDSRLTIIHRDLKASNILLDADMN 475
Query: 418 PKISDFGLARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYG 477
PK++DFGLAR F DQ T R++GT+GYMAPEYA+ G FSVKSDV+SFGVLVLEII G
Sbjct: 476 PKLADFGLARIFGMDQTQENTSRIVGTFGYMAPEYAIHGQFSVKSDVYSFGVLVLEIISG 535
Query: 478 KRNGDFFLSE--HNLET 492
K+N F+ ++ H+L T
Sbjct: 536 KKNNSFYETDGAHDLVT 552
>At4g23130 receptor-like protein kinase 6 (RLK6)
Length = 659
Score = 324 bits (831), Expect = 6e-89
Identities = 205/550 (37%), Positives = 286/550 (51%), Gaps = 94/550 (17%)
Query: 16 FLFTLT----------NAQSPYYRLNFCQNSTEKTNNTSYRSNVRNLLLWINTDSATGSV 65
FLF LT Q P Y + C N + N+ Y SN++ LL +++++A S+
Sbjct: 9 FLFLLTFFIGSLRVSAQLQDPTYVGHVCTNRISR--NSIYFSNLQTLLTSLSSNNAYFSL 66
Query: 66 SNHIAINSNKTNSNDNDGDVVYGFYDCRGDVTGSFCQFCINAAVRAIAQCCPNGVSAMIW 125
+H + + + D+V+G Y C+GD++ C+ C+ A + CP G +I
Sbjct: 67 GSH-------SLTKGQNSDMVFGLYLCKGDLSPESCRECVIFAAKDTRSRCPGGKEFLIQ 119
Query: 126 YDICVIGYSDHNTSGKISVTPSWNLTGTKNTKDSTELEKAVDDMRNLIGRVTAEA----N 181
YD C++GYSD N +VT + +T + + ++ D + +L+ + EA +
Sbjct: 120 YDECMLGYSDRNIFMD-TVTTTTIITWNTQKVTADQSDRFNDAVLSLMKKSAEEAANSTS 178
Query: 182 SNWAVGEFDWSDTEKRYGWVQCNSDLTKDGCRYCLETMLDKVPQCCGTKVKWAVVSPSCG 241
+AV + D+S ++ Y VQC DLT + C CL+ + + KV + PSC
Sbjct: 179 KKFAVKKSDFSSSQSLYASVQCIPDLTSEDCVMCLQ---QSIKELYFNKVGGRFLVPSCN 235
Query: 242 MEIDDNKFYNFQTE----------------SPPSLPNPGG-----LLLRTITPKS----- 275
+ FY E S PS P G +++ + P +
Sbjct: 236 SRYEVYPFYKETIEGTVLPPPVSAPPLPLVSTPSFPPGKGKNSTVIIIAIVVPVAISVLI 295
Query: 276 ----FRDHVPREDSFNGDLPT-----------IPLTVIQQSTDNFSESFKLGEGGFGPVY 320
F H + D P VI+ +TD FS KLG+GGFG VY
Sbjct: 296 CVAVFSFHASKRAKKTYDTPEEDDITTAGSLQFDFKVIEAATDKFSMCNKLGQGGFGQVY 355
Query: 321 KGTLPDGTEIAAKRLSETSGQGLEEFKNEVIFIAKLQHRNLVKLL--------------- 365
KGTLP+G ++A KRLS+TSGQG +EFKNEV+ +AKLQHRNLVKLL
Sbjct: 356 KGTLPNGVQVAVKRLSKTSGQGEKEFKNEVVVVAKLQHRNLVKLLGFCLEREEKILVYEF 415
Query: 366 -----------DEEKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDD 414
D LDW R II GIARG+LYLH+DS L +IHRDLKA N+LLD
Sbjct: 416 VSNKSLDYFLFDSRMQSQLDWTTRYKIIGGIARGILYLHQDSRLTIIHRDLKAGNILLDA 475
Query: 415 EMNPKISDFGLARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEI 474
+MNPK++DFG+AR FE DQ T+RV+GTYGYM+PEYAM G FS+KSDV+SFGVLVLEI
Sbjct: 476 DMNPKVADFGMARIFEIDQTEAHTRRVVGTYGYMSPEYAMYGQFSMKSDVYSFGVLVLEI 535
Query: 475 IYGKRNGDFF 484
I G++N +
Sbjct: 536 ISGRKNSSLY 545
>At4g23160 putative protein
Length = 1240
Score = 313 bits (801), Expect = 2e-85
Identities = 200/533 (37%), Positives = 282/533 (52%), Gaps = 70/533 (13%)
Query: 8 LSFILLHIFLFTLT---NAQSPYYRLNFCQNSTEKTNNTSYRSNVRNLLLWINTDSATGS 64
+ FI L +F F + +AQ+P+Y + C N T ++N++Y +N++ LL + +A+ S
