

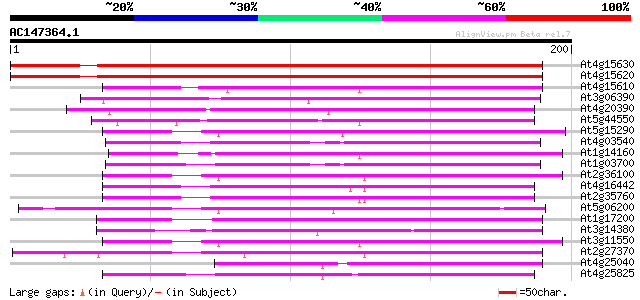
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147364.1 - phase: 0
(200 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g15630 unknown protein 179 1e-45
At4g15620 unknown protein (At4g15620) 174 2e-44
At4g15610 unknown protein 96 2e-20
At3g06390 unknown protein 96 2e-20
At4g20390 putative protein 89 2e-18
At5g44550 unknown protein 89 2e-18
At5g15290 unknown protein 84 4e-17
At4g03540 hypothetical protein 75 2e-14
At1g14160 unknown protein 75 2e-14
At1g03700 putative protein 74 7e-14
At2g36100 unknown protein 66 1e-11
At4g16442 unknown protein 62 2e-10
At2g35760 unknown protein 62 3e-10
At5g06200 putative protein 60 1e-09
At1g17200 unknown protein 57 5e-09
At3g14380 unknown protein 56 1e-08
At3g11550 unknown protein 55 2e-08
At2g27370 unknown protein 55 3e-08
At4g25040 unknown protein 54 6e-08
At4g25825 unknown protein 49 2e-06
>At4g15630 unknown protein
Length = 190
Score = 179 bits (453), Expect = 1e-45
Identities = 94/190 (49%), Positives = 127/190 (66%), Gaps = 6/190 (3%)
Query: 1 MEGRGKAIIDGVEGNYGRDELAMKKPPAGGGSCELVLRFLGFVLTLAAAIVVGTDKQTTI 60
ME K +DG+E G+ E +K EL +R L VLT+ AA V+G KQT +
Sbjct: 1 MEHESKNKVDGMEMEKGKKESGSRK------GLELTMRVLALVLTMVAATVLGVAKQTKV 54
Query: 61 VPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKSKVFWGT 120
VPIK++ +LPPLNV+ +AK YLSAFVY + ANAIAC Y AIS+++ ++++GK
Sbjct: 55 VPIKLIPTLPPLNVSTTAKASYLSAFVYNISANAIACGYTAISIVIVMISKGKRSKSLLM 114
Query: 121 LITIFDALMVALLFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLG 180
+ I D +MVALLFS GAA AIG++G GN HV WKKVC VF K+C+Q A S+ ++ +
Sbjct: 115 AVLIGDLMMVALLFSSTGAAGAIGLMGRHGNKHVMWKKVCGVFGKFCNQAAVSVAITLIA 174
Query: 181 SLVFLLLVVL 190
S+VF+LLVVL
Sbjct: 175 SVVFMLLVVL 184
>At4g15620 unknown protein (At4g15620)
Length = 190
Score = 174 bits (442), Expect = 2e-44
Identities = 91/190 (47%), Positives = 128/190 (66%), Gaps = 6/190 (3%)
Query: 1 MEGRGKAIIDGVEGNYGRDELAMKKPPAGGGSCELVLRFLGFVLTLAAAIVVGTDKQTTI 60
ME GK ++G+E G+ EL +K EL +R L +LT+AAA V+G KQT +
Sbjct: 1 MEHEGKNNMNGMEMEKGKRELGSRK------GVELTMRVLALILTMAAATVLGVAKQTKV 54
Query: 61 VPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKSKVFWGT 120
V IK++ +LPPL++ +AK YLSAFVY + NAIAC Y AIS+ + +++RG+
Sbjct: 55 VSIKLIPTLPPLDITTTAKASYLSAFVYNISVNAIACGYTAISIAILMISRGRRSKKLLM 114
Query: 121 LITIFDALMVALLFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLG 180
++ + D +MVALLFSG GAA+AIG++G GN HV WKKVC VF K+CH+ A S+ L+ L
Sbjct: 115 VVLLGDLVMVALLFSGTGAASAIGLMGLHGNKHVMWKKVCGVFGKFCHRAAPSLPLTLLA 174
Query: 181 SLVFLLLVVL 190
++VF+ LVVL
Sbjct: 175 AVVFMFLVVL 184
>At4g15610 unknown protein
Length = 193
Score = 95.5 bits (236), Expect = 2e-20
Identities = 55/160 (34%), Positives = 90/160 (55%), Gaps = 9/160 (5%)
Query: 34 ELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAV-SAKWHYLSAFVYFLVA 92
