

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147428.5 - phase: 0
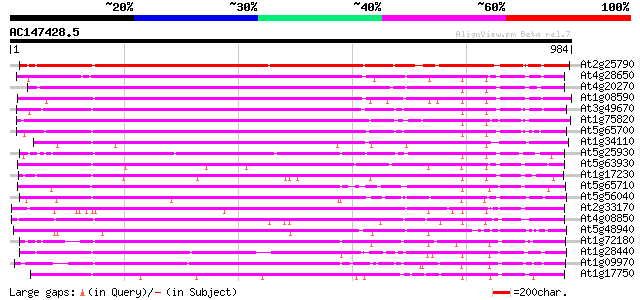
(984 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g25790 putative receptor-like protein kinase 872 0.0
At4g28650 receptor protein kinase-like protein 506 e-143
At4g20270 CLV1 receptor kinase like protein 502 e-142
At1g08590 putative receptor kinase, CLV1 498 e-141
At3g49670 receptor protein kinase - like protein 496 e-140
At1g75820 receptor kinase (CLV1) 491 e-139
At5g65700 receptor protein kinase-like protein 486 e-137
At1g34110 putative protein 466 e-131
At5g25930 receptor-like protein kinase - like 461 e-129
At5g63930 receptor-like protein kinase 458 e-129
At1g17230 putative leucine-rich receptor protein kinase 436 e-122
At5g65710 receptor protein kinase-like protein 430 e-120
At5g56040 receptor protein kinase-like protein 428 e-120
At2g33170 putative receptor-like protein kinase 428 e-120
At4g08850 receptor protein kinase like protein 426 e-119
At5g48940 receptor protein kinase-like protein 424 e-118
At1g72180 leucine-rich receptor-like protein kinase, putative 417 e-116
At1g28440 unknown protein 417 e-116
At1g09970 leucine-rich repeat receptor-like kinase At1g09970 412 e-115
At1g17750 hypothetical protein 401 e-112
>At2g25790 putative receptor-like protein kinase
Length = 960
Score = 872 bits (2254), Expect = 0.0
Identities = 486/976 (49%), Positives = 646/976 (65%), Gaps = 55/976 (5%)
Query: 18 LFMFMLNFHSTHGEQEFELLLSFKASIKFDPLNFLSNWVNTSSDTICKWHGITCDNWSHV 77
LF LNF H E ELLLSFK+SI+ DPL LS+W +S++ +C W G+ C+N S V
Sbjct: 17 LFFLFLNFSCLHAN-ELELLLSFKSSIQ-DPLKHLSSWSYSSTNDVCLWSGVVCNNISRV 74
Query: 78 NTVSLSGKNISGEV-SSSIFQLPHVTNLDLSNNQLVGEIVFNSPFLSS--LLYLNLSNNN 134
++ LSGKN+SG++ +++ F+LP + ++LSNN L G I + SS L YLNLSNNN
Sbjct: 75 VSLDLSGKNMSGQILTAATFRLPFLQTINLSNNNLSGPIPHDIFTTSSPSLRYLNLSNNN 134
Query: 135 LTGPLPQSLFSSSFINLETLDLSNNMFSGKIPDQIGLLSSLTYVDLGGNVLVGKIPNSIT 194
+G +P+ NL TLDLSNNMF+G+I + IG+ S+L +DLGGNVL G +P +
Sbjct: 135 FSGSIPRGFLP----NLYTLDLSNNMFTGEIYNDIGVFSNLRVLDLGGNVLTGHVPGYLG 190
Query: 195 NLTSLESLTLASNQLIGEIPTKICLMKRLKWIYLGYNNLSGEIPKNIGNLVSLNHLNLVY 254
NL+ LE LTLASNQL G +P ++ MK LKWIYLGYNNLSGEIP IG L SLNHL+LVY
Sbjct: 191 NLSRLEFLTLASNQLTGGVPVELGKMKNLKWIYLGYNNLSGEIPYQIGGLSSLNHLDLVY 250
Query: 255 NNLTGPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFNLKNLISLDLSDNYLSGEISNLVV 314
NNL+GPIP SLG+L L+Y+FLY NKL+G IP SIF+L+NLISLD SDN LSGEI LV
Sbjct: 251 NNLSGPIPPSLGDLKKLEYMFLYQNKLSGQIPPSIFSLQNLISLDFSDNSLSGEIPELVA 310
Query: 315 NLQKLEILHLFSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIHNNLTILDLSS 374
+Q LEILHLFSNN TGKIP +TSLP L+VLQLWSN+ +G IP LG HNNLT+LDLS+
Sbjct: 311 QMQSLEILHLFSNNLTGKIPEGVTSLPRLKVLQLWSNRFSGGIPANLGKHNNLTVLDLST 370
Query: 375 NNLTGKIPNSLCASKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDNNLSGKLPLEIT 434
NNLTGK+P++LC S +L K+ILFSNSL +IP L C++LERVRLQ+N SGKLP T
Sbjct: 371 NNLTGKLPDTLCDSGHLTKLILFSNSLDSQIPPSLGMCQSLERVRLQNNGFSGKLPRGFT 430
Query: 435 QLPQIYLLDISGNKFSGRINDRKWNMPSLQMLNLANNNFSGDLPNSFGGNKVEGLDLSQN 494
+L + LD+S N G IN W+MP L+ML+L+ N F G+LP+ +++ LDLS+N
Sbjct: 431 KLQLVNFLDLSNNNLQGNIN--TWDMPQLEMLDLSVNKFFGELPDFSRSKRLKKLDLSRN 488
Query: 495 QFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRLNGEIPEKLAK 554
+ SG + G PE++ L L+ N + G P EL C LV+LDLSHN GEIP A+
Sbjct: 489 KISGVVPQGLMTFPEIMDLDLSENEITGVIPRELSSCKNLVNLDLSHNNFTGEIPSSFAE 548
Query: 555 MPVLGLLDISENQFSGEIPKNLGSVESLVEVNISYNHFHGVLPSTEAFSAINASLVTGN- 613
VL LD+S NQ SGEIPKNLG++ESLV+VNIS+N HG LP T AF AINA+ V GN
Sbjct: 549 FQVLSDLDLSCNQLSGEIPKNLGNIESLVQVNISHNLLHGSLPFTGAFLAINATAVEGNI 608
Query: 614 KLCDGDGDVSNGLPPCKSYNQMNSTRLFVLICFVLTA-LVVLVG--TVVIFVLRMNKSFE 670
LC + ++GL PCK + ++ +++I A L VLV +V+ R + E
Sbjct: 609 DLCSENS--ASGLRPCKVVRKRSTKSWWLIITSTFAAFLAVLVSGFFIVLVFQRTHNVLE 666
Query: 671 VRRVVENEDGT-WEVIFFDYKASKFVTIEDVLSSVKEGKVITKGRNWVSYEGKCVSNEMQ 729
V++ VE EDGT WE FFD K K T+ +LSS+K+ V+ N +
Sbjct: 667 VKK-VEQEDGTKWETQFFDSKFMKSFTVNTILSSLKDQNVLVD------------KNGVH 713
Query: 730 FVVKEISDTNSVSVSFWDDTVTFGKKVRHENIVKIMGMFRCGKRGYLVYEFVEGKSLREI 789
FVVKE+ +S+ D K H+NI+KI+ R YL++E VEGK L ++
Sbjct: 714 FVVKEVKKYDSLPEMISD----MRKLSDHKNILKIVATCRSETVAYLIHEDVEGKRLSQV 769
Query: 790 MHGLSWLRRWKIALGIAKAINFLHCECLWFGLGSEVSPETVLVDGKGVPRLKLDSPGIVV 849
+ GLSW RR KI GI +A+ FLHC C + +SPE +++D PRL L PG++
Sbjct: 770 LSGLSWERRRKIMKGIVEALRFLHCRCSPAVVAGNLSPENIVIDVTDEPRLCLGLPGLLC 829
Query: 850 TPVMGVKGFVSSAYVAPEERNGKDVTEKSEIYGFGVILIELLTGR---NSVDIEAWNGIH 906
+ +AY+APE R K++T KS+IYGFG++L+ LLTG+ ++ DIE +G++
Sbjct: 830 ---------MDAAYMAPETREHKEMTSKSDIYGFGILLLHLLTGKCSSSNEDIE--SGVN 878
Query: 907 YKNNIVEWARYCYSDCHLDTWIDSVVMKGEDSSTYQNDIVETMNLALHCTANDPTTRPCA 966
++V+WARY YS+CH+DTWIDS + D+S +Q +IV MNLAL CTA DP RPC
Sbjct: 879 --GSLVKWARYSYSNCHIDTWIDSSI----DTSVHQREIVHVMNLALKCTAIDPQERPCT 932
Query: 967 RDILKALETVHCNTAT 982
++L+ALE+ ++++
Sbjct: 933 NNVLQALESTSSSSSS 948
>At4g28650 receptor protein kinase-like protein
Length = 1013
Score = 506 bits (1302), Expect = e-143
Identities = 326/1000 (32%), Positives = 524/1000 (51%), Gaps = 65/1000 (6%)
Query: 13 LNFICLFMFMLNFHSTHGE-------QEFELLLSFKASIKFDPLNFLSNWVNTSSDTICK 65
+ I LF++ ST E +LLS K+++ DPLNFL +W + + C
Sbjct: 3 MKIIVLFLYYCYIGSTSSVLASIDNVNELSVLLSVKSTL-VDPLNFLKDWKLSDTSDHCN 61
Query: 66 WHGITCDNWSHVNTVSLSGKNISGEVSSSIFQLPHVTNLDLSNNQLVGEIVFNSPFLSSL 125
W G+ C++ +V + L+G N++G++S SI QL + + ++S N + + P L S+
Sbjct: 62 WTGVRCNSNGNVEKLDLAGMNLTGKISDSISQLSSLVSFNISCNGFESLLPKSIPPLKSI 121
Query: 126 LYLNLSNNNLTGPLPQSLFSSSFINLETLDLSNNMFSGKIPDQIGLLSSLTYVDLGGNVL 185
++S N+ +G L LFS+ + L L+ S N SG + + +G L SL +DL GN
Sbjct: 122 ---DISQNSFSGSL--FLFSNESLGLVHLNASGNNLSGNLTEDLGNLVSLEVLDLRGNFF 176
Query: 186 VGKIPNSITNLTSLESLTLASNQLIGEIPTKICLMKRLKWIYLGYNNLSGEIPKNIGNLV 245
G +P+S NL L L L+ N L GE+P+ + + L+ LGYN G IP GN+
Sbjct: 177 QGSLPSSFKNLQKLRFLGLSGNNLTGELPSVLGQLPSLETAILGYNEFKGPIPPEFGNIN 236
Query: 246 SLNHLNLVYNNLTGPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFNLKNLISLDLSDNYL 305
SL +L+L L+G IP LG L +L+ L LY N TG IP+ I ++ L LD SDN L
Sbjct: 237 SLKYLDLAIGKLSGEIPSELGKLKSLETLLLYENNFTGTIPREIGSITTLKVLDFSDNAL 296
Query: 306 SGEISNLVVNLQKLEILHLFSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIHN 365
+GEI + L+ L++L+L N +G IP I+SL LQVL+LW+N L+GE+P LG ++
Sbjct: 297 TGEIPMEITKLKNLQLLNLMRNKLSGSIPPAISSLAQLQVLELWNNTLSGELPSDLGKNS 356
Query: 366 NLTILDLSSNNLTGKIPNSLCASKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDNNL 425
L LD+SSN+ +G+IP++LC NL K+ILF+N+ G+IP L++C++L RVR+Q+N L
Sbjct: 357 PLQWLDVSSNSFSGEIPSTLCNKGNLTKLILFNNTFTGQIPATLSTCQSLVRVRMQNNLL 416
Query: 426 SGKLPLEITQLPQIYLLDISGNKFSGRINDRKWNMPSLQMLNLANNNFSGDLPNS-FGGN 484
+G +P+ +L ++ L+++GN+ SG I + SL ++ + N LP++ +
Sbjct: 417 NGSIPIGFGKLEKLQRLELAGNRLSGGIPGDISDSVSLSFIDFSRNQIRSSLPSTILSIH 476
Query: 485 KVEGLDLSQNQFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRL 544
++ ++ N SG + F++ P L L L++N L G P + C KLVSL+L +N L
Sbjct: 477 NLQAFLVADNFISGEVPDQFQDCPSLSNLDLSSNTLTGTIPSSIASCEKLVSLNLRNNNL 536
Query: 545 NGEIPEKLAKMPVLGLLDISENQFSGEIPKNLGSVESLVEVNISYNHFHGVLPSTEAFSA 604
GEIP ++ M L +LD+S N +G +P+++G+ +L +N+SYN G +P
Sbjct: 537 TGEIPRQITTMSALAVLDLSNNSLTGVLPESIGTSPALELLNVSYNKLTGPVPINGFLKT 596
Query: 605 INASLVTGNK-LCDGDGDVSNGLPPCKSYNQMNST------RLFVLICFVLTALVVLVGT 657
IN + GN LC G LPPC + + S+ + V + A V+ +G
Sbjct: 597 INPDDLRGNSGLCGG------VLPPCSKFQRATSSHSSLHGKRIVAGWLIGIASVLALGI 650
Query: 658 VVIFVLRMNKSFEVRRVVENE---DGTWEVIFFDYKASKFVTIEDVLSSVKEGKVITKGR 714
+ I + K + +E G W + F T D+L+ +KE +I G
Sbjct: 651 LTIVTRTLYKKWYSNGFCGDETASKGEWPWRLMAFHRLGF-TASDILACIKESNMIGMGA 709
Query: 715 NWVSYEGKCVSNEMQFVVKEI----SDTNSVSVSFWDDTVTFGKKVRHENIVKIMGMFRC 770
+ Y+ + + VK++ +D + + V K+RH NIV+++G
Sbjct: 710 TGIVYKAEMSRSSTVLAVKKLWRSAADIEDGTTGDFVGEVNLLGKLRHRNIVRLLGFLYN 769
Query: 771 GKRGYLVYEFVEGKSLREIMHG--------LSWLRRWKIALGIAKAINFLHCECLWFGLG 822
K +VYEF+ +L + +HG + W+ R+ IALG+A + +LH +C +
Sbjct: 770 DKNMMIVYEFMLNGNLGDAIHGKNAAGRLLVDWVSRYNIALGVAHGLAYLHHDCHPPVIH 829
Query: 823 SEVSPETVLVDGK--------GVPRLKLDSPGIVVTPVMGVKGFVSSAYVAPEERNGKDV 874
++ +L+D G+ R+ + V+ V G S Y+APE V
Sbjct: 830 RDIKSNNILLDANLDARIADFGLARM-MARKKETVSMVAG-----SYGYIAPEYGYTLKV 883
Query: 875 TEKSEIYGFGVILIELLTGRNSVDIEAWNGIHYKNNIVEWARYCYSD-CHLDTWIDSVVM 933
EK +IY +GV+L+ELLTGR ++ E + +IVEW R D L+ +D V
Sbjct: 884 DEKIDIYSYGVVLLELLTGRRPLEPEFGESV----DIVEWVRRKIRDNISLEEALDPNV- 938
Query: 934 KGEDSSTYQNDIVETMNLALHCTANDPTTRPCARDILKAL 973
+ Q +++ + +AL CT P RP RD++ L
Sbjct: 939 --GNCRYVQEEMLLVLQIALLCTTKLPKDRPSMRDVISML 976
>At4g20270 CLV1 receptor kinase like protein
Length = 992
Score = 502 bits (1293), Expect = e-142
Identities = 340/976 (34%), Positives = 527/976 (53%), Gaps = 59/976 (6%)
Query: 32 QEFELLLSFKASI-KFDPLNFLSNWVNTSSDTICKWHGITCDNWSH-VNTVSLSGKNISG 89
++ +L+S K S +DP L +W + +++C W G++CDN + + + LS NISG
Sbjct: 33 RQANVLISLKQSFDSYDPS--LDSWNIPNFNSLCSWTGVSCDNLNQSITRLDLSNLNISG 90
Query: 90 EVSSSIFQL-PHVTNLDLSNNQLVGEIVFNSPFLSSLLYLNLSNNNLTGPLPQSLFSSSF 148
+S I +L P + LD+S+N GE+ LS L LN+S+N G L FS
Sbjct: 91 TISPEISRLSPSLVFLDISSNSFSGELPKEIYELSGLEVLNISSNVFEGELETRGFSQ-M 149
Query: 149 INLETLDLSNNMFSGKIPDQIGLLSSLTYVDLGGNVLVGKIPNSITNLTSLESLTLASNQ 208
L TLD +N F+G +P + L+ L ++DLGGN G+IP S + SL+ L+L+ N
Sbjct: 150 TQLVTLDAYDNSFNGSLPLSLTTLTRLEHLDLGGNYFDGEIPRSYGSFLSLKFLSLSGND 209
Query: 209 LIGEIPTKICLMKRLKWIYLGY-NNLSGEIPKNIGNLVSLNHLNLVYNNLTGPIPESLGN 267
L G IP ++ + L +YLGY N+ G IP + G L++L HL+L +L G IP LGN