Sbjct: 602 ICFIFLFLFSFLTSFKASAQNPFYLNHDCPNRTTYSSNSTYSTNLKTLLSSFASRNASYS 661
Query: 65 VSNHIAINSNKTNSNDNDGDVVYGFYDCRGDVTGSFCQFCINAAVRAIAQCCPNGVSAMI 124
I + +T D V G + CRGD++ C C+ +V CPN A+
Sbjct: 662 TGFQ-NIRAGQTP------DRVTGLFLCRGDLSPEVCSNCVAFSVNESLTRCPNQREAVF 714
Query: 125 WYDICVIGYSDHNT------SGKISVTPSWNLTGTKNTKDSTELEKAVDDMRNLIGRVTA 178
+Y+ C++ YS N G++ + N++ +N +D + V N A
Sbjct: 715 YYEECILRYSHKNFLSTVTYEGELIMRNPNNISSIQNQRD--QFIDLVQSNMNQAANEAA 772
Query: 179 EANSNWAVGEFDWSDTEKRYGWVQCNSDLTKDGCRYCLETMLDKVPQC--CGTKVKWAVV 236
++ ++ + + + + YG VQC DL + C CL + ++++ G + W
Sbjct: 773 NSSRKFSTIKTELTSLQTLYGLVQCTPDLARQDCFSCLTSSINRMMPLFRIGARQFW--- 829
Query: 237 SPSCGMEIDDNKFYN---FQTESPPSL--------PNP------GGLLLRTITPKSFRDH 279
PSC + FYN T SPP L +P G L T K+F
Sbjct: 830 -PSCNSRYELYAFYNETAIGTPSPPPLFPGSTPPLTSPSIPALVGYCFLAQRTKKTFDTA 888
Query: 280 VPRE---DSFNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLS 336
E D D + IQ +T++F+ES K+G GGFG VYKGT +G E+A KRLS
Sbjct: 889 SASEVGDDMATADSLQLDYRTIQTATNDFAESNKIGRGGFGEVYKGTFSNGKEVAVKRLS 948
Query: 337 ETSGQGLEEFKNEVIFIAKLQHRNLVKLL--------------------------DEEKH 370
+ S QG EFK EV+ +AKLQHRNLV+LL D K
Sbjct: 949 KNSRQGEAEFKTEVVVVAKLQHRNLVRLLGFSLQGEERILVYEYMPNKSLDCLLFDPTKQ 1008
Query: 371 KHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFE 430
LDW R +II GIARG+LYLH+DS L +IHRDLKASN+LLD ++NPKI+DFG+AR F
Sbjct: 1009 TQLDWMQRYNIIGGIARGILYLHQDSRLTIIHRDLKASNILLDADINPKIADFGMARIFG 1068
Query: 431 KDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDF 483
DQ T R++GTYGYMAPEYAM G FS+KSDV+SFGVLVLEII G++N F
Sbjct: 1069 LDQTQDNTSRIVGTYGYMAPEYAMHGQFSMKSDVYSFGVLVLEIISGRKNSSF 1121
>At4g23200 serine /threonine kinase - like protein
Length = 648
Score = 277 bits (708), Expect = 1e-74
Identities = 184/541 (34%), Positives = 263/541 (48%), Gaps = 72/541 (13%)
Query: 16 FLFTLTNAQSPYYRLNFCQNSTEKTNNTSYRSNVRNLLLWINTDSATGSVSNHIAINSNK 75
F T T+ S L C N+T N++Y +N R +L + ++ V++H +
Sbjct: 8 FFLTSTSLVSALQTLP-CINTTYFIPNSTYDTNRRVILSLLPSN-----VTSHFGFFNGS 61
Query: 76 TNSNDNDGDVVYGFYDCRGDVTGSFCQFCINAAVRAIAQCCPNGVSAMIWY---DICVIG 132
N VY C C C+ +A + + C +A+IW IC+I
Sbjct: 62 IGQAPNR---VYAVGMCLPGTEEESCIGCLLSASNTLLETCLTEENALIWIANRTICMIR 118
Query: 133 YSDHNTSGKISVTPSWNLTGTKNTK-DSTELEKAVDDM-RNLIGRVTAEANSNWAVGEFD 190
YSD + G + P K + TE + + ++ ++ ++ W+ ++
Sbjct: 119 YSDTSFVGSFELEPHREFLSIHGYKTNETEFNTVWSRLTQRMVQEASSSTDATWSGAKYY 178
Query: 191 WSD------TEKRYGWVQCNSDLTKDGCRYCLETMLDKVPQCCGTKVKWAVVSPSCGMEI 244
+D ++ Y +QC DL+ C CL + CC + ++V SC
Sbjct: 179 TADVAALPDSQTLYAMMQCTPDLSPAECNLCLTESVVNYQSCCLGRQGGSIVRLSCAFRA 238