++VLRF+ F TL + +V+ T KQT + LP + + +A++ A +YF+VA
Sbjct: 30 QVVLRFVLFAATLTSIVVMVTSKQTKNI------FLPGTPIRIPAAEFTNSPALIYFVVA 83
Query: 93 NAIACTYGAISMLLTLLNRGKSKVFWGTLIT--IFDALMVALLFSGNGAATAIGVLGYQG 150
++AC Y +S +T+ K L+ I DA+MV ++ S GA + LG +G
Sbjct: 84 LSVACFYSIVSTFVTVSAFKKHSCSAVLLLNLAIMDAVMVGIVASATGAGGGVAYLGLKG 143
Query: 151 NSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVL 190
N VRW K+C+++DK+C V +I +S S+V LLL ++
Sbjct: 144 NKEVRWGKICHIYDKFCRHVGGAIAVSLFASVVLLLLSII 183
>At3g06390 unknown protein
Length = 199
Score = 95.5 bits (236), Expect = 2e-20
Identities = 58/172 (33%), Positives = 91/172 (52%), Gaps = 12/172 (6%)
Query: 26 PPAGGGS------CELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAK 79
PP+ S +++ R L F TL A IV+ T QT + + V S P VSA+
Sbjct: 24 PPSSAASFLDCRKIDIITRVLLFSATLTALIVMVTSDQTEMTQLPGVSSPAP----VSAE 79
Query: 80 WHYLSAFVYFLVANAIACTYGAISML--LTLLNRGKSKVFWGTLITIFDALMVALLFSGN 137
++ AF+YF+VA +A Y IS L ++LL + + + + D +M+ +L S
Sbjct: 80 FNDSPAFIYFVVALVVASFYALISTLVSISLLLKPEFTAQFSIYLASLDMVMLGILASAT 139
Query: 138 GAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVV 189
G A + + +GN V W K+CNV+DK+C +A S+ LS SL+ L+L +
Sbjct: 140 GTAGGVAYIALKGNEEVGWNKICNVYDKFCRYIATSLALSLFASLLLLVLSI 191
>At4g20390 putative protein
Length = 197
Score = 89.0 bits (219), Expect = 2e-18
Identities = 53/173 (30%), Positives = 93/173 (53%), Gaps = 7/173 (4%)
Query: 21 LAMKKPPAGGGSCE-----LVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVA 75
+A +K GG+ + L LR F+ TLAAAIV+ +K+T + + + ++P +
Sbjct: 1 MAREKIVVAGGTTKSWKLLLGLRIFAFMATLAAAIVMSLNKETKTLVVATIGTVP-IKAT 59
Query: 76 VSAKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRG-KSKVFWGTLITIFDALMVALLF 134
++AK+ + AFV+F++AN + + + +++ + +R + K I I D L L+
Sbjct: 60 LTAKFQHTPAFVFFVIANVMVSFHNLLMIVVQIFSRKLEYKGLRLLSIAILDMLNATLVS 119
Query: 135 SGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLL 187
+ AA + LG GN H +W KVC+ F YC A +II + G ++ LL+
Sbjct: 120 AAANAAVFVAELGKNGNKHAKWNKVCDRFTTYCDHGAGAIIAAFAGVILMLLV 172
>At5g44550 unknown protein
Length = 197
Score = 88.6 bits (218), Expect = 2e-18
Identities = 56/163 (34%), Positives = 93/163 (56%), Gaps = 8/163 (4%)
Query: 30 GGSCELVL--RFLGFVLTLAAAIVVGTDKQT-TIVPIKVVDSLPPLNVAVSAKWHYLSAF 86
G SC+++L R L F TL+AAIV+G +K+T T + KV ++ P+ +AK+ + AF
Sbjct: 13 GKSCKILLGLRLLAFSATLSAAIVMGLNKETKTFIVGKVGNT--PIQATFTAKFDHTPAF 70
Query: 87 VYFLVANAIACTYGAISMLLTLLNRGKSKVFWGTLIT--IFDALMVALLFSGNGAATAIG 144
V+F+VANA+ + + + L + GK + L++ I D L V L+ + AA +
Sbjct: 71 VFFVVANAMVSFHNLLMIALQIFG-GKMEFTGFRLLSVAILDMLNVTLISAAANAAAFMA 129
Query: 145 VLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLL 187
+G GN H RW K+C+ F YC A ++I + G ++ L++
Sbjct: 130 EVGKNGNKHARWDKICDRFATYCDHGAGALIAAFAGVILMLII 172
>At5g15290 unknown protein
Length = 187