Sbjct: 210 LRGRIPNELANITTLVQLYLGYYNDYRGGIPADFGRLINLVHLDLANCSLKGSIPAELGN 269
Query: 268 LTNLQYLFLYLNKLTGPIPKSIFNLKNLISLDLSDNYLSGEISNLVVNLQKLEILHLFSN 327
L NL+ LFL N+LTG +P+ + N+ +L +LDLS+N+L GEI + LQKL++ +LF N
Sbjct: 270 LKNLEVLFLQTNELTGSVPRELGNMTSLKTLDLSNNFLEGEIPLELSGLQKLQLFNLFFN 329
Query: 328 NFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIHNNLTILDLSSNNLTGKIPNSLCA 387
G+IP ++ LP LQ+L+LW N TG+IP LG + NL +DLS+N LTG IP SLC
Sbjct: 330 RLHGEIPEFVSELPDLQILKLWHNNFTGKIPSKLGSNGNLIEIDLSTNKLTGLIPESLCF 389
Query: 388 SKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDNNLSGKLPLEITQLPQIYLLDISGN 447
+ L +ILF+N L G +P+ L C+ L R RL N L+ KLP + LP + LL++ N
Sbjct: 390 GRRLKILILFNNFLFGPLPEDLGQCEPLWRFRLGQNFLTSKLPKGLIYLPNLSLLELQNN 449
Query: 448 KFSGRINDRK---WNMPSLQMLNLANNNFSGDLPNSFGG-NKVEGLDLSQNQFSGYIQIG 503
+G I + + SL +NL+NN SG +P S ++ L L N+ SG I
Sbjct: 450 FLTGEIPEEEAGNAQFSSLTQINLSNNRLSGPIPGSIRNLRSLQILLLGANRLSGQIPGE 509
Query: 504 FKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRLNGEIPEKLAKMPVLGLLDI 563
+L L+++ ++ NN GKFP E C L LDLSHN+++G+IP +++++ +L L++
Sbjct: 510 IGSLKSLLKIDMSRNNFSGKFPPEFGDCMSLTYLDLSHNQISGQIPVQISQIRILNYLNV 569
Query: 564 SENQFSGEIPKNLGSVESLVEVNISYNHFHGVLPSTEAFSAINASLVTGNK-LCDGDGDV 622
S N F+ +P LG ++SL + S+N+F G +P++ FS N + GN LC +
Sbjct: 570 SWNSFNQSLPNELGYMKSLTSADFSHNNFSGSVPTSGQFSYFNNTSFLGNPFLCGFSSNP 629
Query: 623 SNG---LPPCKSYNQMNS-TRLFVLICFVLTALVVLVGTVVIF-VLRMNKSFEVRRVVEN 677
NG + NQ N+ +R + F L + L+G ++F VL + K+ RR+ +N
Sbjct: 630 CNGSQNQSQSQLLNQNNARSRGEISAKFKLFFGLGLLGFFLVFVVLAVVKN---RRMRKN 686
Query: 678 EDGTWEVIFFDYKASKFVTIEDVLSSVKEGKVITKGRNWVSYEGKCVSNEMQFVVKEISD 737
W++I F + E +L VKE VI KG + Y+G + E V K ++
Sbjct: 687 NPNLWKLIGFQKLGFR---SEHILECVKENHVIGKGGRGIVYKGVMPNGEEVAVKKLLTI 743
Query: 738 T--NSVSVSFWDDTVTFGKKVRHENIVKIMGMFRCGKRGYLVYEFVEGKSLREIMHG--- 792
T +S + T G ++RH NIV+++ LVYE++ SL E++HG
Sbjct: 744 TKGSSHDNGLAAEIQTLG-RIRHRNIVRLLAFCSNKDVNLLVYEYMPNGSLGEVLHGKAG 802
Query: 793 --LSWLRRWKIALGIAKAINFLHCECLWFGLGSEVSPETVLVDGK--------GVPRLKL 842
L W R +IAL AK + +LH +C + +V +L+ + G+ + +
Sbjct: 803 VFLKWETRLQIALEAAKGLCYLHHDCSPLIIHRDVKSNNILLGPEFEAHVADFGLAKFMM 862
Query: 843 DSPGI--VVTPVMGVKGFVSSAYVAPEERNGKDVTEKSEIYGFGVILIELLTGRNSVDIE 900
G ++ + G S Y+APE + EKS++Y FGV+L+EL+TGR VD
Sbjct: 863 QDNGASECMSSIAG-----SYGYIAPEYAYTLRIDEKSDVYSFGVVLLELITGRKPVDNF 917
Query: 901 AWNGIHYKNNIVEWARYCYSDCHLDTWIDSVVMKGEDSSTYQNDIVETMNL---ALHCTA 957
GI +IV+W++ ++C+ V+K D + E M L A+ C
Sbjct: 918 GEEGI----DIVQWSK-IQTNCNRQG-----VVKIIDQRLSNIPLAEAMELFFVAMLCVQ 967
Query: 958 NDPTTRPCARDILKAL 973
RP R++++ +
Sbjct: 968 EHSVERPTMREVVQMI 983
>At1g08590 putative receptor kinase, CLV1
Length = 1029
Score = 498 bits (1281), Expect = e-141
Identities = 338/1024 (33%), Positives = 530/1024 (51%), Gaps = 78/1024 (7%)
Query: 15 FICLFMFMLNFHSTHGEQEFELLLSFKASIKFDPLNFLSNWVNTSSDTI------CKWHG 68
+I +F T E E+LL+FK+ + FDP N L +W + T C W G
Sbjct: 12 YIGFALFPFVSSETFQNSEQEILLAFKSDL-FDPSNNLQDWKRPENATTFSELVHCHWTG 70
Query: 69 ITCDNWSHVNTVSLSGKNISGEVSSSIFQLPHVTNLDLSNNQLVGEIVFNSPFLSSLLYL 128
+ CD +V + LS N+SG VS I P + LDLSNN + + L+SL +
Sbjct: 71 VHCDANGYVAKLLLSNMNLSGNVSDQIQSFPSLQALDLSNNAFESSLPKSLSNLTSLKVI 130
Query: 129 NLSNNNLTGPLPQSLFSSSFINLETLDLSNNMFSGKIPDQIGLLSSLTYVDLGGNVLVGK 188
++S N+ G P L ++ L ++ S+N FSG +P+ +G ++L +D G G
Sbjct: 131 DVSVNSFFGTFPYGLGMAT--GLTHVNASSNNFSGFLPEDLGNATTLEVLDFRGGYFEGS 188
Query: 189 IPNSITNLTSLESLTLASNQLIGEIPTKICLMKRLKWIYLGYNNLSGEIPKNIGNLVSLN 248
+P+S NL +L+ L L+ N G++P I + L+ I LGYN GEIP+ G L L
Sbjct: 189 VPSSFKNLKNLKFLGLSGNNFGGKVPKVIGELSSLETIILGYNGFMGEIPEEFGKLTRLQ 248
Query: 249 HLNLVYNNLTGPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFNLKNLISLDLSDNYLSGE 308
+L+L NLTG IP SLG L L ++LY N+LTG +P+ + + +L+ LDLSDN ++GE
Sbjct: 249 YLDLAVGNLTGQIPSSLGQLKQLTTVYLYQNRLTGKLPRELGGMTSLVFLDLSDNQITGE 308
Query: 309 ISNLVVNLQKLEILHLFSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIHNNLT 368
I V L+ L++L+L N TG IP+ I LP+L+VL+LW N L G +P LG ++ L
Sbjct: 309 IPMEVGELKNLQLLNLMRNQLTGIIPSKIAELPNLEVLELWQNSLMGSLPVHLGKNSPLK 368
Query: 369 ILDLSSNNLTGKIPNSLCASKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDNNLSGK 428
LD+SSN L+G IP+ LC S+NL K+ILF+NS G+IP+ + SC TL RVR+Q N++SG
Sbjct: 369 WLDVSSNKLSGDIPSGLCYSRNLTKLILFNNSFSGQIPEEIFSCPTLVRVRIQKNHISGS 428
Query: 429 LPLEITQLPQIYLLDISGNKFSGRINDRKWNMPSLQMLNLANNNFSGDLPNSFGGNKVEG 488
+P LP + L+++ N +G+I D SL ++++ N+ S + F ++
Sbjct: 429 IPAGSGDLPMLQHLELAKNNLTGKIPDDIALSTSLSFIDISFNHLSSLSSSIFSSPNLQT 488
Query: 489 LDLSQNQFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRLNGEI 548
S N F+G I ++ P L L L+ N+ G PE + KLVSL+L N+L GEI
Sbjct: 489 FIASHNNFAGKIPNQIQDRPSLSVLDLSFNHFSGGIPERIASFEKLVSLNLKSNQLVGEI 548
Query: 549 PEKLAKMPVLGLLDISENQFSGEIPKNLGSVESLVEVNISYNHFHGVLPSTEAFSAIN-A 607
P+ LA M +L +LD+S N +G IP +LG+ +L +N+S+N G +PS F+AI+
Sbjct: 549 PKALAGMHMLAVLDLSNNSLTGNIPADLGASPTLEMLNVSFNKLDGPIPSNMLFAAIDPK 608
Query: 608 SLVTGNKLCDGDGDVSNGLPPCK-----SYNQMNSTRLFV---LICFVL-TALVVLVGTV 658
LV N LC G LPPC S N R+ V + F++ T+++V +G +
Sbjct: 609 DLVGNNGLCGG------VLPPCSKSLALSAKGRNPGRIHVNHAVFGFIVGTSVIVAMGMM 662
Query: 659 VI--------FVLRMNKSFE-VRRVVENEDGTWEVIFFDYKASKFVTIEDVLSSVKEGKV 709
+ + L N + E + E+ W ++ F T D+LS +KE +
Sbjct: 663 FLAGRWIYTRWDLYSNFAREYIFCKKPREEWPWRLVAFQRLC---FTAGDILSHIKESNI 719
Query: 710 ITKGRNWVSYEGKCVSNEMQFV-VKEI----SDTNSVSVSFWDD--------TVTFGKKV 756
I G + Y+ + + + V VK++ S N + ++ V +
Sbjct: 720 IGMGAIGIVYKAEVMRRPLLTVAVKKLWRSPSPQNDIEDHHQEEDEEDDILREVNLLGGL 779
Query: 757 RHENIVKIMGMFRCGKRGYLVYEFVEGKSLREIMHG-------LSWLRRWKIALGIAKAI 809
RH NIVKI+G + +VYE++ +L +H WL R+ +A+G+ + +
Sbjct: 780 RHRNIVKILGYVHNEREVMMVYEYMPNGNLGTALHSKDEKFLLRDWLSRYNVAVGVVQGL 839
Query: 810 NFLHCECLWFGLGSEVSPETVLVDGK--------GVPRLKLDSPGIVVTPVMGVKGFVSS 861
N+LH +C + ++ +L+D G+ ++ L V+ V G S
Sbjct: 840 NYLHNDCYPPIIHRDIKSNNILLDSNLEARIADFGLAKMMLHK-NETVSMVAG-----SY 893
Query: 862 AYVAPEERNGKDVTEKSEIYGFGVILIELLTGRNSVDIEAWNGIHYKNNIVEW-ARYCYS 920
Y+APE + EKS+IY GV+L+EL+TG+ +D + I ++VEW R
Sbjct: 894 GYIAPEYGYTLKIDEKSDIYSLGVVLLELVTGKMPIDPSFEDSI----DVVEWIRRKVKK 949
Query: 921 DCHLDTWIDSVVMKGEDSSTYQNDIVETMNLALHCTANDPTTRPCARDILKALETVHCNT 980
+ L+ ID+ + D +++ + +AL CTA P RP RD++ L
Sbjct: 950 NESLEEVIDASI--AGDCKHVIEEMLLALRIALLCTAKLPKDRPSIRDVITMLAEAKPRR 1007
Query: 981 ATLC 984
++C
Sbjct: 1008 KSVC 1011
>At3g49670 receptor protein kinase - like protein
Length = 1002
Score = 496 bits (1278), Expect = e-140
Identities = 334/992 (33%), Positives = 512/992 (50%), Gaps = 56/992 (5%)
Query: 13 LNFICLFMFMLNFHSTHGEQ------EFELLLSFKASIKFDPLN-FLSNWVNTSSDTICK 65
+ + L + +L H +H E LLS K+S D + L++W S T C
Sbjct: 1 MKLLLLLLLLLLLHISHSFTVAKPITELHALLSLKSSFTIDEHSPLLTSW--NLSTTFCS 58
Query: 66 WHGITCD-NWSHVNTVSLSGKNISGEVSSSIFQLPHVTNLDLSNNQLVGEIVFNSPFLSS 124
W G+TCD + HV ++ LSG N+SG +SS + LP + NL L+ NQ+ G I L
Sbjct: 59 WTGVTCDVSLRHVTSLDLSGLNLSGTLSSDVAHLPLLQNLSLAANQISGPIPPQISNLYE 118
Query: 125 LLYLNLSNNNLTGPLPQSLFSSSFINLETLDLSNNMFSGKIPDQIGLLSSLTYVDLGGNV 184
L +LNLSNN G P L SS +NL LDL NN +G +P + L+ L ++ LGGN
Sbjct: 119 LRHLNLSNNVFNGSFPDEL-SSGLVNLRVLDLYNNNLTGDLPVSLTNLTQLRHLHLGGNY 177
Query: 185 LVGKIPNSITNLTSLESLTLASNQLIGEIPTKICLMKRLKWIYLGYNN-LSGEIPKNIGN 243
GKIP + LE L ++ N+L G+IP +I + L+ +Y+GY N +P IGN
Sbjct: 178 FSGKIPATYGTWPVLEYLAVSGNELTGKIPPEIGNLTTLRELYIGYYNAFENGLPPEIGN 237
Query: 244 LVSLNHLNLVYNNLTGPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFNLKNLISLDLSDN 303
L L + LTG IP +G L L LFL +N TG I + + + +L S+DLS+N
Sbjct: 238 LSELVRFDAANCGLTGEIPPEIGKLQKLDTLFLQVNAFTGTITQELGLISSLKSMDLSNN 297
Query: 304 YLSGEISNLVVNLQKLEILHLFSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGI 363
+GEI L+ L +L+LF N G IP I +P L+VLQLW N TG IPQ LG
Sbjct: 298 MFTGEIPTSFSQLKNLTLLNLFRNKLYGAIPEFIGEMPELEVLQLWENNFTGSIPQKLGE 357
Query: 364 HNNLTILDLSSNNLTGKIPNSLCASKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDN 423
+ L ILDLSSN LTG +P ++C+ L +I N L G IP L C++L R+R+ +N
Sbjct: 358 NGRLVILDLSSNKLTGTLPPNMCSGNRLMTLITLGNFLFGSIPDSLGKCESLTRIRMGEN 417
Query: 424 NLSGKLPLEITQLPQIYLLDISGNKFSGRINDRKWNMP-SLQMLNLANNNFSGDLPNSFG 482
L+G +P E+ LP++ +++ N +G + + L ++L+NN SG LP + G
Sbjct: 418 FLNGSIPKELFGLPKLSQVELQDNYLTGELPISGGGVSGDLGQISLSNNQLSGSLPAAIG 477
Query: 483 G-NKVEGLDLSQNQFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSH 541
+ V+ L L N+FSG I L +L +L ++N G+ E+ +C L +DLS
Sbjct: 478 NLSGVQKLLLDGNKFSGSIPPEIGRLQQLSKLDFSHNLFSGRIAPEISRCKLLTFVDLSR 537
Query: 542 NRLNGEIPEKLAKMPVLGLLDISENQFSGEIPKNLGSVESLVEVNISYNHFHGVLPSTEA 601
N L+G+IP +L M +L L++S N G IP + S++SL V+ SYN+ G++PST
Sbjct: 538 NELSGDIPNELTGMKILNYLNLSRNHLVGSIPVTIASMQSLTSVDFSYNNLSGLVPSTGQ 597
Query: 602 FSAINASLVTGNK-LCDGDGDVSNGLPPCKSYNQMNSTRLFVLICFVLTALVVLVGTVVI 660
FS N + GN LC L PC + + +L L +L ++V
Sbjct: 598 FSYFNYTSFVGNSHLC------GPYLGPCGKGTHQSHVKPLSATTKLLLVLGLLFCSMVF 651
Query: 661 FVLRMNKSFEVRRVVENEDGTWEVIFFDYKASKFVTIEDVLSSVKEGKVITKGRNWVSYE 720
++ + K+ +R +E W + F T +DVL S+KE +I KG + Y+
Sbjct: 652 AIVAIIKARSLRNA--SEAKAWRLTAFQ---RLDFTCDDVLDSLKEDNIIGKGGAGIVYK 706
Query: 721 GKCVSNEMQFV--VKEISDTNSVSVSFWDDTVTFGKKVRHENIVKIMGMFRCGKRGYLVY 778
G ++ V + +S +S F + T G ++RH +IV+++G + LVY
Sbjct: 707 GTMPKGDLVAVKRLATMSHGSSHDHGFNAEIQTLG-RIRHRHIVRLLGFCSNHETNLLVY 765
Query: 779 EFVEGKSLREIMHG-----LSWLRRWKIALGIAKAINFLHCECLWFGLGSEVSPETVLVD 833
E++ SL E++HG L W R+KIAL AK + +LH +C + +V +L+D
Sbjct: 766 EYMPNGSLGEVLHGKKGGHLHWNTRYKIALEAAKGLCYLHHDCSPLIVHRDVKSNNILLD 825