Query: 245 DDNKF---YNFQTESPPSLPNPGGLLLRTITPKSFRDHVPREDSFNGD------------ 289
+ F + T P S P P + K + FNG+
Sbjct: 239 ELYPFGGAFTVMTARPLSQPPPSLIKKGEFFAKFMSNSQEPRKVFNGNYCCNCCSHYSGR 298
Query: 290 -----------LPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSET 338
+ I+ +T+NF+++ KLG+GGFG VYKGTL +GTE+A KRLS+T
Sbjct: 299 YHLLAGITTLHFQQLDFKTIEVATENFAKTNKLGQGGFGEVYKGTLVNGTEVAVKRLSKT 358
Query: 339 SGQGLEEFKNEVIFIAKLQHRNLVKLL--------------------------DEEKHKH 372
S QG +EFKNEV+ +AKLQHRNLVKLL D K
Sbjct: 359 SEQGAQEFKNEVVLVAKLQHRNLVKLLGYCLEPEEKILVYEFVPNKSLDYFLFDPTKQGQ 418
Query: 373 LDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKD 432
LDW R +II GI RG+LYLH+DS L +IHRDLKASN+LLD +M PKI+DFG+AR D
Sbjct: 419 LDWTKRYNIIGGITRGILYLHQDSRLTIIHRDLKASNILLDADMIPKIADFGMARISGID 478
Query: 433 QCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNLET 492
Q TKR+ GT+GYM PEY + G FS+KSDV+SFGVL+LEII GK+N F+ ++ E
Sbjct: 479 QSVANTKRIAGTFGYMPPEYVIHGQFSMKSDVYSFGVLILEIICGKKNRSFYQADTKAEN 538
Query: 493 L 493
L
Sbjct: 539 L 539
>At4g03230 putative receptor kinase
Length = 852
Score = 261 bits (667), Expect = 6e-70
Identities = 142/238 (59%), Positives = 164/238 (68%), Gaps = 26/238 (10%)
Query: 282 REDSFNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQ 341
++DS D+P+ L I +T NFS + KLG+GGFGPVYKG P EIA KRLS SGQ
Sbjct: 509 QDDSQGIDVPSFELETILYATSNFSNANKLGQGGFGPVYKGMFPGDQEIAVKRLSRCSGQ 568
Query: 342 GLEEFKNEVIFIAKLQHRNLVKLLD-----EEK--------HK-------------HLDW 375
GLEEFKNEV+ IAKLQHRNLV+LL EEK HK LDW
Sbjct: 569 GLEEFKNEVVLIAKLQHRNLVRLLGYCVAGEEKLLLYEYMPHKSLDFFIFDRKLCQRLDW 628
Query: 376 KLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCH 435
K+R +II GIARGLLYLH+DS LR+IHRDLK SN+LLD+EMNPKISDFGLAR F +
Sbjct: 629 KMRCNIILGIARGLLYLHQDSRLRIIHRDLKTSNILLDEEMNPKISDFGLARIFGGSETS 688
Query: 436 TKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNLETL 493
T RV+GTYGYM+PEYA+ GLFS KSDVFSFGV+V+E I GKRN F E +L L
Sbjct: 689 ANTNRVVGTYGYMSPEYALEGLFSFKSDVFSFGVVVIETISGKRNTGFHEPEKSLSLL 746
>At4g27300 putative receptor protein kinase
Length = 815
Score = 261 bits (666), Expect = 8e-70
Identities = 142/253 (56%), Positives = 169/253 (66%), Gaps = 29/253 (11%)
Query: 267 LLRTITPKSFRDHVPREDSFNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPD 326
+++ ++FR + ED DLP I +TD+FS LG GGFGPVYKG L D
Sbjct: 465 IMKRYRGENFRKGIEEEDL---DLPIFDRKTISIATDDFSYVNFLGRGGFGPVYKGKLED 521
Query: 327 GTEIAAKRLSETSGQGLEEFKNEVIFIAKLQHRNLVKLL--------------------- 365
G EIA KRLS SGQG+EEFKNEV IAKLQHRNLV+LL
Sbjct: 522 GQEIAVKRLSANSGQGVEEFKNEVKLIAKLQHRNLVRLLGCCIQGEECMLIYEYMPNKSL 581
Query: 366 -----DEEKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKI 420