Score = 84.3 bits (207), Expect = 4e-17
Identities = 53/168 (31%), Positives = 89/168 (52%), Gaps = 13/168 (7%)
Query: 34 ELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLN--VAVSAKWHYLSAFVYFLV 91
E +LR + F T+ +AI++GT +T LP + A+++ L A +F+V
Sbjct: 29 EFILRIVAFFNTIGSAILMGTTHET----------LPFFTQFIRFQAEYNDLPALTFFVV 78
Query: 92 ANAIACTYGAISMLLTLLNRGKSKVF-WGTLITIFDALMVALLFSGNGAATAIGVLGYQG 150
ANA+ Y +S+ L ++ K K L+ I D M+ LL SG +A AI L + G
Sbjct: 79 ANAVVSGYLILSLTLAFVHIVKRKTQNTRILLIILDVAMLGLLTSGASSAAAIVYLAHNG 138
Query: 151 NSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVLLPILRSRR 198
N+ W +C F+ +C +++ S+I S + ++ +LL++L I SRR
Sbjct: 139 NNKTNWFAICQQFNSFCERISGSLIGSFIAIVLLILLILLSAIALSRR 186
>At4g03540 hypothetical protein
Length = 164
Score = 75.5 bits (184), Expect = 2e-14
Identities = 44/155 (28%), Positives = 79/155 (50%), Gaps = 16/155 (10%)
Query: 35 LVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVANA 94
LVLR F LAA IV+ T ++ L +++ AK+ ++AF YF++ANA
Sbjct: 11 LVLRLAAFGAALAALIVMITSRERASF----------LAISLEAKYTDMAAFKYFVIANA 60
Query: 95 IACTYGAISMLLTLLNRGKSKVFWGTLITIFDALMVALLFSGNGAATAIGVLGYQGNSHV 154
+ Y + + L K + W + + D +M LL S AA A+ +G +GN++
Sbjct: 61 VVSVYSFLVLFLP-----KESLLW-KFVVVLDLVMTMLLTSSLSAALAVAQVGKKGNANA 114
Query: 155 RWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVV 189
W +C K+C Q+ ++I + ++++LL++
Sbjct: 115 GWLPICGQVPKFCDQITGALIAGFVALVLYVLLLL 149
>At1g14160 unknown protein
Length = 209
Score = 75.5 bits (184), Expect = 2e-14
Identities = 48/163 (29%), Positives = 88/163 (53%), Gaps = 9/163 (5%)
Query: 36 VLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVANAI 95
VLR T+ +A+ +GT ++ + SL L V + K+ L ++F+VANAI
Sbjct: 54 VLRLFAVFGTIGSALAMGTTHESVV-------SLSQL-VLLKVKYSDLPTLMFFVVANAI 105
Query: 96 ACTYGAISMLLTLLNRGKSKVFWGTLIT-IFDALMVALLFSGNGAATAIGVLGYQGNSHV 154
+ Y +S+ +++ + ++ +I + D +M+AL+ SG AATA L ++GN+
Sbjct: 106 SGGYLVLSLPVSIFHIFSTQAKTSRIILLVVDTVMLALVSSGASAATATVYLAHEGNTTA 165
Query: 155 RWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVLLPILRSR 197
W +C FD +C +++ S+I S ++ +L+V+ I SR
Sbjct: 166 NWPPICQQFDGFCERISGSLIGSFCAVILLMLIVINSAISLSR 208
>At1g03700 putative protein
Length = 164
Score = 73.6 bits (179), Expect = 7e-14
Identities = 41/155 (26%), Positives = 78/155 (49%), Gaps = 16/155 (10%)
Query: 35 LVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVANA 94
LVLRF F L A I + T ++ + + +++ AK+ L+AF YF++ANA
Sbjct: 11 LVLRFAAFCAALGAVIAMITSRERSSFFV----------ISLVAKYSDLAAFKYFVIANA 60
Query: 95 IACTYGAISMLLTLLNRGKSKVFWGTLITIFDALMVALLFSGNGAATAIGVLGYQGNSHV 154
I Y + + L K + W + + D ++ LL S AA A+ +G +GN++
Sbjct: 61 IVTVYSFLVLFLP-----KESLLW-KFVVVLDLMVTMLLTSSLSAAVAVAQVGKRGNANA 114
Query: 155 RWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVV 189
W +C ++C Q+ ++I + ++++ L++
Sbjct: 115 GWLPICGQVPRFCDQITGALIAGLVALVLYVFLLI 149