Query: 834 GK--------GVPRLKLDS-PGIVVTPVMGVKGFVSSAYVAPEERNGKDVTEKSEIYGFG 884
G+ + DS ++ + G S Y+APE V EKS++Y FG
Sbjct: 826 SNFEAHVADFGLAKFLQDSGTSECMSAIAG-----SYGYIAPEYAYTLKVDEKSDVYSFG 880
Query: 885 VILIELLTGRNSVDIEAWNGIHYKNNIVEWARYCYSDCHLDTWIDSVVMKGEDSSTYQND 944
V+L+EL+TG+ V E +G+ +IV+W R +D + D + + ++ SS ++
Sbjct: 881 VVLLELITGKKPVG-EFGDGV----DIVQWVR-SMTDSNKDCVLKVIDLR--LSSVPVHE 932
Query: 945 IVETMNLALHCTANDPTTRPCARDILKALETV 976
+ +AL C RP R++++ L +
Sbjct: 933 VTHVFYVALLCVEEQAVERPTMREVVQILTEI 964
>At1g75820 receptor kinase (CLV1)
Length = 980
Score = 491 bits (1264), Expect = e-139
Identities = 326/997 (32%), Positives = 521/997 (51%), Gaps = 55/997 (5%)
Query: 13 LNFICLFMFMLNFHSTHGEQEFELLLSFKASIKFDPLNFLSNWVNTSS-DTICKWHGITC 71
L F+ L++F F + E+LL+ K+S+ + L +W+++SS D C + G++C
Sbjct: 10 LLFLHLYLF---FSPCFAYTDMEVLLNLKSSMIGPKGHGLHDWIHSSSPDAHCSFSGVSC 66
Query: 72 DNWSHVNTVSLSGKNISGEVSSSIFQLPHVTNLDLSNNQLVGEIVFNSPFLSSLLYLNLS 131
D+ + V ++++S + G +S I L H+ NL L+ N GE+ L+SL LN+S
Sbjct: 67 DDDARVISLNVSFTPLFGTISPEIGMLTHLVNLTLAANNFTGELPLEMKSLTSLKVLNIS 126
Query: 132 NN-NLTGPLPQSLFSSSFINLETLDLSNNMFSGKIPDQIGLLSSLTYVDLGGNVLVGKIP 190
NN NLTG P + + ++LE LD NN F+GK+P ++ L L Y+ GGN G+IP
Sbjct: 127 NNGNLTGTFPGEILKA-MVDLEVLDTYNNNFNGKLPPEMSELKKLKYLSFGGNFFSGEIP 185
Query: 191 NSITNLTSLESLTLASNQLIGEIPTKICLMKRLKWIYLGY-NNLSGEIPKNIGNLVSLNH 249
S ++ SLE L L L G+ P + +K L+ +Y+GY N+ +G +P G L L
Sbjct: 186 ESYGDIQSLEYLGLNGAGLSGKSPAFLSRLKNLREMYIGYYNSYTGGVPPEFGGLTKLEI 245
Query: 250 LNLVYNNLTGPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFNLKNLISLDLSDNYLSGEI 309
L++ LTG IP SL NL +L LFL++N LTG IP + L +L SLDLS N L+GEI
Sbjct: 246 LDMASCTLTGEIPTSLSNLKHLHTLFLHINNLTGHIPPELSGLVSLKSLDLSINQLTGEI 305
Query: 310 SNLVVNLQKLEILHLFSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIHNNLTI 369
+NL + +++LF NN G+IP I LP L+V ++W N T ++P LG + NL
Sbjct: 306 PQSFINLGNITLINLFRNNLYGQIPEAIGELPKLEVFEVWENNFTLQLPANLGRNGNLIK 365
Query: 370 LDLSSNNLTGKIPNSLCASKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDNNLSGKL 429
LD+S N+LTG IP LC + L +IL +N G IP+ L CK+L ++R+ N L+G +
Sbjct: 366 LDVSDNHLTGLIPKDLCRGEKLEMLILSNNFFFGPIPEELGKCKSLTKIRIVKNLLNGTV 425
Query: 430 PLEITQLPQIYLLDISGNKFSGRINDRKWNMPSLQMLNLANNNFSGDLPNSFGG-NKVEG 488
P + LP + +++++ N FSG + + L + L+NN FSG++P + G ++
Sbjct: 426 PAGLFNLPLVTIIELTDNFFSGEL-PVTMSGDVLDQIYLSNNWFSGEIPPAIGNFPNLQT 484
Query: 489 LDLSQNQFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRLNGEI 548
L L +N+F G I L L ++ + NN+ G P+ + +C+ L+S+DLS NR+NGEI
Sbjct: 485 LFLDRNRFRGNIPREIFELKHLSRINTSANNITGGIPDSISRCSTLISVDLSRNRINGEI 544
Query: 549 PEKLAKMPVLGLLDISENQFSGEIPKNLGSVESLVEVNISYNHFHGVLPSTEAFSAINAS 608
P+ + + LG L+IS NQ +G IP +G++ SL +++S+N G +P F N +
Sbjct: 545 PKGINNVKNLGTLNISGNQLTGSIPTGIGNMTSLTTLDLSFNDLSGRVPLGGQFLVFNET 604
Query: 609 LVTGNKLCDGDGDVSNGLPPCKSYNQMNSTRLFVLICFVLTALVVLVGTVVIFVL--RMN 666
GN VS P ++ + N T LF V+T + + G ++I V +MN
Sbjct: 605 SFAGNTYLCLPHRVSCPTRPGQT-SDHNHTALFSPSRIVITVIAAITGLILISVAIRQMN 663
Query: 667 KSFEVRRVVENEDGTWEVIFF---DYKASKFVTIEDVLSSVKEGKVITKGRNWVSYEGKC 723
K + + W++ F D+K+ EDVL +KE +I KG + Y G
Sbjct: 664 KKKNQKSL------AWKLTAFQKLDFKS------EDVLECLKEENIIGKGGAGIVYRGS- 710
Query: 724 VSNEMQFVVKEI--SDTNSVSVSFWDDTVTFGKKVRHENIVKIMGMFRCGKRGYLVYEFV 781
+ N + +K + T F + T G ++RH +IV+++G L+YE++
Sbjct: 711 MPNNVDVAIKRLVGRGTGRSDHGFTAEIQTLG-RIRHRHIVRLLGYVANKDTNLLLYEYM 769
Query: 782 EGKSLREIMHG-----LSWLRRWKIALGIAKAINFLHCECLWFGLGSEVSPETVLVDGK- 835
SL E++HG L W R ++A+ AK + +LH +C L +V +L+D
Sbjct: 770 PNGSLGELLHGSKGGHLQWETRHRVAVEAAKGLCYLHHDCSPLILHRDVKSNNILLDSDF 829
Query: 836 -------GVPRLKLDSPGIVVTPVMGVKGFVSSAYVAPEERNGKDVTEKSEIYGFGVILI 888
G+ + +D G + + G S Y+APE V EKS++Y FGV+L+
Sbjct: 830 EAHVADFGLAKFLVD--GAASECMSSIAG--SYGYIAPEYAYTLKVDEKSDVYSFGVVLL 885
Query: 889 ELLTGRNSVDIEAWNGIHYKNNIVEWARYCYSDC--HLDTWIDSVVMKGEDSSTYQNDIV 946
EL+ G+ V E G+ +IV W R + D I ++ + ++
Sbjct: 886 ELIAGKKPVG-EFGEGV----DIVRWVRNTEEEITQPSDAAIVVAIVDPRLTGYPLTSVI 940
Query: 947 ETMNLALHCTANDPTTRPCARDILKALETVHCNTATL 983
+A+ C + RP R+++ L + A L
Sbjct: 941 HVFKIAMMCVEEEAAARPTMREVVHMLTNPPKSVANL 977
>At5g65700 receptor protein kinase-like protein
Length = 1003
Score = 486 bits (1250), Expect = e-137
Identities = 328/992 (33%), Positives = 519/992 (52%), Gaps = 52/992 (5%)
Query: 13 LNFICLFMFMLN----FHSTHGEQEFELLLSFKASIKF---DPLNFLSNWVNTSSDTICK 65
+ L +F+L+ F ++ EF LLS K S+ D + LS+W ++S C
Sbjct: 1 MKLFLLLLFLLHISHTFTASRPISEFRALLSLKTSLTGAGDDKNSPLSSWKVSTS--FCT 58
Query: 66 WHGITCD-NWSHVNTVSLSGKNISGEVSSSIFQLPHVTNLDLSNNQLVGEIVFNSPFLSS 124
W G+TCD + HV ++ LSG N+SG +S + L + NL L+ N + G I LS
Sbjct: 59 WIGVTCDVSRRHVTSLDLSGLNLSGTLSPDVSHLRLLQNLSLAENLISGPIPPEISSLSG 118
Query: 125 LLYLNLSNNNLTGPLPQSLFSSSFINLETLDLSNNMFSGKIPDQIGLLSSLTYVDLGGNV 184
L +LNLSNN G P + SS +NL LD+ NN +G +P + L+ L ++ LGGN
Sbjct: 119 LRHLNLSNNVFNGSFPDEI-SSGLVNLRVLDVYNNNLTGDLPVSVTNLTQLRHLHLGGNY 177
Query: 185 LVGKIPNSITNLTSLESLTLASNQLIGEIPTKICLMKRLKWIYLGYNNLSGE-IPKNIGN 243
GKIP S + +E L ++ N+L+G+IP +I + L+ +Y+GY N + +P IGN
Sbjct: 178 FAGKIPPSYGSWPVIEYLAVSGNELVGKIPPEIGNLTTLRELYIGYYNAFEDGLPPEIGN 237
Query: 244 LVSLNHLNLVYNNLTGPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFNLKNLISLDLSDN 303
L L + LTG IP +G L L LFL +N +GP+ + L +L S+DLS+N
Sbjct: 238 LSELVRFDGANCGLTGEIPPEIGKLQKLDTLFLQVNVFSGPLTWELGTLSSLKSMDLSNN 297
Query: 304 YLSGEISNLVVNLQKLEILHLFSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGI 363
+GEI L+ L +L+LF N G+IP I LP L+VLQLW N TG IPQ LG
Sbjct: 298 MFTGEIPASFAELKNLTLLNLFRNKLHGEIPEFIGDLPELEVLQLWENNFTGSIPQKLGE 357
Query: 364 HNNLTILDLSSNNLTGKIPNSLCASKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDN 423
+ L ++DLSSN LTG +P ++C+ L +I N L G IP L C++L R+R+ +N
Sbjct: 358 NGKLNLVDLSSNKLTGTLPPNMCSGNKLETLITLGNFLFGSIPDSLGKCESLTRIRMGEN 417
Query: 424 NLSGKLPLEITQLPQIYLLDISGNKFSGRINDRKWNMPSLQMLNLANNNFSGDLPNSFGG 483
L+G +P + LP++ +++ N SG + +L ++L+NN SG LP + G
Sbjct: 418 FLNGSIPKGLFGLPKLTQVELQDNYLSGELPVAGGVSVNLGQISLSNNQLSGPLPPAIGN 477
Query: 484 -NKVEGLDLSQNQFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHN 542
V+ L L N+F G I L +L ++ ++N G+ E+ +C L +DLS N
Sbjct: 478 FTGVQKLLLDGNKFQGPIPSEVGKLQQLSKIDFSHNLFSGRIAPEISRCKLLTFVDLSRN 537
Query: 543 RLNGEIPEKLAKMPVLGLLDISENQFSGEIPKNLGSVESLVEVNISYNHFHGVLPSTEAF 602
L+GEIP ++ M +L L++S N G IP ++ S++SL ++ SYN+ G++P T F
Sbjct: 538 ELSGEIPNEITAMKILNYLNLSRNHLVGSIPGSISSMQSLTSLDFSYNNLSGLVPGTGQF 597
Query: 603 SAINASLVTGNKLCDGD--GDVSNGLPPCKSYNQMNSTRLFVLICFVLTALVVLVGTVVI 660
S N + GN G G +G+ K +Q +S +L L +LV ++
Sbjct: 598 SYFNYTSFLGNPDLCGPYLGPCKDGV--AKGGHQSHSKGPLSASMKLLLVLGLLVCSIAF 655
Query: 661 FVLRMNKSFEVRRVVENEDGTWEVIFFDYKASKFVTIEDVLSSVKEGKVITKGRNWVSYE 720
V+ + K+ +++ +E W + F T +DVL S+KE +I KG + Y+
Sbjct: 656 AVVAIIKARSLKKA--SESRAWRLTAFQ---RLDFTCDDVLDSLKEDNIIGKGGAGIVYK 710
Query: 721 GKCVSNEMQFV--VKEISDTNSVSVSFWDDTVTFGKKVRHENIVKIMGMFRCGKRGYLVY 778
G + ++ V + +S +S F + T G ++RH +IV+++G + LVY
Sbjct: 711 GVMPNGDLVAVKRLAAMSRGSSHDHGFNAEIQTLG-RIRHRHIVRLLGFCSNHETNLLVY 769
Query: 779 EFVEGKSLREIMHG-----LSWLRRWKIALGIAKAINFLHCECLWFGLGSEVSPETVLVD 833
E++ SL E++HG L W R+KIAL AK + +LH +C + +V +L+D
Sbjct: 770 EYMPNGSLGEVLHGKKGGHLHWDTRYKIALEAAKGLCYLHHDCSPLIVHRDVKSNNILLD 829
Query: 834 GK--------GVPRLKLDS-PGIVVTPVMGVKGFVSSAYVAPEERNGKDVTEKSEIYGFG 884
G+ + DS ++ + G S Y+APE V EKS++Y FG
Sbjct: 830 SNFEAHVADFGLAKFLQDSGTSECMSAIAG-----SYGYIAPEYAYTLKVDEKSDVYSFG 884
Query: 885 VILIELLTGRNSVDIEAWNGIHYKNNIVEWARYCYSDCHLDTWIDSVVMKGEDSSTYQND 944
V+L+EL+TGR V E +G+ +IV+W R +D + D+ + V+ SS ++
Sbjct: 885 VVLLELVTGRKPVG-EFGDGV----DIVQWVRK-MTDSNKDSVLK--VLDPRLSSIPIHE 936
Query: 945 IVETMNLALHCTANDPTTRPCARDILKALETV 976
+ +A+ C RP R++++ L +
Sbjct: 937 VTHVFYVAMLCVEEQAVERPTMREVVQILTEI 968
>At1g34110 putative protein
Length = 1049
Score = 466 bits (1200), Expect = e-131
Identities = 322/1036 (31%), Positives = 517/1036 (49%), Gaps = 123/1036 (11%)
Query: 43 SIKFDPLNFLSNWVNTSSDTICKWHGITCDNWSHVNTVS--------------------- 81
S+K + S+W + T C W+GITC + V +VS
Sbjct: 17 SLKRPSPSLFSSW-DPQDQTPCSWYGITCSADNRVISVSIPDTFLNLSSIPDLSSLSSLQ 75
Query: 82 ---LSGKNISGEVSSSIFQLPHVTNLDLSNNQLVGEIVFNSPFLSSLLYLNLSNNNLTGP 138
LS N+SG + S +L H+ LDLS+N L G I LS+L +L L+ N L+G
Sbjct: 76 FLNLSSTNLSGPIPPSFGKLTHLRLLDLSSNSLSGPIPSELGRLSTLQFLILNANKLSGS 135
Query: 139 LPQSLFSSSFINLETLDLSNNMFSGKIPDQIGLLSSLTYVDLGGNV-------------- 184
+P + S+ L+ L L +N+ +G IP G L SL LGGN
Sbjct: 136 IPSQI--SNLFALQVLCLQDNLLNGSIPSSFGSLVSLQQFRLGGNTNLGGPIPAQLGFLK 193
Query: 185 -----------LVGKIPNSITNLTSLESLTLASNQLIGEIPTKICLMKRLKWIYLGYNNL 233
L G IP++ NL +L++L L ++ G IP ++ L L+ +YL N L
Sbjct: 194 NLTTLGFAASGLSGSIPSTFGNLVNLQTLALYDTEISGTIPPQLGLCSELRNLYLHMNKL 253
Query: 234 SGEIPKNIGNLVSLNHLNLVYNNLTGPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFNLK 293
+G IPK +G L + L L N+L+G IP + N ++L + N LTG IP + L
Sbjct: 254 TGSIPKELGKLQKITSLLLWGNSLSGVIPPEISNCSSLVVFDVSANDLTGDIPGDLGKLV 313
Query: 294 NLISLDLSDNYLSGEISNLVVNLQKLEILHLFSNNFTGKIPNTITSLPHLQVLQLWSNKL 353
L L LSDN +G+I + N L L L N +G IP+ I +L LQ LW N +
Sbjct: 314 WLEQLQLSDNMFTGQIPWELSNCSSLIALQLDKNKLSGSIPSQIGNLKSLQSFFLWENSI 373
Query: 354 TGEIPQTLGIHNNLTILDLSSNNLTGKIPNSLCASKNLHKIILFSNSLKGEIPKGLTSCK 413
+G IP + G +L LDLS N LTG+IP L + K L K++L NSL G +PK + C+
Sbjct: 374 SGTIPSSFGNCTDLVALDLSRNKLTGRIPEELFSLKRLSKLLLLGNSLSGGLPKSVAKCQ 433
Query: 414 TLERVRLQDNNLSGKLPLEITQLPQIYLLDISGNKFSGRINDRKWNMPSLQMLNLANNNF 473
+L R+R+ +N LSG++P EI +L + LD+ N FSG + N+ L++L++ NN