DE + LDWK R++II G+ARG+LYLH+DS LR+IHRDLKA NVLLD++MNPKI
Sbjct: 582 DFFIFDERRSTELDWKKRMNIINGVARGILYLHQDSRLRIIHRDLKAGNVLLDNDMNPKI 641
Query: 421 SDFGLARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRN 480
SDFGLA++F DQ + T RV+GTYGYM PEYA+ G FSVKSDVFSFGVLVLEII GK N
Sbjct: 642 SDFGLAKSFGGDQSESSTNRVVGTYGYMPPEYAIDGHFSVKSDVFSFGVLVLEIITGKTN 701
Query: 481 GDFFLSEHNLETL 493
F ++H+L L
Sbjct: 702 RGFRHADHDLNLL 714
>At4g21390 serine/threonine kinase - like protein
Length = 849
Score = 260 bits (665), Expect = 1e-69
Identities = 138/226 (61%), Positives = 158/226 (69%), Gaps = 26/226 (11%)
Query: 289 DLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKN 348
+LP L I +T++F + +LG GGFGPVYKG L DG EIA KRLS SGQG++EFKN
Sbjct: 513 ELPVFSLNAIAIATNDFCKENELGRGGFGPVYKGVLEDGREIAVKRLSGKSGQGVDEFKN 572
Query: 349 EVIFIAKLQHRNLVKLL--------------------------DEEKHKHLDWKLRLSII 382
E+I IAKLQHRNLV+LL DE K +DWKLR SII
Sbjct: 573 EIILIAKLQHRNLVRLLGCCFEGEEKMLVYEYMPNKSLDFFLFDETKQALIDWKLRFSII 632
Query: 383 KGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVI 442
+GIARGLLYLH DS LR+IHRDLK SNVLLD EMNPKISDFG+AR F +Q T RV+
Sbjct: 633 EGIARGLLYLHRDSRLRIIHRDLKVSNVLLDAEMNPKISDFGMARIFGGNQNEANTVRVV 692
Query: 443 GTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEH 488
GTYGYM+PEYAM GLFSVKSDV+SFGVL+LEI+ GKRN SEH
Sbjct: 693 GTYGYMSPEYAMEGLFSVKSDVYSFGVLLLEIVSGKRNTSLRSSEH 738
>At4g27290 putative receptor like kinase
Length = 772
Score = 259 bits (662), Expect = 2e-69
Identities = 137/252 (54%), Positives = 168/252 (66%), Gaps = 26/252 (10%)
Query: 268 LRTITPKSFRDHVPREDSFNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDG 327
+ T+ +S R +++ + +LP + L + ++T FS KLG+GGFGPVYKGTL G
Sbjct: 417 IETLQRESSRVSSRKQEEEDLELPFLDLDTVSEATSGFSAGNKLGQGGFGPVYKGTLACG 476
Query: 328 TEIAAKRLSETSGQGLEEFKNEVIFIAKLQHRNLVKLL---------------------- 365
E+A KRLS TS QG+EEFKNE+ IAKLQHRNLVK+L
Sbjct: 477 QEVAVKRLSRTSRQGVEEFKNEIKLIAKLQHRNLVKILGYCVDEEERMLIYEYQPNKSLD 536
Query: 366 ----DEEKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKIS 421
D+E+ + LDW R+ IIKGIARG+LYLHEDS LR+IHRDLKASNVLLD +MN KIS
Sbjct: 537 SFIFDKERRRELDWPKRVEIIKGIARGMLYLHEDSRLRIIHRDLKASNVLLDSDMNAKIS 596
Query: 422 DFGLARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNG 481
DFGLAR D+ T RV+GTYGYM+PEY + G FS+KSDVFSFGVLVLEI+ G+RN
Sbjct: 597 DFGLARTLGGDETEANTTRVVGTYGYMSPEYQIDGYFSLKSDVFSFGVLVLEIVSGRRNR 656
Query: 482 DFFLSEHNLETL 493
F EH L L
Sbjct: 657 GFRNEEHKLNLL 668
>At1g11330 receptor-like protein kinase, putative
Length = 840
Score = 254 bits (648), Expect = 1e-67