>At2g36100 unknown protein
Length = 206
Score = 65.9 bits (159), Expect = 1e-11
Identities = 43/167 (25%), Positives = 79/167 (46%), Gaps = 13/167 (7%)
Query: 34 ELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLN--VAVSAKWHYLSAFVYFLV 91
+ +LR +T+ AA V+ T ++T LP + A + L AF YF++
Sbjct: 49 DFLLRLAAIAVTIGAASVMYTAEET----------LPFFTQFLQFQAGYDDLPAFQYFVI 98
Query: 92 ANAIACTYGAISMLLTLLNRGKSKVFWGTLITIF-DALMVALLFSGNGAATAIGVLGYQG 150
A A+ +Y +S+ ++++ + LI + D L+V L S AA +I L + G
Sbjct: 99 AVAVVASYLVLSLPFSIVSIVRPHAVAPRLILLICDTLVVTLNTSAAAAAASITYLAHNG 158
Query: 151 NSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVLLPILRSR 197
N W +C F +C V+ +++ + L F++L+++ I R
Sbjct: 159 NQSTNWLPICQQFGDFCQNVSTAVVADSIAILFFIVLIIISAIALKR 205
>At4g16442 unknown protein
Length = 182
Score = 62.0 bits (149), Expect = 2e-10
Identities = 42/158 (26%), Positives = 74/158 (46%), Gaps = 14/158 (8%)
Query: 34 ELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVAN 93
EL+LR V L A I+V TD + ++ + +AK+ + A V+ +VAN
Sbjct: 12 ELLLRCSISVFALLALILVVTDTEVKLI----------FTIKKTAKYTDMKAVVFLVVAN 61
Query: 94 AIACTYGAISMLLTLLNRGKSKVFWGT-LITIF---DALMVALLFSGNGAATAIGVLGYQ 149
IA Y + + ++ K KV + L F D M L + A GV+ +
Sbjct: 62 GIAAVYSLLQSVRCVVGTMKGKVLFSKPLAWAFFSGDQAMAYLNVAAIAATAESGVIARE 121
Query: 150 GNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLL 187
G ++W +VC ++ K+C+Q+A + + L S+ + +
Sbjct: 122 GEEDLQWMRVCTMYGKFCNQMAIGVSSALLASIAMVFV 159
>At2g35760 unknown protein
Length = 201
Score = 61.6 bits (148), Expect = 3e-10
Identities = 43/158 (27%), Positives = 71/158 (44%), Gaps = 14/158 (8%)
Query: 34 ELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVAN 93
EL+LR L VL L AAI++ TD Q + + AK+ + A V +V N
Sbjct: 31 ELILRCLVCVLALVAAILIATDVQVREI----------FMIQKKAKFTDMKALVLLVVVN 80
Query: 94 AIACTYGAISMLLTLLNRGKSKVFWGTLIT--IF--DALMVALLFSGNGAATAIGVLGYQ 149
IA Y + + ++ K +V + + IF D + L +G AA
Sbjct: 81 GIAAGYSLVQAVRCVVGLMKGRVLFSKPLAWAIFFGDQAVAYLCVAGVAAAAQSAAFAKL 140
Query: 150 GNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLL 187
G ++W K+CN++ K+C+QV I + + +L+
Sbjct: 141 GEPELQWMKICNMYGKFCNQVGEGIASALFACIGMVLI 178
>At5g06200 putative protein
Length = 202
Score = 59.7 bits (143), Expect = 1e-09
Identities = 50/191 (26%), Positives = 86/191 (44%), Gaps = 18/191 (9%)
Query: 4 RGKAIIDGVEGNYGRDELAMKKPPAGGGSCELVLRFLGFVLTLAAAIVVGTDKQTTIVPI 63
+GKA + G+ RD G + +LR V LAAA +GT +T
Sbjct: 19 KGKAPLLGL----ARDHTGSGGYKRGLSIFDFLLRLAAIVAALAAAATMGTSDET----- 69
Query: 64 KVVDSLPPLN--VAVSAKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKS-KVFWGT 120
LP + A + L F +F+VA AI Y +S+ +++ + V
Sbjct: 70 -----LPFFTQFLQFEASYDDLPTFQFFVVAIAIVAGYLVLSLPFSVVTIVRPLAVAPRL 124
Query: 121 LITIFDALMVALLFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLG 180
L+ + D +AL + AA AI L + GN++ W +C F +C + + +++ S