Sbjct: 434 SLVRLRVGENQLSGQIPKEIGELQNLVFLDLYMNHFSGGLPYEISNITVLELLDVHNNYI 493
Query: 474 SGDLPNSFGG-NKVEGLDLSQNQFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCN 532
+GD+P G +E LDLS+N F+G I + F NL L +L LNNN L G+ P+ +
Sbjct: 494 TGDIPAQLGNLVNLEQLDLSRNSFTGNIPLSFGNLSYLNKLILNNNLLTGQIPKSIKNLQ 553
Query: 533 KLVSLDLSHNRLNGEIPEKLAKMPVLGL-LDISENQFSGEIP------------------ 573
KL LDLS+N L+GEIP++L ++ L + LD+S N F+G IP
Sbjct: 554 KLTLLDLSYNSLSGEIPQELGQVTSLTINLDLSYNTFTGNIPETFSDLTQLQSLDLSSNS 613
Query: 574 -----KNLGSVESLVEVNISYNHFHGVLPSTEAFSAIN-ASLVTGNKLCDGDGDVSNGLP 627
K LGS+ SL +NIS N+F G +PST F I+ S + LC ++
Sbjct: 614 LHGDIKVLGSLTSLASLNISCNNFSGPIPSTPFFKTISTTSYLQNTNLCHSLDGIT---- 669
Query: 628 PCKSY----NQMNSTRLFVLICFVLTALVVLVGTVVIFVLRMNKSFEVRRVVENEDGTWE 683
C S+ N + S ++ L +L ++ + + + +LR N ++ + + T E
Sbjct: 670 -CSSHTGQNNGVKSPKIVALTAVILASITIAILAAWLLILRNNHLYKTSQNSSSSPSTAE 728
Query: 684 VIFFDYKASKF----VTIEDVLSSVKEGKVITKGRNWVSYEGKCVSNEMQFVVK--EISD 737
+ + F +T+ ++++S+ + VI KG + + Y+ + + ++ V K + D
Sbjct: 729 DFSYPWTFIPFQKLGITVNNIVTSLTDENVIGKGCSGIVYKAEIPNGDIVAVKKLWKTKD 788
Query: 738 TNSVSVSFWDD---TVTFGKKVRHENIVKIMGMFRCGKRGYLVYEFVEGKSLREIMHG-- 792
N S D + +RH NIVK++G L+Y + +L++++ G
Sbjct: 789 NNEEGESTIDSFAAEIQILGNIRHRNIVKLLGYCSNKSVKLLLYNYFPNGNLQQLLQGNR 848
Query: 793 -LSWLRRWKIALGIAKAINFLHCECLWFGLGSEVSPETVLVDGK--------GVPRLKLD 843
L W R+KIA+G A+ + +LH +C+ L +V +L+D K G+ +L ++
Sbjct: 849 NLDWETRYKIAIGAAQGLAYLHHDCVPAILHRDVKCNNILLDSKYEAILADFGLAKLMMN 908
Query: 844 SPGIVVTPVMGVKGFVSSAYVAPEERNGKDVTEKSEIYGFGVILIELLTGRNSVDIEAWN 903
SP + ++ E ++TEKS++Y +GV+L+E+L+GR++V+ + +
Sbjct: 909 SP-----------NYHNAMSRVAEYGYTMNITEKSDVYSYGVVLLEILSGRSAVEPQIGD 957
Query: 904 GIHYKNNIVEWARYCYSDCHLDTWIDSVVMKGEDSSTYQNDIVETMNLALHCTANDPTTR 963
G+H IVEW + + V ++G Q ++++T+ +A+ C P R
Sbjct: 958 GLH----IVEWVKKKMGTFEPALSVLDVKLQGLPDQIVQ-EMLQTLGIAMFCVNPSPVER 1012
Query: 964 PCARDILKALETVHCN 979
P ++++ L V C+
Sbjct: 1013 PTMKEVVTLLMEVKCS 1028
>At5g25930 receptor-like protein kinase - like
Length = 1005
Score = 461 bits (1185), Expect = e-129
Identities = 337/999 (33%), Positives = 509/999 (50%), Gaps = 86/999 (8%)
Query: 18 LFMFM----LNFHSTHGEQEFELLLSFKASIKFDPLNFLSNWVNTSSDTICKWHGITCDN 73
LF F+ L+ S +Q LL+ K + P L W NTSS C W ITC
Sbjct: 9 LFFFLTSIPLSVFSQFNDQS--TLLNLKRDLGDPPS--LRLWNNTSSP--CNWSEITCTA 62
Query: 74 WSHVNTVSLSGKNISGEVSSSIFQLPHVTNLDLSNNQLVGEIVFNSPFLSSLLYLNLSNN 133
+V ++ +N +G V ++I L ++ LDLS N GE + L YL+LS N
Sbjct: 63 -GNVTGINFKNQNFTGTVPTTICDLSNLNFLDLSFNYFAGEFPTVLYNCTKLQYLDLSQN 121
Query: 134 NLTGPLPQSLFSSSFINLETLDLSNNMFSGKIPDQIGLLSSLTYVDLGGNVLVGKIPNSI 193
L G LP + S L+ LDL+ N FSG IP +G +S L ++L + G P+ I
Sbjct: 122 LLNGSLPVDIDRLS-PELDYLDLAANGFSGDIPKSLGRISKLKVLNLYQSEYDGTFPSEI 180
Query: 194 TNLTSLESLTLASNQLI--GEIPTKICLMKRLKWIYLGYNNLSGEI-PKNIGNLVSLNHL 250
+L+ LE L LA N +IP + +K+LK+++L NL GEI P N+ L H+
Sbjct: 181 GDLSELEELRLALNDKFTPAKIPIEFGKLKKLKYMWLEEMNLIGEISPVVFENMTDLEHV 240
Query: 251 NLVYNNLTGPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFNLKNLISLDLSDNYLSGEIS 310
+L NNLTG IP+ L L NL +L+ N LTG IPKSI + NL+ LDLS N L+G I
Sbjct: 241 DLSVNNLTGRIPDVLFGLKNLTEFYLFANGLTGEIPKSI-SATNLVFLDLSANNLTGSIP 299
Query: 311 NLVVNLQKLEILHLFSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIHNNLTIL 370
+ NL KL++L+LF+N TG+IP I LP L+ ++++NKLTGEIP +G+H+ L
Sbjct: 300 VSIGNLTKLQVLNLFNNKLTGEIPPVIGKLPGLKEFKIFNNKLTGEIPAEIGVHSKLERF 359
Query: 371 DLSSNNLTGKIPNSLCASKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDNNLSGKLP 430
++S N LTGK+P +LC L ++++SN+L GEIP+ L C TL V+LQ+N+ SGK P
Sbjct: 360 EVSENQLTGKLPENLCKGGKLQGVVVYSNNLTGEIPESLGDCGTLLTVQLQNNDFSGKFP 419
Query: 431 LEITQLPQIYLLDISGNKFSGRINDR-KWNMPSLQMLNLANNNFSGDLPNSFG-GNKVEG 488
I +Y L +S N F+G + + WNM ++ + NN FSG++P G + +
Sbjct: 420 SRIWNASSMYSLQVSNNSFTGELPENVAWNMSRIE---IDNNRFSGEIPKKIGTWSSLVE 476
Query: 489 LDLSQNQFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRLNGEI 548
NQFSG +L L+ + L+ N+L G+ P+E+ L++L LS N+L+GEI
Sbjct: 477 FKAGNNQFSGEFPKELTSLSNLISIFLDENDLTGELPDEIISWKSLITLSLSKNKLSGEI 536
Query: 549 PEKLAKMPVLGLLDISENQFSGEIPKNLGSVESLVEVNISYNHFHGVLPSTEAFSAINAS 608
P L +P L LD+SENQFSG IP +GS++ L N+S N G +P A S
Sbjct: 537 PRALGLLPRLLNLDLSENQFSGGIPPEIGSLK-LTTFNVSSNRLTGGIPEQLDNLAYERS 595
Query: 609 LVTGNKLCDGDGDVSNGLPPCKSYNQMN---STRLFVLICFVLTALVVLVGTVVIFVLRM 665
+ + LC + +S LP C+ + + ++ +I + L+ + V FV+R
Sbjct: 596 FLNNSNLCADNPVLS--LPDCRKQRRGSRGFPGKILAMILVIAVLLLTITLFVTFFVVRD 653
Query: 666 NKSFEVRRVVENEDGTWEVIFF---DYKASKFVTIEDVLSSVKEGKVITKGRNWVSYEGK 722
+ RR +E TW++ F D+ S D++S++ E VI G + Y+
Sbjct: 654 YTRKQRRRGLE----TWKLTSFHRVDFAES------DIVSNLMEHYVIGSGGSGKVYKIF 703
Query: 723 CVSNEMQFVVKEISDTNSVSVSFWDD---TVTFGKKVRHENIVKIMGMFRCGKRGYLVYE 779
S+ VK I D+ + + V +RH NIVK++ LVYE
Sbjct: 704 VESSGQCVAVKRIWDSKKLDQKLEKEFIAEVEILGTIRHSNIVKLLCCISREDSKLLVYE 763
Query: 780 FVEGKSLREIMHG-----------LSWLRRWKIALGIAKAINFLHCECLWFGLGSEVSPE 828
++E +SL + +HG L+W +R IA+G A+ + ++H +C + +V
Sbjct: 764 YLEKRSLDQWLHGKKKGGTVEANNLTWSQRLNIAVGAAQGLCYMHHDCTPAIIHRDVKSS 823
Query: 829 TVLVDGK--------GVPRL--KLDSPGIVVTPVMGVKGFVSSAYVAPEERNGKDVTEKS 878
+L+D + G+ +L K + ++ V G S Y+APE V EK
Sbjct: 824 NILLDSEFNAKIADFGLAKLLIKQNQEPHTMSAVAG-----SFGYIAPEYAYTSKVDEKI 878
Query: 879 EIYGFGVILIELLTGRNSVDIEAWNGIHYKNNIVEWARYCYSDCHLDTWIDSVVMKGEDS 938
++Y FGV+L+EL+TGR E NG + N+ +W+ Y + D
Sbjct: 879 DVYSFGVVLLELVTGR-----EGNNGDEH-TNLADWSWKHYQS-------GKPTAEAFDE 925
Query: 939 STYQNDIVETM----NLALHCTANDPTTRPCARDILKAL 973
+ E M L L CT P+ RP +++L L
Sbjct: 926 DIKEASTTEAMTTVFKLGLMCTNTLPSHRPSMKEVLYVL 964
>At5g63930 receptor-like protein kinase
Length = 1102
Score = 458 bits (1179), Expect = e-129
Identities = 344/1090 (31%), Positives = 533/1090 (48%), Gaps = 154/1090 (14%)
Query: 15 FICLFMFMLNFHSTHGEQEFELLLSFKASIKFDPLNFLSNWVNTSSDTICKWHGITCDNW 74
FI L + +L +T E + LL K+ D L NW N++ C W G+ C N+
Sbjct: 12 FISLLLILLISETTGLNLEGQYLLEIKSKF-VDAKQNLRNW-NSNDSVPCGWTGVMCSNY 69
Query: 75 S---HVNTVSLSGKNISGEVSSSIFQLPHVTNLDLSNNQLVGEIVFNSPFLSSLLYLNLS 131
S V +++LS +SG++S SI L H+ LDLS N L G+I SSL L L+
Sbjct: 70 SSDPEVLSLNLSSMVLSGKLSPSIGGLVHLKQLDLSYNGLSGKIPKEIGNCSSLEILKLN 129
Query: 132 NNNLTGPLPQSLFSSSFINLETLDLSNNMFSGKIPDQIGLLSSLTYVDLGGNVLVGKIPN 191
NN G +P + ++LE L + NN SG +P +IG L SL+ + N + G++P
Sbjct: 130 NNQFDGEIPVEI--GKLVSLENLIIYNNRISGSLPVEIGNLLSLSQLVTYSNNISGQLPR 187
Query: 192 SITNLTSLES------------------------LTLASNQLIGEIPTKICLMKRLKWIY 227
SI NL L S L LA NQL GE+P +I ++K+L +
Sbjct: 188 SIGNLKRLTSFRAGQNMISGSLPSEIGGCESLVMLGLAQNQLSGELPKEIGMLKKLSQVI 247
Query: 228 LGYNNLSGEIPKNIGNLVSLNHLNLVYNNLTGPIPESLGNLTNLQYLFLYLNKLTGPIPK 287
L N SG IP+ I N SL L L N L GPIP+ LG+L +L++L+LY N L G IP+
Sbjct: 248 LWENEFSGFIPREISNCTSLETLALYKNQLVGPIPKELGDLQSLEFLYLYRNGLNGTIPR 307
Query: 288 SIFNLKNLISLDLSDNYLSGEISNLVVNLQKLEILHLFSNNFTGKIPNTITSLPHLQ--- 344
I NL I +D S+N L+GEI + N++ LE+L+LF N TG IP +++L +L
Sbjct: 308 EIGNLSYAIEIDFSENALTGEIPLELGNIEGLELLYLFENQLTGTIPVELSTLKNLSKLD 367
Query: 345 ---------------------VLQLWSNKLTGEIPQTLGIHNNLTILDLSSNNLTGKIPN 383
+LQL+ N L+G IP LG +++L +LD+S N+L+G+IP+
Sbjct: 368 LSINALTGPIPLGFQYLRGLFMLQLFQNSLSGTIPPKLGWYSDLWVLDMSDNHLSGRIPS 427
Query: 384 SLCASKNLHKIILFSNSLKGEIPKGLTSCKT----------------------------- 414
LC N+ + L +N+L G IP G+T+CKT
Sbjct: 428 YLCLHSNMIILNLGTNNLSGNIPTGITTCKTLVQLRLARNNLVGRFPSNLCKQVNVTAIE 487
Query: 415 -------------------LERVRLQDNNLSGKLPLEITQLPQIYLLDISGNKFSGRIND 455
L+R++L DN +G+LP EI L Q+ L+IS NK +G +
Sbjct: 488 LGQNRFRGSIPREVGNCSALQRLQLADNGFTGELPREIGMLSQLGTLNISSNKLTGEVPS 547
Query: 456 RKWNMPSLQMLNLANNNFSGDLPNSFGG-NKVEGLDLSQNQFSGYIQIGFKNLPELVQLK 514
+N LQ L++ NNFSG LP+ G ++E L LS N SG I + NL L +L+
Sbjct: 548 EIFNCKMLQRLDMCCNNFSGTLPSEVGSLYQLELLKLSNNNLSGTIPVALGNLSRLTELQ 607
Query: 515 LNNNNLFGKFPEELFQCNKL-VSLDLSHNRLNGEIPEKLAKMPVLGLLDISENQFSGEIP 573
+ N G P EL L ++L+LS+N+L GEIP +L+ + +L L ++ N SGEIP
Sbjct: 608 MGGNLFNGSIPRELGSLTGLQIALNLSYNKLTGEIPPELSNLVMLEFLLLNNNNLSGEIP 667
Query: 574 KNLGSVESLVEVNISYNHFHGVLPSTEAFSAINASLVTGNKLCD---GDGDVSNGLPPCK 630
+ ++ SL+ N SYN G +P S +S + LC + P +
Sbjct: 668 SSFANLSSLLGYNFSYNSLTGPIPLLRNISM--SSFIGNEGLCGPPLNQCIQTQPFAPSQ 725
Query: 631 SYNQ---MNSTRLFVLICFVLTALVVLVGTVVIFVLRMNKSFEVRRVVEN-EDGTWEVIF 686
S + M S+++ + V+ + +++ ++++++R VR V + +DG +
Sbjct: 726 STGKPGGMRSSKIIAITAAVIGGVSLMLIALIVYLMRR----PVRTVASSAQDGQPSEMS 781
Query: 687 FD--YKASKFVTIEDVLSSV---KEGKVITKGRNWVSYEGKCVSNEMQFVVK-----EIS 736
D + + T +D++++ E V+ +G Y+ + V K E
Sbjct: 782 LDIYFPPKEGFTFQDLVAATDNFDESFVVGRGACGTVYKAVLPAGYTLAVKKLASNHEGG 841
Query: 737 DTNSVSVSFWDDTVTFGKKVRHENIVKIMGMFRCGKRGYLVYEFVEGKSLREIMH----G 792
+ N+V SF + +T G +RH NIVK+ G L+YE++ SL EI+H
Sbjct: 842 NNNNVDNSFRAEILTLG-NIRHRNIVKLHGFCNHQGSNLLLYEYMPKGSLGEILHDPSCN 900
Query: 793 LSWLRRWKIALGIAKAINFLHCECLWFGLGSEVSPETVLVDGK--------GVPRLKLDS 844
L W +R+KIALG A+ + +LH +C ++ +L+D K G+ ++ +D
Sbjct: 901 LDWSKRFKIALGAAQGLAYLHHDCKPRIFHRDIKSNNILLDDKFEAHVGDFGLAKV-IDM 959
Query: 845 P-GIVVTPVMGVKGFVSSAYVAPEERNGKDVTEKSEIYGFGVILIELLTGRNSVDIEAWN 903
P ++ + G S Y+APE VTEKS+IY +GV+L+ELLTG+ V
Sbjct: 960 PHSKSMSAIAG-----SYGYIAPEYAYTMKVTEKSDIYSYGVVLLELLTGKAPV-----Q 1009
Query: 904 GIHYKNNIVEWARYCYSDCHLDTWIDSVVMKGEDSSTYQNDIVETMNLALHCTANDPTTR 963
I ++V W R L + + + ED + ++ + +AL CT+ P R
Sbjct: 1010 PIDQGGDVVNWVRSYIRRDALSSGVLDARLTLEDERIVSH-MLTVLKIALLCTSVSPVAR 1068
Query: 964 PCARDILKAL 973
P R ++ L
Sbjct: 1069 PSMRQVVLML 1078
>At1g17230 putative leucine-rich receptor protein kinase
Length = 1133
Score = 436 bits (1120), Expect = e-122