Identities = 133/231 (57%), Positives = 156/231 (66%), Gaps = 26/231 (11%)
Query: 289 DLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKN 348
+LP V+ STD+FS KLG+GGFGPVYKG LP+G EIA KRLS SGQGLEE N
Sbjct: 506 ELPLFEFQVLATSTDSFSLRNKLGQGGFGPVYKGKLPEGQEIAVKRLSRKSGQGLEELMN 565
Query: 349 EVIFIAKLQHRNLVKLL--------------------------DEEKHKHLDWKLRLSII 382
EV+ I+KLQHRNLVKLL D K K LDWK R +I+
Sbjct: 566 EVVVISKLQHRNLVKLLGCCIEGEERMLVYEYMPKKSLDAYLFDPMKQKILDWKTRFNIM 625
Query: 383 KGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVI 442
+GI RGLLYLH DS L++IHRDLKASN+LLD+ +NPKISDFGLAR F ++ T+RV+
Sbjct: 626 EGICRGLLYLHRDSRLKIIHRDLKASNILLDENLNPKISDFGLARIFRANEDEANTRRVV 685
Query: 443 GTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNLETL 493
GTYGYM+PEYAM G FS KSDVFS GV+ LEII G+RN E+NL L
Sbjct: 686 GTYGYMSPEYAMEGFFSEKSDVFSLGVIFLEIISGRRNSSSHKEENNLNLL 736
>At1g11410 putative brassinosteroid insensitive protein; similar to
EST gb|W43102
Length = 840
Score = 253 bits (645), Expect = 2e-67
Identities = 133/234 (56%), Positives = 162/234 (68%), Gaps = 26/234 (11%)
Query: 283 EDSFNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQG 342
+ S + +LP L+ I +T+NF+ KLG GGFGPVYKG L +G EIA KRLS++SGQG
Sbjct: 496 DKSRSRELPLFELSTIATATNNFAFQNKLGAGGFGPVYKGVLQNGMEIAVKRLSKSSGQG 555
Query: 343 LEEFKNEVIFIAKLQHRNLVKLLD--------------------------EEKHKHLDWK 376
+EEFKNEV I+KLQHRNLV++L EE+ LDW
Sbjct: 556 MEEFKNEVKLISKLQHRNLVRILGCCVEFEEKMLVYEYLPNKSLDYFIFHEEQRAELDWP 615
Query: 377 LRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHT 436
R+ II+GI RG+LYLH+DS LR+IHRDLKASNVLLD+EM PKI+DFGLAR F +Q
Sbjct: 616 KRMGIIRGIGRGILYLHQDSRLRIIHRDLKASNVLLDNEMIPKIADFGLARIFGGNQIEG 675
Query: 437 KTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNL 490
T RV+GTYGYM+PEYAM G FS+KSDV+SFGVL+LEII GKRN F+ NL
Sbjct: 676 STNRVVGTYGYMSPEYAMDGQFSIKSDVYSFGVLILEIITGKRNSAFYEESLNL 729
>At4g21380 receptor-like serine/threonine protein kinase ARK3
Length = 850
Score = 249 bits (635), Expect = 3e-66
Identities = 131/246 (53%), Positives = 169/246 (68%), Gaps = 27/246 (10%)
Query: 275 SFRDHVPREDSFNG-DLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAK 333
S R H+ RE++ + +LP + + +T+NFS + KLG+GGFG VYKG L DG E+A K
Sbjct: 495 SSRRHISRENNTDDLELPLMEFEEVAMATNNFSNANKLGQGGFGIVYKGKLLDGQEMAVK 554
Query: 334 RLSETSGQGLEEFKNEVIFIAKLQHRNLVKLL--------------------------DE 367
RLS+TS QG +EFKNEV IA+LQH NLV+LL D+
Sbjct: 555 RLSKTSVQGTDEFKNEVKLIARLQHINLVRLLACCVDAGEKMLIYEYLENLSLDSHLFDK 614
Query: 368 EKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLAR 427
++ L+W++R II GIARGLLYLH+DS R+IHRDLKASN+LLD M PKISDFG+AR