Identities = 330/1081 (30%), Positives = 517/1081 (47%), Gaps = 150/1081 (13%)
Query: 16 ICLFMFMLNFHSTHGEQEFELLLSFKASIKFDPLNFLSNWVNTSSDTICKWHGITCDNWS 75
+C F F+L +E +LL FKA + D +L++W S+ C W GI C +
Sbjct: 13 LCSFSFIL---VRSLNEEGRVLLEFKAFLN-DSNGYLASWNQLDSNP-CNWTGIACTHLR 67
Query: 76 HVNTVSLSGKNISGEVSSSIFQLPHVTNLDLSNNQLVGEIVFNSPFLSSLLYLNLSNNNL 135
V +V L+G N+SG +S I +L + L++S N + G I + SL L+L N
Sbjct: 68 TVTSVDLNGMNLSGTLSPLICKLHGLRKLNVSTNFISGPIPQDLSLCRSLEVLDLCTNRF 127
Query: 136 TGPLPQSLFSSSFINLETLDLSNNMFSGKIPDQIGLLSSLTYVDLGGNVLVGKIPNSITN 195
G +P L + I L+ L L N G IP QIG LSSL + + N L G IP S+
Sbjct: 128 HGVIPIQL--TMIITLKKLYLCENYLFGSIPRQIGNLSSLQELVIYSNNLTGVIPPSMAK 185
Query: 196 LT------------------------SLESLTLASNQLIGEIPTKICLMKRLKWIYLGYN 231
L SL+ L LA N L G +P ++ ++ L + L N
Sbjct: 186 LRQLRIIRAGRNGFSGVIPSEISGCESLKVLGLAENLLEGSLPKQLEKLQNLTDLILWQN 245
Query: 232 NLSGEIPKNIGNLVSLNHLNLVYNNLTGPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFN 291
LSGEIP ++GN+ L L L N TG IP +G LT ++ L+LY N+LTG IP+ I N
Sbjct: 246 RLSGEIPPSVGNISRLEVLALHENYFTGSIPREIGKLTKMKRLYLYTNQLTGEIPREIGN 305
Query: 292 LKNLISLDLSDNYLSGEISNLVVNLQKLEILHLFSN------------------------ 327
L + +D S+N L+G I ++ L++LHLF N
Sbjct: 306 LIDAAEIDFSENQLTGFIPKEFGHILNLKLLHLFENILLGPIPRELGELTLLEKLDLSIN 365
Query: 328 NFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIHNNLTILDLSSNNLTGKIPNSLCA 387
G IP + LP+L LQL+ N+L G+IP +G ++N ++LD+S+N+L+G IP C
Sbjct: 366 RLNGTIPQELQFLPYLVDLQLFDNQLEGKIPPLIGFYSNFSVLDMSANSLSGPIPAHFCR 425
Query: 388 SKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDNNLSGKLPLEITQLPQIYLLDISGN 447
+ L + L SN L G IP+ L +CK+L ++ L DN L+G LP+E+ L + L++ N
Sbjct: 426 FQTLILLSLGSNKLSGNIPRDLKTCKSLTKLMLGDNQLTGSLPIELFNLQNLTALELHQN 485
Query: 448 KFSGRINDRKWNMPSLQMLNLANNNFSGDLPNSFG-----------GNKVEG-------- 488
SG I+ + +L+ L LANNNF+G++P G N++ G
Sbjct: 486 WLSGNISADLGKLKNLERLRLANNNFTGEIPPEIGNLTKIVGFNISSNQLTGHIPKELGS 545
Query: 489 ------LDLSQNQFSGYI--QIG----------------------FKNLPELVQLKLNNN 518
LDLS N+FSGYI ++G F +L L++L+L N
Sbjct: 546 CVTIQRLDLSGNKFSGYIAQELGQLVYLEILRLSDNRLTGEIPHSFGDLTRLMELQLGGN 605
Query: 519 NLFGKFPEELFQCNKL-VSLDLSHNRLNGEIPEKLAKMPVLGLLDISENQFSGEIPKNLG 577
L P EL + L +SL++SHN L+G IP+ L + +L +L +++N+ SGEIP ++G
Sbjct: 606 LLSENIPVELGKLTSLQISLNISHNNLSGTIPDSLGNLQMLEILYLNDNKLSGEIPASIG 665
Query: 578 SVESLVEVNISYNHFHGVLPSTEAFSAINASLVTGNK-LCDGDGDVSNGLPPCKS----- 631
++ SL+ NIS N+ G +P T F +++S GN LC+ L P
Sbjct: 666 NLMSLLICNISNNNLVGTVPDTAVFQRMDSSNFAGNHGLCNSQRSHCQPLVPHSDSKLNW 725
Query: 632 -YNQMNSTRLFVLICFVLTALVVLVGTVVIFVLRMNKSFEVRRVVENEDGTWEVIFFDYK 690
N ++ + C V+ ++ ++ + + ++ + V + + + +F K
Sbjct: 726 LINGSQRQKILTITCIVIGSVFLITFLGLCWTIKRREPAFVALEDQTKPDVMDSYYFPKK 785
Query: 691 ASKFVTIEDVLSSVKEGKVITKGRNWVSYEGKCVSNEMQFVVKEISDTN---SVSVSFWD 747
+ + D + E V+ +G Y+ + E+ VK+++ S SF
Sbjct: 786 GFTYQGLVDATRNFSEDVVLGRGACGTVYKAEMSGGEV-IAVKKLNSRGEGASSDNSFRA 844
Query: 748 DTVTFGKKVRHENIVKIMGMFRCGKRGYLVYEFVEGKSLREIMHG------LSWLRRWKI 801
+ T G K+RH NIVK+ G L+YE++ SL E + L W R++I
Sbjct: 845 EISTLG-KIRHRNIVKLYGFCYHQNSNLLLYEYMSKGSLGEQLQRGEKNCLLDWNARYRI 903
Query: 802 ALGIAKAINFLHCECLWFGLGSEVSPETVLVDGK--------GVPRLKLDSPGIVVTPVM 853
ALG A+ + +LH +C + ++ +L+D + G+ +L S ++ V
Sbjct: 904 ALGAAEGLCYLHHDCRPQIVHRDIKSNNILLDERFQAHVGDFGLAKLIDLSYSKSMSAVA 963
Query: 854 GVKGFVSSAYVAPEERNGKDVTEKSEIYGFGVILIELLTGRNSVDIEAWNGIHYKNNIVE 913
G S Y+APE VTEK +IY FGV+L+EL+TG+ V + ++V
Sbjct: 964 G-----SYGYIAPEYAYTMKVTEKCDIYSFGVVLLELITGKPPV-----QPLEQGGDLVN 1013
Query: 914 WARYCYSDCHLDTWIDSVVMKGEDSSTYQNDIVETMNL----ALHCTANDPTTRPCARDI 969
W R + I ++ M T V M+L AL CT+N P +RP R++
Sbjct: 1014 WVRR-----SIRNMIPTIEMFDARLDTNDKRTVHEMSLVLKIALFCTSNSPASRPTMREV 1068
Query: 970 L 970
+
Sbjct: 1069 V 1069
>At5g65710 receptor protein kinase-like protein
Length = 976
Score = 430 bits (1106), Expect = e-120
Identities = 316/1009 (31%), Positives = 506/1009 (49%), Gaps = 95/1009 (9%)
Query: 15 FICLFMFMLN-FHSTHGEQEFELLLSFKASIKFDPLNFLSNWVNTSSD-TICKWHGITCD 72
F L + +L+ F + E+L K + FDP L +WV T + + C W GITC
Sbjct: 8 FFFLSLLLLSCFLQVSSNGDAEILSRVKKTRLFDPDGNLQDWVITGDNRSPCNWTGITCH 67
Query: 73 ----NWSHVNTVSLSGKNISGEVSSSIFQLPHVTNLDLSNNQLVGEIVFNSPF--LSSLL 126
+ V T+ LSG NISG ++ + N+ LS N L G I ++P S L
Sbjct: 68 IRKGSSLAVTTIDLSGYNISGGFPYGFCRIRTLINITLSQNNLNGTID-SAPLSLCSKLQ 126
Query: 127 YLNLSNNNLTGPLPQSLFSSSFINLETLDLSNNMFSGKIPDQIGLLSSLTYVDLGGNVLV 186
L L+ NN +G LP+ FS F L L+L +N+F+G+IP G L++L ++L GN L
Sbjct: 127 NLILNQNNFSGKLPE--FSPEFRKLRVLELESNLFTGEIPQSYGRLTALQVLNLNGNPLS 184
Query: 187 GKIPNSITNLTSLESLTLASNQLI-GEIPTKICLMKRLKWIYLGYNNLSGEIPKNIGNLV 245
G +P + LT L L LA IP+ + + L + L ++NL GEIP +I NLV
Sbjct: 185 GIVPAFLGYLTELTRLDLAYISFDPSPIPSTLGNLSNLTDLRLTHSNLVGEIPDSIMNLV 244
Query: 246 SLNHLNLVYNNLTGPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFNLKNLISLDLSDNYL 305
L +L+L N+LTG IPES+G L ++ + LY N+L+G +P+SI NL L + D+S N L
Sbjct: 245 LLENLDLAMNSLTGEIPESIGRLESVYQIELYDNRLSGKLPESIGNLTELRNFDVSQNNL 304
Query: 306 SGEISNLVVNLQKLEILHLFSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIHN 365
+GE+ + LQ + +L N FTG +P+ + P+L ++++N TG +P+ LG +
Sbjct: 305 TGELPEKIAALQLISF-NLNDNFFTGGLPDVVALNPNLVEFKIFNNSFTGTLPRNLGKFS 363
Query: 366 NLTILDLSSNNLTGKIPNSLCASKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDNNL 425
++ D+S+N +G++P LC + L KII FSN L GEIP+ C +L +R+ DN L
Sbjct: 364 EISEFDVSTNRFSGELPPYLCYRRKLQKIITFSNQLSGEIPESYGDCHSLNYIRMADNKL 423
Query: 426 SGKLPLEITQLPQIYLLDISGNKFSGRINDRKWNMPSLQMLNLANNNFSGDLPNSFGGNK 485
SG++P +LP L + N+ G I L L ++ NNFSG +P +
Sbjct: 424 SGEVPARFWELPLTRLELANNNQLQGSIPPSISKARHLSQLEISANNFSGVIPVKLCDLR 483
Query: 486 -VEGLDLSQNQFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRL 544
+ +DLS+N F G I L L ++++ N L G+ P + C +L L+LS+NRL
Sbjct: 484 DLRVIDLSRNSFLGSIPSCINKLKNLERVEMQENMLDGEIPSSVSSCTELTELNLSNNRL 543
Query: 545 NGEIPEKLAKMPVLGLLDISENQFSGEIPKNLGSVESLVEVNISYNHFHGVLPSTEAFSA 604
G IP +L +PVL LD+S NQ +GEIP L ++ L + N+S N +G
Sbjct: 544 RGGIPPELGDLPVLNYLDLSNNQLTGEIPAELLRLK-LNQFNVSDNKLYG---------- 592
Query: 605 INASLVTGNKLCDGDGDVSNGLPPCKSYNQMNSTRLFVLICFVLTALVVLVGTVVIFVLR 664
N +L N + + PC+S + ++C +V L G +V ++
Sbjct: 593 -NPNLCAPN---------LDPIRPCRSKRETRYILPISILC-----IVALTGALVWLFIK 637
Query: 665 MNKSFEVRRVVENEDGTWEVIFFDYKASKFVTIEDVLSSVKEGKVITKGRNWVSYEGKCV 724
F+ + N+ ++ + F T ED+ + E +I G + + Y K
Sbjct: 638 TKPLFKRKPKRTNKITIFQRVGF--------TEEDIYPQLTEDNIIGSGGSGLVYRVKLK 689
Query: 725 SNEMQFVVKEISDTNSVSVS---FWDDTVTFGKKVRHENIVKIMGMFRCGKRGYLVYEFV 781
S + V K +T + S F + T G +VRH NIVK++ + +LVYEF+
Sbjct: 690 SGQTLAVKKLWGETGQKTESESVFRSEVETLG-RVRHGNIVKLLMCCNGEEFRFLVYEFM 748
Query: 782 EGKSLREIMHG---------LSWLRRWKIALGIAKAINFLHCECLWFGLGSEVSPETVLV 832
E SL +++H L W R+ IA+G A+ +++LH + + + +V +L+
Sbjct: 749 ENGSLGDVLHSEKEHRAVSPLDWTTRFSIAVGAAQGLSYLHHDSVPPIVHRDVKSNNILL 808
Query: 833 DGKGVPRL----------KLDSPGIVVTPVMGVKGFVSSAYVAPEERNGKDVTEKSEIYG 882
D + PR+ + D+ G+ + V G S Y+APE V EKS++Y
Sbjct: 809 DHEMKPRVADFGLAKPLKREDNDGVSDVSMSCVAG--SYGYIAPEYGYTSKVNEKSDVYS 866
Query: 883 FGVILIELLTGRNSVDIEAWNGIHYKNNIVEWARYCYSDCHLDTWIDSVVMKGEDSSTYQ 942
FGV+L+EL+TG+ D + +IV++A + C+ + M + Y+
Sbjct: 867 FGVVLLELITGKRPND----SSFGENKDIVKFAMEA-ALCYPSPSAEDGAMNQDSLGNYR 921
Query: 943 N-----------------DIVETMNLALHCTANDPTTRPCARDILKALE 974
+ +I + +++AL CT++ P RP R +++ L+
Sbjct: 922 DLSKLVDPKMKLSTREYEEIEKVLDVALLCTSSFPINRPTMRKVVELLK 970
>At5g56040 receptor protein kinase-like protein
Length = 1090
Score = 428 bits (1101), Expect = e-120
Identities = 332/1066 (31%), Positives = 517/1066 (48%), Gaps = 139/1066 (13%)
Query: 17 CLFMFMLNFHST--------HGEQEFELLLSFKASIKFDPLNFLSNWVNTSSDTICKWHG 68
C F+F+L FHS+ +++ LLS+K+ + + LS+W + S+ C+W G
Sbjct: 8 CFFLFLL-FHSSLFFSIPCFSIDEQGLALLSWKSQLNISG-DALSSWKASESNP-CQWVG 64
Query: 69 ITCDNWSHVNTV-------------------------SLSGKNISGEVSSSIFQLPHVTN 103
I C+ V+ + SL+ N++G + + L +
Sbjct: 65 IKCNERGQVSEIQLQVMDFQGPLPATNLRQIKSLTLLSLTSVNLTGSIPKELGDLSELEV 124
Query: 104 LDLSNNQLVGEIVFNSPFLSSLLYLNLSNNNLTGPLPQSLFSSSFINLETLDLSNNMFSG 163
LDL++N L GEI + L L L+L+ NNL G +P L + +NL L L +N +G
Sbjct: 125 LDLADNSLSGEIPVDIFKLKKLKILSLNTNNLEGVIPSEL--GNLVNLIELTLFDNKLAG 182
Query: 164 KIPDQIGLLSSLTYVDLGGNV-LVGKIPNSITNLTSLESLTLASNQLIGEIPTKICLMKR 222
+IP IG L +L GGN L G++P I N SL +L LA L G +P I +K+
Sbjct: 183 EIPRTIGELKNLEIFRAGGNKNLRGELPWEIGNCESLVTLGLAETSLSGRLPASIGNLKK 242
Query: 223 LKWIYLGYNNLSGEIPKNIGNLVSLNHLNLVYNNLTGPIPESLGNLTNLQYLFLYLNKL- 281
++ I L + LSG IP IGN L +L L N+++G IP S+G L LQ L L+ N L
Sbjct: 243 VQTIALYTSLLSGPIPDEIGNCTELQNLYLYQNSISGSIPVSMGRLKKLQSLLLWQNNLV 302
Query: 282 -----------------------TGPIPKSIFNLKNLISLDLSDNYLSGEISNLVVNLQK 318
TG IP+S NL NL L LS N LSG I + N K
Sbjct: 303 GKIPTELGTCPELFLVDLSENLLTGNIPRSFGNLPNLQELQLSVNQLSGTIPEELANCTK 362
Query: 319 LEILHLFSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIHNNLTILDLSSNNLT 378
L L + +N +G+IP I L L + W N+LTG IP++L L +DLS NNL+
Sbjct: 363 LTHLEIDNNQISGEIPPLIGKLTSLTMFFAWQNQLTGIIPESLSQCQELQAIDLSYNNLS 422
Query: 379 GKIPNSLCASKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDNNLSGKLPLEITQLPQ 438
G IPN + +NL K++L SN L G IP + +C L R+RL N L+G +P EI L
Sbjct: 423 GSIPNGIFEIRNLTKLLLLSNYLSGFIPPDIGNCTNLYRLRLNGNRLAGNIPAEIGNLKN 482
Query: 439 IYLLDISGNKFSGRINDRKWNMPSLQMLNLANNNFSGDLPNSFGGNKVEGLDLSQNQFSG 498
+ +DIS N+ G I SL+ ++L +N +G LP + ++ +DLS N +G
Sbjct: 483 LNFIDISENRLIGNIPPEISGCTSLEFVDLHSNGLTGGLPGTL-PKSLQFIDLSDNSLTG 541