Sbjct: 615 SRNSKLNWQMRFDIINGIARGLLYLHQDSRFRIIHRDLKASNILLDKYMTPKISDFGMAR 674
Query: 428 AFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSE 487
F +D+ T++V+GTYGYM+PEYAM G+FS+KSDVFSFGVL+LEII KRN F+ S+
Sbjct: 675 IFGRDETEANTRKVVGTYGYMSPEYAMDGIFSMKSDVFSFGVLLLEIISSKRNKGFYNSD 734
Query: 488 HNLETL 493
+L L
Sbjct: 735 RDLNLL 740
>At1g11340 receptor kinase, putative
Length = 901
Score = 249 bits (635), Expect = 3e-66
Identities = 134/243 (55%), Positives = 162/243 (66%), Gaps = 28/243 (11%)
Query: 274 KSFRDHVPREDSFNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAK 333
+SFR ++ + N +LP L I +T+NFS KLG GGFGPVYKG L + EIA K
Sbjct: 554 ESFR--FEQDKARNRELPLFDLNTIVAATNNFSSQNKLGAGGFGPVYKGVLQNRMEIAVK 611
Query: 334 RLSETSGQGLEEFKNEVIFIAKLQHRNLVKLLD--------------------------E 367
RLS SGQG+EEFKNEV I+KLQHRNLV++L E
Sbjct: 612 RLSRNSGQGMEEFKNEVKLISKLQHRNLVRILGCCVELEEKMLVYEYLPNKSLDYFIFHE 671
Query: 368 EKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLAR 427
E+ LDW R+ I++GIARG+LYLH+DS LR+IHRDLKASN+LLD EM PKISDFG+AR
Sbjct: 672 EQRAELDWPKRMEIVRGIARGILYLHQDSRLRIIHRDLKASNILLDSEMIPKISDFGMAR 731
Query: 428 AFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSE 487
F +Q T RV+GT+GYMAPEYAM G FS+KSDV+SFGVL+LEII GK+N F
Sbjct: 732 IFGGNQMEGCTSRVVGTFGYMAPEYAMEGQFSIKSDVYSFGVLMLEIITGKKNSAFHEES 791
Query: 488 HNL 490
NL
Sbjct: 792 SNL 794
>At1g61610
Length = 842
Score = 248 bits (633), Expect = 5e-66
Identities = 129/226 (57%), Positives = 153/226 (67%), Gaps = 26/226 (11%)
Query: 289 DLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKN 348
DLP + +T +F+E KLG+GGFG VYKG +G EIA KRLS S QGLEEFKN
Sbjct: 509 DLPIFSFDSVASATGDFAEENKLGQGGFGTVYKGNFSEGREIAVKRLSGKSKQGLEEFKN 568
Query: 349 EVIFIAKLQHRNLVKLL--------------------------DEEKHKHLDWKLRLSII 382
E++ IAKLQHRNLV+LL DE K LDW+ R +I
Sbjct: 569 EILLIAKLQHRNLVRLLGCCIEDNEKMLLYEYMPNKSLDRFLFDESKQGSLDWRKRWEVI 628
Query: 383 KGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVI 442
GIARGLLYLH DS L++IHRDLKASN+LLD EMNPKISDFG+AR F Q H T RV+
Sbjct: 629 GGIARGLLYLHRDSRLKIIHRDLKASNILLDTEMNPKISDFGMARIFNYRQDHANTIRVV 688
Query: 443 GTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEH 488
GTYGYMAPEYAM G+FS KSDV+SFGVL+LEI+ G++N F ++H
Sbjct: 689 GTYGYMAPEYAMEGIFSEKSDVYSFGVLILEIVSGRKNVSFRGTDH 734
>At1g11350 putative receptor-like protein kinase gi|4008010; similar
to EST gb|AI999419.1
Length = 830
Score = 247 bits (630), Expect = 1e-65
Identities = 127/227 (55%), Positives = 156/227 (67%), Gaps = 26/227 (11%)
Query: 289 DLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKN 348
+LP V+ +T+NFS + KLG+GGFG VYKG L +G +IA KRLS TSGQG+EEF N
Sbjct: 496 ELPLFEFQVLAVATNNFSITNKLGQGGFGAVYKGRLQEGLDIAVKRLSRTSGQGVEEFVN 555
Query: 349 EVIFIAKLQHRNLVKLL--------------------------DEEKHKHLDWKLRLSII 382
EV+ I+KLQHRNLV+LL D K + LDWK R +II
Sbjct: 556 EVVVISKLQHRNLVRLLGFCIEGEERMLVYEFMPENCLDAYLFDPVKQRLLDWKTRFNII 615
Query: 383 KGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVI 442
GI RGL+YLH DS L++IHRDLKASN+LLD+ +NPKISDFGLAR F+ ++ T RV+
Sbjct: 616 DGICRGLMYLHRDSRLKIIHRDLKASNILLDENLNPKISDFGLARIFQGNEDEVSTVRVV 675
Query: 443 GTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHN 489
GTYGYMAPEYAM GLFS KSDVFS GV++LEI+ G+RN F+ N
Sbjct: 676 GTYGYMAPEYAMGGLFSEKSDVFSLGVILLEIVSGRRNSSFYNDGQN 722
>At1g11300 putative s-locus protein kinase (PPC:1.7.2)
Length = 820
Score = 245 bits (626), Expect = 4e-65
Identities = 129/220 (58%), Positives = 151/220 (68%), Gaps = 26/220 (11%)
Query: 289 DLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKN 348
+LP V+ +T+NFS KLG+GGFGPVYKG L +G EIA KRLS SGQGLEE N
Sbjct: 493 ELPLFEFQVLAAATNNFSLRNKLGQGGFGPVYKGKLQEGQEIAVKRLSRASGQGLEELVN 552
Query: 349 EVIFIAKLQHRNLVKLL--------------------------DEEKHKHLDWKLRLSII 382
EV+ I+KLQHRNLVKLL D + K LDWK R +II
Sbjct: 553 EVVVISKLQHRNLVKLLGCCIAGEERMLVYEFMPKKSLDYYLFDSRRAKLLDWKTRFNII 612
Query: 383 KGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVI 442
GI RGLLYLH DS LR+IHRDLKASN+LLD+ + PKISDFGLAR F ++ T+RV+
Sbjct: 613 NGICRGLLYLHRDSRLRIIHRDLKASNILLDENLIPKISDFGLARIFPGNEDEANTRRVV 672
Query: 443 GTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGD 482
GTYGYMAPEYAM GLFS KSDVFS GV++LEII G+RN +
Sbjct: 673 GTYGYMAPEYAMGGLFSEKSDVFSLGVILLEIISGRRNSN 712
>At1g65790 receptor kinase, putative
Length = 843
Score = 244 bits (624), Expect = 6e-65
Identities = 129/231 (55%), Positives = 157/231 (67%), Gaps = 26/231 (11%)
Query: 289 DLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKN 348
+LP + L + +T+NFS KLG+GGFG VYKG L DG EIA KRLS+ S QG +EF N
Sbjct: 503 ELPLLELEALATATNNFSNDNKLGQGGFGIVYKGRLLDGKEIAVKRLSKMSSQGTDEFMN 562
Query: 349 EVIFIAKLQHRNLVKLL--------------------------DEEKHKHLDWKLRLSII 382
EV IAKLQH NLV+LL D+ + +L+W+ R II
Sbjct: 563 EVRLIAKLQHINLVRLLGCCVDKGEKMLIYEYLENLSLDSHLFDQTRSSNLNWQKRFDII 622
Query: 383 KGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVI 442
GIARGLLYLH+DS R+IHRDLKASNVLLD M PKISDFG+AR F +++ T+RV+
Sbjct: 623 NGIARGLLYLHQDSRCRIIHRDLKASNVLLDKNMTPKISDFGMARIFGREETEANTRRVV 682
Query: 443 GTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNLETL 493
GTYGYM+PEYAM G+FS+KSDVFSFGVL+LEII GKRN F+ S +L L
Sbjct: 683 GTYGYMSPEYAMDGIFSMKSDVFSFGVLLLEIISGKRNKGFYNSNRDLNLL 733
>At1g11295 putative s-locus protein kinase (PPC:1.7.2)