Query: 499 YIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRLNGEIPEKLAKMPVL 558
+ G +L EL +L L N G+ P E+ C L L+L N GEIP +L ++P L
Sbjct: 542 SLPTGIGSLTELTKLNLAKNRFSGEIPREISSCRSLQLLNLGDNGFTGEIPNELGRIPSL 601
Query: 559 GL-LDISENQFSGEIPK------NLGS-----------------VESLVEVNISYNHFHG 594
+ L++S N F+GEIP NLG+ +++LV +NIS+N F G
Sbjct: 602 AISLNLSCNHFTGEIPSRFSSLTNLGTLDVSHNKLAGNLNVLADLQNLVSLNISFNEFSG 661
Query: 595 VLPSTEAFSAINASLVTGNKLCDGDGDVSNGLPPCKSYNQMNSTRLFVLICFVLTALVVL 654
LP+T F + S++ NK NG+ + + + V + ++ A VVL
Sbjct: 662 ELPNTLFFRKLPLSVLESNKGLFISTRPENGI------QTRHRSAVKVTMSILVAASVVL 715
Query: 655 VGTVVIFVLRMNKSFEVRRVVENEDGTWEVIFFDYKASKFVTIEDVLSSVKEGKVITKGR 714
V++ V + K+ + E D +WEV Y+ F +I+D++ ++ VI G
Sbjct: 716 ---VLMAVYTLVKAQRITGKQEELD-SWEVTL--YQKLDF-SIDDIVKNLTSANVIGTGS 768
Query: 715 NWVSYEGKCVSNEMQFVVKEISDTNSVSVSFWDDTVTFGKKVRHENIVKIMGMFRCGKRG 774
+ V Y S E V K S + +F + T G +RH NI++++G
Sbjct: 769 SGVVYRVTIPSGETLAVKKMWS--KEENRAFNSEINTLG-SIRHRNIIRLLGWCSNRNLK 825
Query: 775 YLVYEFVEGKSLREIMH-------GLSWLRRWKIALGIAKAINFLHCECLWFGLGSEVSP 827
L Y+++ SL ++H G W R+ + LG+A A+ +LH +CL L +V
Sbjct: 826 LLFYDYLPNGSLSSLLHGAGKGSGGADWEARYDVVLGVAHALAYLHHDCLPPILHGDVKA 885
Query: 828 ETVL-----------------VDGKGVPRLKLDSPGIVVTPVMGVKGFVSSAYVAPEERN 870
VL V G+GV DS + P + S Y+APE +
Sbjct: 886 MNVLLGSRFESYLADFGLAKIVSGEGV--TDGDSSKLSNRPPLA----GSYGYMAPEHAS 939
Query: 871 GKDVTEKSEIYGFGVILIELLTGRNSVDIEAWNGIHYKNNIVEWARYCYSDCHLDTWIDS 930
+ +TEKS++Y +GV+L+E+LTG++ +D + G H +V+W R + I
Sbjct: 940 MQHITEKSDVYSYGVVLLEVLTGKHPLDPDLPGGAH----LVQWVRDHLAGKKDPREILD 995
Query: 931 VVMKGEDSSTYQNDIVETMNLALHCTANDPTTRPCARDILKALETV 976
++G + +++++T+ ++ C +N + RP +DI+ L+ +
Sbjct: 996 PRLRGR-ADPIMHEMLQTLAVSFLCVSNKASDRPMMKDIVAMLKEI 1040
>At2g33170 putative receptor-like protein kinase
Length = 1124
Score = 428 bits (1101), Expect = e-120
Identities = 344/1110 (30%), Positives = 526/1110 (46%), Gaps = 159/1110 (14%)
Query: 3 KETPATFSKFLNFICLFMFMLNFHSTHGEQEFELLLSFKASIKFDPLNFLSNWVNTSSDT 62
KE+ + F L + L ++ ++ G+ LL K D LN L NW N +T
Sbjct: 10 KESKSMFVGVLFLLTLLVWTSESLNSDGQ----FLLELKNRGFQDSLNRLHNW-NGIDET 64
Query: 63 ICKWHGITCDNWSH--------VNTVSLSGKNISGEVSSSIFQLPHVTNLDLSNNQLVGE 114
C W G+ C + V ++ LS N+SG VS SI L ++ L+L+ N L G+
Sbjct: 65 PCNWIGVNCSSQGSSSSSNSLVVTSLDLSSMNLSGIVSPSIGGLVNLVYLNLAYNALTGD 124
Query: 115 I--------------VFNSPF----------LSSLLYLNLSNN----------------- 133
I + N+ F LS L N+ NN
Sbjct: 125 IPREIGNCSKLEVMFLNNNQFGGSIPVEINKLSQLRSFNICNNKLSGPLPEEIGDLYNLE 184
Query: 134 -------NLTGPLPQSL---------------FSSSF-------INLETLDLSNNMFSGK 164
NLTGPLP+SL FS + +NL+ L L+ N SG+
Sbjct: 185 ELVAYTNNLTGPLPRSLGNLNKLTTFRAGQNDFSGNIPTEIGKCLNLKLLGLAQNFISGE 244
Query: 165 IPDQIGLLSSLTYVDLGGNVLVGKIPNSITNLTSLESLTLASNQLIGEIPTKICLMKRLK 224
+P +IG+L L V L N G IP I NLTSLE+L L N L+G IP++I MK LK
Sbjct: 245 LPKEIGMLVKLQEVILWQNKFSGFIPKDIGNLTSLETLALYGNSLVGPIPSEIGNMKSLK 304
Query: 225 WIYLGYNNLSGEIPKNIGNLVSLNHLNLVYNNLTGPIPESLGNLTNLQYLFLYLNKLTGP 284
+YL N L+G IPK +G L + ++ N L+G IP L ++ L+ L+L+ NKLTG
Sbjct: 305 KLYLYQNQLNGTIPKELGKLSKVMEIDFSENLLSGEIPVELSKISELRLLYLFQNKLTGI 364
Query: 285 IPKSIFNLKNLISLDLSDNYLSGEISNLVVNLQKLEILHLFSNNFTGKIPNTITSLPHLQ 344
IP + L+NL LDLS N L+G I NL + L LF N+ +G IP + L
Sbjct: 365 IPNELSKLRNLAKLDLSINSLTGPIPPGFQNLTSMRQLQLFHNSLSGVIPQGLGLYSPLW 424
Query: 345 VLQLWSNKLTGEIPQTLGIHNNLTILDLSS------------------------NNLTGK 380
V+ N+L+G+IP + +NL +L+L S N LTG+
Sbjct: 425 VVDFSENQLSGKIPPFICQQSNLILLNLGSNRIFGNIPPGVLRCKSLLQLRVVGNRLTGQ 484
Query: 381 IPNSLCASKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDNNLSGKLPLEITQLPQIY 440
P LC NL I L N G +P + +C+ L+R+ L N S LP EI++L +
Sbjct: 485 FPTELCKLVNLSAIELDQNRFSGPLPPEIGTCQKLQRLHLAANQFSSNLPNEISKLSNLV 544
Query: 441 LLDISGNKFSGRINDRKWNMPSLQMLNLANNNFSGDLPNSFGG-NKVEGLDLSQNQFSGY 499
++S N +G I N LQ L+L+ N+F G LP G +++E L LS+N+FSG
Sbjct: 545 TFNVSSNSLTGPIPSEIANCKMLQRLDLSRNSFIGSLPPELGSLHQLEILRLSENRFSGN 604
Query: 500 IQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKL-VSLDLSHNRLNGEIPEKLAKMPVL 558
I NL L +L++ N G P +L + L ++++LS+N +GEIP ++ + +L
Sbjct: 605 IPFTIGNLTHLTELQMGGNLFSGSIPPQLGLLSSLQIAMNLSYNDFSGEIPPEIGNLHLL 664
Query: 559 GLLDISENQFSGEIPKNLGSVESLVEVNISYNHFHGVLPSTEAFSAINASLVTGNK-LCD 617
L ++ N SGEIP ++ SL+ N SYN+ G LP T+ F + + GNK LC
Sbjct: 665 MYLSLNNNHLSGEIPTTFENLSSLLGCNFSYNNLTGQLPHTQIFQNMTLTSFLGNKGLCG 724
Query: 618 G---DGDVSNGLPPCKSYNQMNST---RLFVLICFVLTALVVLVGTVVIFVLRMNKSFEV 671
G D S+ P S + S R+ +++ V+ + +L+ +V+ LR
Sbjct: 725 GHLRSCDPSHSSWPHISSLKAGSARRGRIIIIVSSVIGGISLLLIAIVVHFLRNPVEPTA 784
Query: 672 RRVVENEDGTWEVIFFDYKASKFVTIEDVLSSVK---EGKVITKGRNWVSYEGKCVSNEM 728
V + E E + +F T++D+L + K + ++ +G Y+ S +
Sbjct: 785 PYVHDKEPFFQESDIYFVPKERF-TVKDILEATKGFHDSYIVGRGACGTVYKAVMPSGKT 843
Query: 729 QFVVK-------EISDTNSVSVSFWDDTVTFGKKVRHENIVKIMGMFRCGKRG----YLV 777
V K +++N+ SF + +T G K+RH NIV++ C +G L+
Sbjct: 844 IAVKKLESNREGNNNNSNNTDNSFRAEILTLG-KIRHRNIVRLYSF--CYHQGSNSNLLL 900
Query: 778 YEFVEGKSLREIMHG-----LSWLRRWKIALGIAKAINFLHCECLWFGLGSEVSPETVLV 832
YE++ SL E++HG + W R+ IALG A+ + +LH +C + ++ +L+
Sbjct: 901 YEYMSRGSLGELLHGGKSHSMDWPTRFAIALGAAEGLAYLHHDCKPRIIHRDIKSNNILI 960
Query: 833 DGK--------GVPRLKLDSPGIVVTPVMGVKGFVSSAYVAPEERNGKDVTEKSEIYGFG 884
D G+ ++ +D P + V V G S Y+APE VTEK +IY FG
Sbjct: 961 DENFEAHVGDFGLAKV-IDMP--LSKSVSAVAG--SYGYIAPEYAYTMKVTEKCDIYSFG 1015
Query: 885 VILIELLTGRNSVDIEAWNGIHYKNNIVEWARYCYSDCHLDTWI-DSVVMKGEDSSTYQN 943
V+L+ELLTG+ V + ++ W R D L + I D + K ED N
Sbjct: 1016 VVLLELLTGKAPV-----QPLEQGGDLATWTRNHIRDHSLTSEILDPYLTKVED-DVILN 1069
Query: 944 DIVETMNLALHCTANDPTTRPCARDILKAL 973
++ +A+ CT + P+ RP R+++ L
Sbjct: 1070 HMITVTKIAVLCTKSSPSDRPTMREVVLML 1099
>At4g08850 receptor protein kinase like protein
Length = 1045
Score = 426 bits (1096), Expect = e-119
Identities = 316/1049 (30%), Positives = 499/1049 (47%), Gaps = 106/1049 (10%)
Query: 3 KETPATFSKFLNFICLFMFMLNFHSTHGEQEFELLLSFKASIKFDPLNF-LSNWVNTSSD 61
KE P L I + +F + +E LL +K++ + LS+WVN ++
Sbjct: 22 KEKPRDLQVLL--IISIVLSCSFAVSATVEEANALLKWKSTFTNQTSSSKLSSWVNPNTS 79
Query: 62 TIC-KWHGITCDNWSHVNTVSLSGKNISGEVSSSIFQ-LPHVTNLDLSNNQLVGEIVFNS 119
+ C W+G+ C S + ++L+ I G F LP++T +DLS N+ G I
Sbjct: 80 SFCTSWYGVACSLGSIIR-LNLTNTGIEGTFEDFPFSSLPNLTFVDLSMNRFSGTISPLW 138
Query: 120 PFLSSLLYLNLSNNNLTGPLPQSLFSSSFINLETLDLSNNMFSGKIPDQIGLLSSLTYVD 179
S L Y +LS N L G +P L S NL+TL L N +G IP +IG L+ +T +
Sbjct: 139 GRFSKLEYFDLSINQLVGEIPPELGDLS--NLDTLHLVENKLNGSIPSEIGRLTKVTEIA 196
Query: 180 LGGNVLVGKIPNSITNLTSLESLTLASNQLIGEIPTKICLMKRLKWIYLGYNNLSGEIPK 239
+ N+L G IP+S NLT L +L L N L G IP++I + L+ + L NNL+G+IP
Sbjct: 197 IYDNLLTGPIPSSFGNLTKLVNLYLFINSLSGSIPSEIGNLPNLRELCLDRNNLTGKIPS 256
Query: 240 NIGNLVSLNHLNLVYNNLTGPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFNLKNLISLD 299
+ GNL ++ LN+ N L+G IP +GN+T L L L+ NKLTGPIP ++ N+K L L
Sbjct: 257 SFGNLKNVTLLNMFENQLSGEIPPEIGNMTALDTLSLHTNKLTGPIPSTLGNIKTLAVLH 316
Query: 300 LSDNYLSGEISNLVVNLQKLEILHLFSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQ 359
L N L+G I + ++ + L + N TG +P++ L L+ L L N+L+G IP
Sbjct: 317 LYLNQLNGSIPPELGEMESMIDLEISENKLTGPVPDSFGKLTALEWLFLRDNQLSGPIPP 376
Query: 360 TLGIHNNLTILDLSSNNLTGKIPNSLCASKNLHKIILFSNSLKGEIPKGLTSCKTLERVR 419
+ LT+L + +NN TG +P+++C L + L N +G +PK L CK+L RVR
Sbjct: 377 GIANSTELTVLQVDTNNFTGFLPDTICRGGKLENLTLDDNHFEGPVPKSLRDCKSLIRVR 436
Query: 420 LQDNNLSGKLPLEITQLPQIYLLDISGNKFSGRIN------------------------D 455
+ N+ SG + P + +D+S N F G+++
Sbjct: 437 FKGNSFSGDISEAFGVYPTLNFIDLSNNNFHGQLSANWEQSQKLVAFILSNNSITGAIPP 496
Query: 456 RKWNMPSLQMLNLANNNFSGDLPNS-----------FGGNKVEG--------------LD 490
WNM L L+L++N +G+LP S GN++ G LD
Sbjct: 497 EIWNMTQLSQLDLSSNRITGELPESISNINRISKLQLNGNRLSGKIPSGIRLLTNLEYLD 556
Query: 491 LSQNQFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRLNGEIPE 550
LS N+FS I NLP L + L+ N+L PE L + ++L LDLS+N+L+GEI
Sbjct: 557 LSSNRFSSEIPPTLNNLPRLYYMNLSRNDLDQTIPEGLTKLSQLQMLDLSYNQLDGEISS 616
Query: 551 KLAKMPVLGLLDISENQFSGEIPKNLGSVESLVEVNISYNHFHGVLPSTEAFSAINASLV 610
+ + L LD+S N SG+IP + + +L V++S+N+ G +P AF
Sbjct: 617 QFRSLQNLERLDLSHNNLSGQIPPSFKDMLALTHVDVSHNNLQGPIPDNAAFRNAPPDAF 676
Query: 611 TGNKLCDGDGDVSNGLPPCKSYNQMNSTRLFVLICFVLT----ALVVLVGTVVIFVLRMN 666
GNK G + + GL PC + S + LI ++L A+++L IF+
Sbjct: 677 EGNKDLCGSVNTTQGLKPCSITSSKKSHKDRNLIIYILVPIIGAIIILSVCAGIFICFRK 736
Query: 667 KSFEVRRVVENEDG--TWEVIFFDYKASKFVTIEDVLSSVKEGKVITKGRNWVSYEGKCV 724
++ ++ ++E G T + FD K ++ I +I G + Y+ K
Sbjct: 737 RTKQIEEHTDSESGGETLSIFSFDGKV-RYQEIIKATGEFDPKYLIGTGGHGKVYKAKLP 795
Query: 725 SNEMQFVVKEISDTNSVSVSFWDDTVTFGK------KVRHENIVKIMGMFRCGKRGYLVY 778
+ M VK++++T S+S F ++RH N+VK+ G + +LVY
Sbjct: 796 NAIM--AVKKLNETTDSSISNPSTKQEFLNEIRALTEIRHRNVVKLFGFCSHRRNTFLVY 853
Query: 779 EFVEGKSLREIMHG------LSWLRRWKIALGIAKAINFLHCECLWFGLGSEVSPETVLV 832
E++E SLR+++ L W +R + G+A A++++H + + ++S +L+
Sbjct: 854 EYMERGSLRKVLENDDEAKKLDWGKRINVVKGVAHALSYMHHDRSPAIVHRDISSGNILL 913
Query: 833 ---------DGKGVPRLKLDSPGIVVTPVMGVKGFVSSAYVAPEERNGKDVTEKSEIYGF 883
D LK DS + V G G YVAPE VTEK ++Y F
Sbjct: 914 GEDYEAKISDFGTAKLLKPDSSN--WSAVAGTYG-----YVAPELAYAMKVTEKCDVYSF 966
Query: 884 GVILIELLTGRNSVDIEAWNGIHYKNNIVEWARYCYSDCHLDTWIDSVVMKGEDSSTYQN 943
GV+ +E++ G + D +V D L S E + +
Sbjct: 967 GVLTLEVIKGEHPGD------------LVSTLSSSPPDATLSLKSISDHRLPEPTPEIKE 1014
Query: 944 DIVETMNLALHCTANDPTTRPCARDILKA 972
+++E + +AL C +DP RP I A
Sbjct: 1015 EVLEILKVALLCLHSDPQARPTMLSISTA 1043
>At5g48940 receptor protein kinase-like protein
Length = 1110
Score = 424 bits (1089), Expect = e-118
Identities = 333/1062 (31%), Positives = 510/1062 (47%), Gaps = 127/1062 (11%)
Query: 8 TFSKFLNFICLFMFMLNFHSTHGEQEFELLLSFKASIKFDPLNFLSNWVNTSSDTICKWH 67
T S F + LF+ ++ E L+S+ S P + S W + SD C+W
Sbjct: 14 TVSHFSITLSLFLAFFISSTSASTNEVSALISWLHSSNSPPPSVFSGWNPSDSDP-CQWP 72
Query: 68 GITCDNWSH-----VNTVS---------------------LSGKNISGEVSSSIFQLPHV 101
ITC + + +N VS +S N++G +SS I +
Sbjct: 73 YITCSSSDNKLVTEINVVSVQLALPFPPNISSFTSLQKLVISNTNLTGAISSEIGDCSEL 132
Query: 102 TNLDLSNNQLVGEIVFNSPFLSSLLYLNLSNNNLTGPLPQSLFSS-SFINLETLD--LSN 158
+DLS+N LVGEI + L +L L L++N LTG +P L S NLE D LS
Sbjct: 133 IVIDLSSNSLVGEIPSSLGKLKNLQELCLNSNGLTGKIPPELGDCVSLKNLEIFDNYLSE 192
Query: 159 NM--------------------FSGKIPDQIGLLSSLTYVDLGGNVLVGKIPNSITNLTS 198
N+ SGKIP++IG +L + L + G +P S+ L+
Sbjct: 193 NLPLELGKISTLESIRAGGNSELSGKIPEEIGNCRNLKVLGLAATKISGSLPVSLGQLSK 252
Query: 199 LESLTLASNQLIGEIPTKICLMKRLKWIYLGYNNLSGEIPKNIGNLVSLNHLNLVYNNLT 258
L+SL++ S L GEIP ++ L ++L N+LSG +PK +G L +L + L NNL
Sbjct: 253 LQSLSVYSTMLSGEIPKELGNCSELINLFLYDNDLSGTLPKELGKLQNLEKMLLWQNNLH 312
Query: 259 GPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFNLKNLISLDLSDNYLSGEISNLVVNLQK 318
GPIPE +G + +L + L +N +G IPKS NL NL L LS N ++G I +++ N K
Sbjct: 313 GPIPEEIGFMKSLNAIDLSMNYFSGTIPKSFGNLSNLQELMLSSNNITGSIPSILSNCTK 372
Query: 319 LEILHLFSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIHNNLTILDLSSNNLT 378
L + +N +G IP I L L + W NKL G IP L NL LDLS N LT
Sbjct: 373 LVQFQIDANQISGLIPPEIGLLKELNIFLGWQNKLEGNIPDELAGCQNLQALDLSQNYLT 432
Query: 379 GKIPNSLCASKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDNNLSGKLPLEITQLPQ 438
G +P L +NL K++L SN++ G IP + +C +L R+RL +N ++G++P I L
Sbjct: 433 GSLPAGLFQLRNLTKLLLISNAISGVIPLEIGNCTSLVRLRLVNNRITGEIPKGIGFLQN 492
Query: 439 IYLLDISGNKFSGRINDRKWNMPSLQMLNLANNNFSGDLPNSFGG-NKVEGLD------- 490
+ LD+S N SG + N LQMLNL+NN G LP S K++ LD
Sbjct: 493 LSFLDLSENNLSGPVPLEISNCRQLQMLNLSNNTLQGYLPLSLSSLTKLQVLDVSSNDLT 552
Query: 491 -----------------LSQNQFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNK 533
LS+N F+G I + L L L++NN+ G PEELF
Sbjct: 553 GKIPDSLGHLISLNRLILSKNSFNGEIPSSLGHCTNLQLLDLSSNNISGTIPEELFDIQD 612
Query: 534 L-VSLDLSHNRLNGEIPEKLAKMPVLGLLDISENQFSGEIPKNLGSVESLVEVNISYNHF 592
L ++L+LS N L+G IPE+++ + L +LDIS N SG++ L +E+LV +NIS+N F
Sbjct: 613 LDIALNLSWNSLDGFIPERISALNRLSVLDISHNMLSGDL-SALSGLENLVSLNISHNRF 671
Query: 593 HGVLPSTEAFSAINASLVTGNKLCDGDGDVSNGLPPCKSYNQ--------MNSTRLFVLI 644
G LP ++ F + + + GN +G S G C N ++S RL + I
Sbjct: 672 SGYLPDSKVFRQLIGAEMEGN-----NGLCSKGFRSCFVSNSSQLTTQRGVHSHRLRIAI 726
Query: 645 CFVLTALVVLVGTVVIFVLRMNKSFEVRRVVENEDGTWEVIFFDYKASKFVTIEDVLSSV 704
+++ VL V+ V+R + E + W F ++ F T+E VL +
Sbjct: 727 GLLISVTAVLAVLGVLAVIRAKQMIRDDNDSETGENLWTWQFTPFQKLNF-TVEHVLKCL 785
Query: 705 KEGKVITKGRNWVSYEGKCVSNEMQFVVK----------EISDTNSVSVSFWDDTVTFGK 754
EG VI KG + + Y+ + + E+ V K E + ++ V SF + T G
Sbjct: 786 VEGNVIGKGCSGIVYKAEMPNREVIAVKKLWPVTVPNLNEKTKSSGVRDSFSAEVKTLG- 844
Query: 755 KVRHENIVKIMGMFRCGKRGYLVYEFVEGKSLREIMHGLSWLRRWKIALGIAKAINFLHC 814
+RH+NIV+ +G L+Y+++ SL ++H S + + KA N L
Sbjct: 845 SIRHKNIVRFLGCCWNKNTRLLMYDYMSNGSLGSLLHERSGVCSLGWEVRDIKANNIL-- 902
Query: 815 ECLWFGLGSEVSPETVLVDGKGVPRLKLDSPGIVVTPVMGVKGFVSSAYVAPEERNGKDV 874
+G + P + G+ +L D G + G S Y+APE +
Sbjct: 903 ------IGPDFEP---YIGDFGLAKLVDD--GDFARSSNTIAG--SYGYIAPEYGYSMKI 949
Query: 875 TEKSEIYGFGVILIELLTGRNSVDIEAWNGIHYKNNIVEWARYCYSDCHLDTWIDSVVMK 934
TEKS++Y +GV+++E+LTG+ +D +G+H IV+W + D + ID +
Sbjct: 950 TEKSDVYSYGVVVLEVLTGKQPIDPTIPDGLH----IVDWVKK-IRDIQV---IDQGLQA 1001
Query: 935 GEDSSTYQNDIVETMNLALHCTANDPTTRPCARDILKALETV 976
+S ++++T+ +AL C P RP +D+ L +
Sbjct: 1002 RPESEV--EEMMQTLGVALLCINPIPEDRPTMKDVAAMLSEI 1041
>At1g72180 leucine-rich receptor-like protein kinase, putative
Length = 977
Score = 417 bits (1071), Expect = e-116
Identities = 311/992 (31%), Positives = 482/992 (48%), Gaps = 89/992 (8%)
Query: 18 LFMFMLNFHSTHGEQEFELLLSFKASIKFDPLNFLSNWVNTSSDTICKWHGITCDNWS-H 76
LF+F N ST E + L FK + D N L +W SD+ C + GITCD S
Sbjct: 22 LFIFPPNVEST---VEKQALFRFKNRLD-DSHNILQSW--KPSDSPCVFRGITCDPLSGE 75
Query: 77 VNTVSLSGKNISGEVSSSIFQLPHVTNLDLSNNQLVGEIVFNSPFLSSLLYLNLSNNNLT 136
V +SL N+SG +S SI L + L L+L +N ++
Sbjct: 76 VIGISLGNVNLSGTISPSISAL------------------------TKLSTLSLPSNFIS 111
Query: 137 GPLPQSLFSSSFINLETLDLSNNMFSGKIPDQIGLLSSLTYVDLGGNVLVGKIPNSITNL 196
G +P + + NL+ L+L++N SG IP+ + L SL +D+ GN L G+ + I N+
Sbjct: 112 GRIPPEIVNCK--NLKVLNLTSNRLSGTIPN-LSPLKSLEILDISGNFLNGEFQSWIGNM 168
Query: 197 TSLESLTLASNQLI-GEIPTKICLMKRLKWIYLGYNNLSGEIPKNIGNLVSLNHLNLVYN 255
L SL L +N G IP I +K+L W++L +NL+G+IP +I +L +L+ ++ N
Sbjct: 169 NQLVSLGLGNNHYEEGIIPESIGGLKKLTWLFLARSNLTGKIPNSIFDLNALDTFDIANN 228
Query: 256 NLTGPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFNLKNLISLDLSDNYLSGEISNLVVN 315
++ P + L NL + L+ N LTG IP I NL L D+S N LSG + +
Sbjct: 229 AISDDFPILISRLVNLTKIELFNNSLTGKIPPEIKNLTRLREFDISSNQLSGVLPEELGV 288
Query: 316 LQKLEILHLFSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIHNNLTILDLSSN 375
L++L + H NNFTG+ P+ L HL L ++ N +GE P +G + L +D+S N
Sbjct: 289 LKELRVFHCHENNFTGEFPSGFGDLSHLTSLSIYRNNFSGEFPVNIGRFSPLDTVDISEN 348
Query: 376 NLTGKIPNSLCASKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDNNLSGKLPLEITQ 435
TG P LC +K L ++ N GEIP+ CK+L R+R+ +N LSG++
Sbjct: 349 EFTGPFPRFLCQNKKLQFLLALQNEFSGEIPRSYGECKSLLRLRINNNRLSGQVVEGFWS 408
Query: 436 LPQIYLLDISGNKFSGRINDRKWNMPSLQMLNLANNNFSGDLPNSFGG-NKVEGLDLSQN 494
LP ++D+S N+ +G ++ + L L L NN FSG +P G +E + LS N
Sbjct: 409 LPLAKMIDLSDNELTGEVSPQIGLSTELSQLILQNNRFSGKIPRELGRLTNIERIYLSNN 468
Query: 495 QFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRLNGEIPEKLAK 554
SG I + +L EL L L NN+L G P+EL C KLV L+L+ N L GEIP L++
Sbjct: 469 NLSGEIPMEVGDLKELSSLHLENNSLTGFIPKELKNCVKLVDLNLAKNFLTGEIPNSLSQ 528
Query: 555 MPVLGLLDISENQFSGEIPKNLGSVESLVEVNISYNHFHGVLPSTEAFSAINASLVTGNK 614
+ L LD S N+ +GEIP +L ++ L +++S N G +P + + K
Sbjct: 529 IASLNSLDFSGNRLTGEIPASLVKLK-LSFIDLSGNQLSGRIPPDLLAVGGSTAFSRNEK 587
Query: 615 LC-DGDGDVSN---GLPPCKSY------NQMNSTRLFVLICFVLTALVVLVGTVVIFVLR 664
LC D + +N GL C Y + ++ T LF+ + V+ LV + + V++
Sbjct: 588 LCVDKENAKTNQNLGLSICSGYQNVKRNSSLDGTLLFLALAIVVVVLVSGLFALRYRVVK 647
Query: 665 MNKSFEVRRVVENEDGTWEVIFFDYKASKFVTIEDVLSSVKEGKVITKGRNWVSYEGKCV 724
+ + R + D W++ F + D + + E VI G Y
Sbjct: 648 IRELDSENRDINKADAKWKIASF----HQMELDVDEICRLDEDHVIGSGSAGKVYRVDLK 703
Query: 725 SNEMQFVVK-------EISDTNSVSVSFWDDTVTFGKKVRHENIVKIMGMFRCGKRGYLV 777
VK E D VSV+ + K+RH N++K+ YLV
Sbjct: 704 KGGGTVAVKWLKRGGGEEGDGTEVSVA----EMEILGKIRHRNVLKLYACLVGRGSRYLV 759
Query: 778 YEFVE--------GKSLREIMHGLSWLRRWKIALGIAKAINFLHCECLWFGLGSEVSPET 829
+EF+E G +++ + L WL+R+KIA+G AK I +LH +C + ++
Sbjct: 760 FEFMENGNLYQALGNNIKGGLPELDWLKRYKIAVGAAKGIAYLHHDCCPPIIHRDIKSSN 819
Query: 830 VLVDGKGVPRL------KLDSPGIVVTPVMGVKGFVSSAYVAPEERNGKDVTEKSEIYGF 883
+L+DG ++ K+ G + V G G Y+APE TEKS++Y F
Sbjct: 820 ILLDGDYESKIADFGVAKVADKGYEWSCVAGTHG-----YMAPELAYSFKATEKSDVYSF 874
Query: 884 GVILIELLTGRNSVDIEAWNGIHYKNNIVEWARYCYSDCHLDTWIDSVVMKGEDSSTY-Q 942
GV+L+EL+TG ++ E G +IV+ Y YS D V+ + STY +
Sbjct: 875 GVVLLELVTGLRPMEDEFGEG----KDIVD---YVYSQIQQDPRNLQNVLDKQVLSTYIE 927
Query: 943 NDIVETMNLALHCTANDPTTRPCARDILKALE 974
++ + + L CT P RP R++++ L+
Sbjct: 928 ESMIRVLKMGLLCTTKLPNLRPSMREVVRKLD 959
>At1g28440 unknown protein
Length = 996
Score = 417 bits (1071), Expect = e-116
Identities = 312/1014 (30%), Positives = 504/1014 (48%), Gaps = 110/1014 (10%)
Query: 18 LFMFMLNFHSTHGEQEFELLLSFKASIKFDPLNFLSNWVNTSSDTICKWHGITC-DNWSH 76
LF+F+L Q+ +L K S+ DP ++LS+W N++ + C+W G++C ++S
Sbjct: 4 LFLFLLFPTVFSLNQDGFILQQVKLSLD-DPDSYLSSW-NSNDASPCRWSGVSCAGDFSS 61
Query: 77 VNTVSLSGKNISGEVSSSIFQLPHVTNLDLSNNQLVGEIVFNSPFLSSLLYLNLSNNNLT 136
V +V LS N++G S I +L ++ +L L NN + + N SL L+LS N LT
Sbjct: 62 VTSVDLSSANLAGPFPSVICRLSNLAHLSLYNNSINSTLPLNIAACKSLQTLDLSQNLLT 121
Query: 137 GPLPQSLFSSSFINLETLDLSNNMFSGKIPDQIGLLSSLTYVDLGGNVLVGKIPNSITNL 196
G LPQ+L + L LDL+ N FSG IP G +L + L N+L G IP + N+
Sbjct: 122 GELPQTL--ADIPTLVHLDLTGNNFSGDIPASFGKFENLEVLSLVYNLLDGTIPPFLGNI 179
Query: 197 TSLESLTLASNQLI-GEIPTKICLMKRLKWIYLGYNNLSGEIPKNIGNLVSLNHLNLVYN 255
++L+ L L+ N IP + + L+ ++L +L G+IP ++G L L L+L N
Sbjct: 180 STLKMLNLSYNPFSPSRIPPEFGNLTNLEVMWLTECHLVGQIPDSLGQLSKLVDLDLALN 239
Query: 256 NLTGPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFNLKNLISLDLSDNYLSGEISNLVVN 315
+L G IP SLG LTN+ + LY N LTG IP + NLK+L LD S N L+G+I + +
Sbjct: 240 DLVGHIPPSLGGLTNVVQIELYNNSLTGEIPPELGNLKSLRLLDASMNQLTGKIPDELCR 299
Query: 316 LQKLEILHLFSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIHNNLTILDLSSN 375
+ LE L+L+ NN G++P +I P+L ++++ N+LTG +P+ LG+++ L LD+S N
Sbjct: 300 VP-LESLNLYENNLEGELPASIALSPNLYEIRIFGNRLTGGLPKDLGLNSPLRWLDVSEN 358
Query: 376 NLTGKIPNSLCASKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDNNLSGKLPLEITQ 435
+G +P LCA L ++++ NS G IP+ L C++L R+RL N SG +P
Sbjct: 359 EFSGDLPADLCAKGELEELLIIHNSFSGVIPESLADCRSLTRIRLAYNRFSGSVPTGF-- 416
Query: 436 LPQIYLLDISGNKFSGRINDRKWNMPSLQMLNLANNNFSGDLPNSFGG-NKVEGLDLSQN 494
W +P + +L L NN+FSG++ S GG + + L LS N
Sbjct: 417 ----------------------WGLPHVNLLELVNNSFSGEISKSIGGASNLSLLILSNN 454
Query: 495 QFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRLNGEIPEKLAK 554
+F+G + +L L QL + N G P+ L +L +LDL N+ +GE+ +
Sbjct: 455 EFTGSLPEEIGSLDNLNQLSASGNKFSGSLPDSLMSLGELGTLDLHGNQFSGELTSGIKS 514
Query: 555 MPVLGLLDISENQFSGEIPKNLGSVE-----------------------SLVEVNISYNH 591
L L++++N+F+G+IP +GS+ L ++N+SYN