Length = 820
Score = 243 bits (621), Expect = 1e-64
Identities = 130/226 (57%), Positives = 150/226 (65%), Gaps = 26/226 (11%)
Query: 289 DLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKN 348
+LP V+ +TDNFS S KLG+GGFGPVYKG L +G EIA KRLS+ SGQGLEE
Sbjct: 493 ELPLFEFQVLATATDNFSLSNKLGQGGFGPVYKGMLLEGQEIAVKRLSQASGQGLEELVT 552
Query: 349 EVIFIAKLQHRNLVKLL--------------------------DEEKHKHLDWKLRLSII 382
EV+ I+KLQHRNLVKL D + K LDW R II
Sbjct: 553 EVVVISKLQHRNLVKLFGCCIAGEERMLVYEFMPKKSLDFYIFDPREAKLLDWNTRFEII 612
Query: 383 KGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVI 442
GI RGLLYLH DS LR+IHRDLKASN+LLD+ + PKISDFGLAR F ++ T+RV+
Sbjct: 613 NGICRGLLYLHRDSRLRIIHRDLKASNILLDENLIPKISDFGLARIFPGNEDEANTRRVV 672
Query: 443 GTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEH 488
GTYGYMAPEYAM GLFS KSDVFS GV++LEII G+RN L H
Sbjct: 673 GTYGYMAPEYAMGGLFSEKSDVFSLGVILLEIISGRRNSHSTLLAH 718
>At1g61370 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 814
Score = 243 bits (620), Expect = 2e-64
Identities = 133/239 (55%), Positives = 160/239 (66%), Gaps = 31/239 (12%)
Query: 275 SFRDHVPREDSFNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKR 334
++R+ + +D D+ TI LT+ T+NFS KLG+GGFGPVYKG L DG EIA KR
Sbjct: 476 AWREQLKPQDVNFFDMQTI-LTI----TNNFSMENKLGQGGFGPVYKGNLQDGKEIAIKR 530
Query: 335 LSETSGQGLEEFKNEVIFIAKLQHRNLVKLL--------------------------DEE 368
LS TSGQGLEEF NE+I I+KLQHRNLV+LL D
Sbjct: 531 LSSTSGQGLEEFMNEIILISKLQHRNLVRLLGCCIEGEEKLLIYEFMANKSLNTFIFDST 590
Query: 369 KHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARA 428
K LDW R II+GIA GLLYLH DS LRV+HRD+K SN+LLD+EMNPKISDFGLAR
Sbjct: 591 KKLELDWPKRFEIIQGIACGLLYLHRDSCLRVVHRDMKVSNILLDEEMNPKISDFGLARM 650
Query: 429 FEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSE 487
F+ Q T+RV+GT GYM+PEYA G+FS KSD+++FGVL+LEII GKR F + E
Sbjct: 651 FQGTQHQANTRRVVGTLGYMSPEYAWTGMFSEKSDIYAFGVLLLEIITGKRISSFTIGE 709
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,896,566
Number of Sequences: 26719
Number of extensions: 547950
Number of successful extensions: 4460
Number of sequences better than 10.0: 1050
Number of HSP's better than 10.0 without gapping: 897
Number of HSP's successfully gapped in prelim test: 153
Number of HSP's that attempted gapping in prelim test: 1499
Number of HSP's gapped (non-prelim): 1444
length of query: 493
length of database: 11,318,596
effective HSP length: 103
effective length of query: 390
effective length of database: 8,566,539
effective search space: 3340950210
effective search space used: 3340950210
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147431.1