Sbjct: 515 WKKLNELNLADNEFTGKIPDEIGSLSVLNYLDLSGNMFSGKIPVSLQSLKLNQLNLSYNR 574
Query: 592 FHGVLPSTEAFSAINASLVTGNKLCDGDGDVSNGLPPCKSYNQMNSTRLFVLI--CFVLT 649
G LP + A S + LC GD+ GL C S N+ L+ FVL
Sbjct: 575 LSGDLPPSLAKDMYKNSFIGNPGLC---GDI-KGL--CGSENEAKKRGYVWLLRSIFVLA 628
Query: 650 ALVVLVGTVVIFVLRMNKSFEVRRVVENEDGTWEVIFFDYKASKFVTIEDVLSSVKEGKV 709
A+V+L G V + ++F+ R +E W ++ F + ++L S+ E V
Sbjct: 629 AMVLLAG--VAWFYFKYRTFKKARAMER--SKWTLMSFHKLG---FSEHEILESLDEDNV 681
Query: 710 ITKGRNWVSYEGKCVSNEMQFV-------VKEISDTNS--------VSVSFWDDTVTFGK 754
I G + Y+ + E V VKE D + +F + T G
Sbjct: 682 IGAGASGKVYKVVLTNGETVAVKRLWTGSVKETGDCDPEKGYKPGVQDEAFEAEVETLG- 740
Query: 755 KVRHENIVKIMGMFRCGKRGYLVYEFVEGKSLREIMHG-----LSWLRRWKIALGIAKAI 809
K+RH+NIVK+ LVYE++ SL +++H L W R+KI L A+ +
Sbjct: 741 KIRHKNIVKLWCCCSTRDCKLLVYEYMPNGSLGDLLHSSKGGMLGWQTRFKIILDAAEGL 800
Query: 810 NFLHCECLWFGLGSEVSPETVLVDGKGVPRL-------KLDSPGIVVTPVMGVKGFVSSA 862
++LH + + + ++ +L+DG R+ +D G + + G S
Sbjct: 801 SYLHHDSVPPIVHRDIKSNNILIDGDYGARVADFGVAKAVDLTGKAPKSMSVIAG--SCG 858
Query: 863 YVAPEERNGKDVTEKSEIYGFGVILIELLTGRNSVDIEAWNGIHYKNNIVEWARYCYSDC 922
Y+APE V EKS+IY FGV+++E++T + VD E + ++V+W
Sbjct: 859 YIAPEYAYTLRVNEKSDIYSFGVVILEIVTRKRPVDPELG-----EKDLVKWVCSTLDQK 913
Query: 923 HLDTWIDSVVMKGEDSSTYQNDIVETMNLALHCTANDPTTRPCARDILKALETV 976
++ ID + S ++ +I + +N+ L CT+ P RP R ++K L+ +
Sbjct: 914 GIEHVIDPKL-----DSCFKEEISKILNVGLLCTSPLPINRPSMRRVVKMLQEI 962
>At1g09970 leucine-rich repeat receptor-like kinase At1g09970
Length = 976
Score = 412 bits (1058), Expect = e-115
Identities = 307/1002 (30%), Positives = 486/1002 (47%), Gaps = 96/1002 (9%)
Query: 9 FSKFLNFICLFMFMLNFHSTHGEQEFELLLSFKASIKFDPLNFLSNWVNTSSDTICKWHG 68
F +F F+ +F S + ++LL K+S L +W S C + G
Sbjct: 11 FHRFSTFLVFSLF-----SVVSSDDLQVLLKLKSSFADSNLAVFDSWKLNSGIGPCSFIG 65
Query: 69 ITCDNWSHVNTVSLSGKNISGEVSSSIFQLPHVTNLDLSNNQLVGEIVFNSPF-LSSLLY 127
+TC++ +V T +DLS L G F+S + SL
Sbjct: 66 VTCNSRGNV------------------------TEIDLSRRGLSGNFPFDSVCEIQSLEK 101
Query: 128 LNLSNNNLTGPLPQSLFSSSFINLETLDLSNNMFSGKIPDQIGLLSSLTYVDLGGNVLVG 187
L+L N+L+G +P L + + +L+ LDL NN+FSG P+ L+ L ++ L + G
Sbjct: 102 LSLGFNSLSGIIPSDLKNCT--SLKYLDLGNNLFSGAFPE-FSSLNQLQFLYLNNSAFSG 158
Query: 188 KIP-NSITNLTSLESLTLASNQL--IGEIPTKICLMKRLKWIYLGYNNLSGEIPKNIGNL 244
P S+ N TSL L+L N + P ++ +K+L W+YL +++G+IP IG+L
Sbjct: 159 VFPWKSLRNATSLVVLSLGDNPFDATADFPVEVVSLKKLSWLYLSNCSIAGKIPPAIGDL 218
Query: 245 VSLNHLNLVYNNLTGPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFNLKNLISLDLSDNY 304
L +L + + LTG IP + LTNL L LY N LTG +P NLKNL LD S N
Sbjct: 219 TELRNLEISDSGLTGEIPSEISKLTNLWQLELYNNSLTGKLPTGFGNLKNLTYLDASTNL 278
Query: 305 LSGEISNLVVNLQKLEILHLFSNNFTGKIPNTITSLPHLQVLQLWSNKLTGEIPQTLGIH 364
L G++S L +L L L +F N F+G+IP L L L++NKLTG +PQ LG
Sbjct: 279 LQGDLSEL-RSLTNLVSLQMFENEFSGEIPLEFGEFKDLVNLSLYTNKLTGSLPQGLGSL 337
Query: 365 NNLTILDLSSNNLTGKIPNSLCASKNLHKIILFSNSLKGEIPKGLTSCKTLERVRLQDNN 424
+ +D S N LTG IP +C + + ++L N+L G IP+ +C TL+R R+ +NN
Sbjct: 338 ADFDFIDASENLLTGPIPPDMCKNGKMKALLLLQNNLTGSIPESYANCLTLQRFRVSENN 397
Query: 425 LSGKLPLEITQLPQIYLLDISGNKFSGRINDRKWNMPSLQMLNLANNNFSGDLPNSFGGN 484
L+G +P + LP++ ++DI N F G I N L L L N S +LP G
Sbjct: 398 LNGTVPAGLWGLPKLEIIDIEMNNFEGPITADIKNGKMLGALYLGFNKLSDELPEEIGDT 457
Query: 485 K-VEGLDLSQNQFSGYIQIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNR 543
+ + ++L+ N+F+G I L L LK+ +N G+ P+ + C+ L ++++ N
Sbjct: 458 ESLTKVELNNNRFTGKIPSSIGKLKGLSSLKMQSNGFSGEIPDSIGSCSMLSDVNMAQNS 517
Query: 544 LNGEIPEKLAKMPVLGLLDISENQFSGEIPKNLGSVESLVEVNISYNHFHGVLPSTEAFS 603
++GEIP L +P L L++S+N+ SG IP++L S+ L +++S N G +P + S
Sbjct: 518 ISGEIPHTLGSLPTLNALNLSDNKLSGRIPESLSSLR-LSLLDLSNNRLSGRIPL--SLS 574
Query: 604 AINASLVTGNKLCDGD-GDVSNGLPPCKSYNQMNSTRLFVLICFVLTALVVLVGTVVIFV 662
+ N S LC + + P +S+ TR+FVL C V L++L V
Sbjct: 575 SYNGSFNGNPGLCSTTIKSFNRCINPSRSH---GDTRVFVL-CIVFGLLILLASLVFFLY 630
Query: 663 LRMNKSFEVRRVVENEDGTWEVIFFDYKASKFVTIEDVLSSVKEGKVITKGRNWVSY--- 719
L+ + E R + + +W + F T +D++ S+KE +I +G Y
Sbjct: 631 LKKTEKKEGRSL---KHESWSIKSF---RKMSFTEDDIIDSIKEENLIGRGGCGDVYRVV 684
Query: 720 --EGK--------CVSNEMQF--VVKEISDTNSVSVSFWDDTVTFGKKVRHENIVKIMGM 767
+GK C S + F + +++ S F + T +RH N+VK+
Sbjct: 685 LGDGKEVAVKHIRCSSTQKNFSSAMPILTEREGRSKEFETEVQTL-SSIRHLNVVKLYCS 743
Query: 768 FRCGKRGYLVYEFVEGKSLREIMH-----GLSWLRRWKIALGIAKAINFLHCECLWFGLG 822
LVYE++ SL +++H L W R+ IALG AK + +LH +
Sbjct: 744 ITSDDSSLLVYEYLPNGSLWDMLHSCKKSNLGWETRYDIALGAAKGLEYLHHGYERPVIH 803
Query: 823 SEVSPETVLVDGKGVPR---------LKLDSPGIVVTPVM-GVKGFVSSAYVAPEERNGK 872
+V +L+D PR L+ + G T V+ G G Y+APE
Sbjct: 804 RDVKSSNILLDEFLKPRIADFGLAKILQASNGGPESTHVVAGTYG-----YIAPEYGYAS 858
Query: 873 DVTEKSEIYGFGVILIELLTGRNSVDIEAWNGIHYKNNIVEWARYCYSDCHLDTWIDSVV 932
VTEK ++Y FGV+L+EL+TG+ ++ E +IV W ++ + +V
Sbjct: 859 KVTEKCDVYSFGVVLMELVTGKKPIEAE----FGESKDIVNWVS---NNLKSKESVMEIV 911
Query: 933 MKGEDSSTYQNDIVETMNLALHCTANDPTTRPCARDILKALE 974
K + Y+ D V+ + +A+ CTA P RP R +++ +E
Sbjct: 912 DK-KIGEMYREDAVKMLRIAIICTARLPGLRPTMRSVVQMIE 952
>At1g17750 hypothetical protein
Length = 1088
Score = 401 bits (1031), Expect = e-112
Identities = 308/1062 (29%), Positives = 498/1062 (46%), Gaps = 147/1062 (13%)
Query: 37 LLSFKASIKFDPLNFLSNWV-NTSSDTICK--WHGITCD-NWSHVNTVSLSGKNISGEVS 92
LLS PL S W NTS T C W G+ CD + + V T++LS +SG++
Sbjct: 34 LLSLLKHFDKVPLEVASTWKENTSETTPCNNNWFGVICDLSGNVVETLNLSASGLSGQLG 93
Query: 93 SSIFQLPHVTNLDLSNNQLVGEIVFNSPFLSSLLYLNLSNNNLTGPLPQSLFSSSFINLE 152
S I +L + LDLS N G + +SL YL+LSNN+ +G +P S NL
Sbjct: 94 SEIGELKSLVTLDLSLNSFSGLLPSTLGNCTSLEYLDLSNNDFSGEVPDIF--GSLQNLT 151
Query: 153 TLDLSNNMFSGKIPDQIGLLSSLTYVDLGGNVLVGKIPNSITNLTSLESLTLASNQLIGE 212
L L N SG IP +G L L + + N L G IP + N + LE L L +N+L G
Sbjct: 152 FLYLDRNNLSGLIPASVGGLIELVDLRMSYNNLSGTIPELLGNCSKLEYLALNNNKLNGS 211
Query: 213 IPTKICLMKRLKWIY------------------------LGYNNLSGEIPKNIGNLVSLN 248
+P + L++ L ++ L +N+ G +P IGN SL+
Sbjct: 212 LPASLYLLENLGELFVSNNSLGGRLHFGSSNCKKLVSLDLSFNDFQGGVPPEIGNCSSLH 271
Query: 249 HLNLVYNNLTGPIPESLGNLTNLQYLFLYLNKLTGPIPKSIFNLKNLISLDLSDNYLSGE 308
L +V NLTG IP S+G L + + L N+L+G IP+ + N +L +L L+DN L GE
Sbjct: 272 SLVMVKCNLTGTIPSSMGMLRKVSVIDLSDNRLSGNIPQELGNCSSLETLKLNDNQLQGE 331
Query: 309 ISNLVVNLQKLEILHLF------------------------SNNFTGKIPNTITSLPHLQ 344
I + L+KL+ L LF +N TG++P +T L HL+
Sbjct: 332 IPPALSKLKKLQSLELFFNKLSGEIPIGIWKIQSLTQMLVYNNTLTGELPVEVTQLKHLK 391
Query: 345 VLQLWSNKLTGEIPQTLGIHNNLTILDLSSNNLTGKIPNSLCASKNLHKIILFSNSLKGE 404
L L++N G+IP +LG++ +L +DL N TG+IP LC + L IL SN L G+
Sbjct: 392 KLTLFNNGFYGDIPMSLGLNRSLEEVDLLGNRFTGEIPPHLCHGQKLRLFILGSNQLHGK 451
Query: 405 IPKGLTSCKTLERVRLQDNNLSGKLPLEITQLPQIYL----------------------- 441
IP + CKTLERVRL+DN LSG LP L Y+
Sbjct: 452 IPASIRQCKTLERVRLEDNKLSGVLPEFPESLSLSYVNLGSNSFEGSIPRSLGSCKNLLT 511
Query: 442 LDISGNKFSGRINDRKWNMPSLQMLNLANNNFSGDLPNSFGG-NKVEGLDLSQNQFSGYI 500
+D+S NK +G I N+ SL +LNL++N G LP+ G ++ D+ N +G I
Sbjct: 512 IDLSQNKLTGLIPPELGNLQSLGLLNLSHNYLEGPLPSQLSGCARLLYFDVGSNSLNGSI 571
Query: 501 QIGFKNLPELVQLKLNNNNLFGKFPEELFQCNKLVSLDLSHNRLNGEIPEKLAKMPVLGL 560
F++ L L L++NN G P+ L + ++L L ++ N G+IP + + L
Sbjct: 572 PSSFRSWKSLSTLVLSDNNFLGAIPQFLAELDRLSDLRIARNAFGGKIPSSVGLLKSLRY 631
Query: 561 -LDISENQFSGEIPKNLGSVESLVEVNISYNHFHGVLPSTEAFSAIN------------- 606
LD+S N F+GEIP LG++ +L +NIS N G L ++ ++N
Sbjct: 632 GLDLSANVFTGEIPTTLGALINLERLNISNNKLTGPLSVLQSLKSLNQVDVSYNQFTGPI 691
Query: 607 -ASLVTGNKLCDGDGD------------VSNGLPPCKSYNQMNSTRLFVLICFVLTALVV 653
+L++ + G+ D + CK ++++ ++ ++ +++
Sbjct: 692 PVNLLSNSSKFSGNPDLCIQASYSVSAIIRKEFKSCKGQVKLSTWKIALIAAGSSLSVLA 751
Query: 654 LVGTVVIFVLRMNKSFEVRRV-VENEDGTWEVIFFDYKASKFVTIEDVLSSVKEGKVITK 712
L+ + + + R + + + E+G ++ +K + D L + +I +
Sbjct: 752 LLFALFLVLCRCKRGTKTEDANILAEEGLSLLL------NKVLAATDNLD---DKYIIGR 802
Query: 713 GRNWVSYEGKCVSNEMQFVVKEISDTNSVSVS--FWDDTVTFGKKVRHENIVKIMGMFRC 770
G + V Y S E ++ VK++ + + + T G VRH N++++ +
Sbjct: 803 GAHGVVYRASLGSGE-EYAVKKLIFAEHIRANQNMKREIETIGL-VRHRNLIRLERFWMR 860
Query: 771 GKRGYLVYEFVEGKSLREIMHG-------LSWLRRWKIALGIAKAINFLHCECLWFGLGS 823
+ G ++Y+++ SL +++H L W R+ IALGI+ + +LH +C +
Sbjct: 861 KEDGLMLYQYMPNGSLHDVLHRGNQGEAVLDWSARFNIALGISHGLAYLHHDCHPPIIHR 920
Query: 824 EVSPETVLVDGKGVPRLK-------LDSPGIVVTPVMGVKGFVSSAYVAPEERNGKDVTE 876
++ PE +L+D P + LD + V G G Y+APE ++
Sbjct: 921 DIKPENILMDSDMEPHIGDFGLARILDDSTVSTATVTGTTG-----YIAPENAYKTVRSK 975
Query: 877 KSEIYGFGVILIELLTGRNSVDIEAWNGIHYKNNIVEWAR-----YCYSDCHLDTWIDSV 931
+S++Y +GV+L+EL+TG+ ++D I NIV W R Y D +D
Sbjct: 976 ESDVYSYGVVLLELVTGKRALDRSFPEDI----NIVSWVRSVLSSYEDEDDTAGPIVDPK 1031
Query: 932 VMKGEDSSTYQNDIVETMNLALHCTANDPTTRPCARDILKAL 973
++ + + ++ +LAL CT P RP RD++K L
Sbjct: 1032 LVDELLDTKLREQAIQVTDLALRCTDKRPENRPSMRDVVKDL 1073
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.137 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,312,127
Number of Sequences: 26719
Number of extensions: 1097833
Number of successful extensions: 28864
Number of sequences better than 10.0: 1058
Number of HSP's better than 10.0 without gapping: 794
Number of HSP's successfully gapped in prelim test: 264
Number of HSP's that attempted gapping in prelim test: 2784
Number of HSP's gapped (non-prelim): 4919
length of query: 984
length of database: 11,318,596
effective HSP length: 109
effective length of query: 875
effective length of database: 8,406,225
effective search space: 7355446875
effective search space used: 7355446875
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC147428.5