Sbjct: 125 LLLVLDTAALALDTAAASAAAAIVYLAHNGNTNTNWLPICQQFGDFCQKTSGAVV-SAFA 183
Query: 181 SLVFLLLVVLL 191
S+ FL ++V++
Sbjct: 184 SVTFLAILVVI 194
>At1g17200 unknown protein
Length = 204
Score = 57.4 bits (137), Expect = 5e-09
Identities = 37/159 (23%), Positives = 69/159 (43%), Gaps = 14/159 (8%)
Query: 32 SCELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLV 91
+ E +LR L +AA +V+ D +T N S + L+AF Y +
Sbjct: 32 TAETMLRLAPVGLCVAALVVMLKDSET--------------NEFGSISYSNLTAFRYLVH 77
Query: 92 ANAIACTYGAISMLLTLLNRGKSKVFWGTLITIFDALMVALLFSGNGAATAIGVLGYQGN 151
AN I Y +S + + R S + D L+ L+ + + + L Y G+
Sbjct: 78 ANGICAGYSLLSAAIAAMPRSSSTMPRVWTFFCLDQLLTYLVLAAGAVSAEVLYLAYNGD 137
Query: 152 SHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVL 190
S + W C+ + +CH+ AS+I++ +++L ++
Sbjct: 138 SAITWSDACSSYGGFCHRATASVIITFFVVCFYIVLSLI 176
>At3g14380 unknown protein
Length = 178
Score = 56.2 bits (134), Expect = 1e-08
Identities = 40/160 (25%), Positives = 73/160 (45%), Gaps = 16/160 (10%)
Query: 32 SCELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLV 91
S E VLR L++ +++ + +S+ N S + + AF+Y +
Sbjct: 22 SAEAVLRVASMALSITGLVIM------------IKNSIS--NEFGSVSYSNIGAFMYLVS 67
Query: 92 ANAIACTYGAISMLLTL-LNRGKSKVFWGTLITIFDALMVALLFSGNGAATAIGVLGYQG 150
AN + Y +S L L L SKV TL + + +L +G +A + L Y G
Sbjct: 68 ANGVCAAYSLLSALAILALPCPISKVQVRTLFLLDQVVTYVVLAAGAVSAETV-YLAYYG 126
Query: 151 NSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVL 190
N + W C+ + +CH S++ + + SL+++LL ++
Sbjct: 127 NIPITWSSACDSYGSFCHNALISVVFTFVVSLLYMLLSLI 166
>At3g11550 unknown protein
Length = 204
Score = 55.5 bits (132), Expect = 2e-08
Identities = 39/167 (23%), Positives = 77/167 (45%), Gaps = 13/167 (7%)
Query: 34 ELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLN--VAVSAKWHYLSAFVYFLV 91
+ +LR V LAAA +GT +T LP + A + L F +F++
Sbjct: 47 DFLLRLAAIVAALAAAATMGTSDET----------LPFFTQFLQFEASYDDLPTFQFFVI 96
Query: 92 ANAIACTYGAISMLLTLLNRGKSKVFWGTLIT-IFDALMVALLFSGNGAATAIGVLGYQG 150
A A+ Y +S+ ++++ + L+ + D ++AL + +A AI L + G
Sbjct: 97 AMALVGGYLVLSLPISVVTILRPLATAPRLLLLVLDTGVLALNTAAASSAAAISYLAHSG 156
Query: 151 NSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVLLPILRSR 197
N + W +C F +C + + +++ + + + F +LVV+ + R
Sbjct: 157 NQNTNWLPICQQFGDFCQKSSGAVVSAFVSVVFFTILVVISGVALKR 203
>At2g27370 unknown protein
Length = 221
Score = 54.7 bits (130), Expect = 3e-08
Identities = 49/199 (24%), Positives = 84/199 (41%), Gaps = 20/199 (10%)
Query: 2 EGRGKAIIDGVEGNYGR---DELAMKKPPAGG----GSCELVLRFLGFVLTLAAAIVVGT 54
+G+GKA + NY R K PAG + VLR + + +AAA + T
Sbjct: 24 KGKGKAHVFAPPMNYNRIMDKHKQEKMSPAGWKRGVAIFDFVLRLIAAITAMAAAAKMAT 83
Query: 55 DKQTTIVPIKVVDSLPPLNVAVSAKWHY--LSAFVYFLVANAIACTYGAISMLLTLLNRG 112
++T LP + + Y L F++ N+I Y +S+ +++
Sbjct: 84 TEET----------LPFFTQFLQFQADYTDLPTMSSFVIVNSIVGGYLTLSLPFSIVCIL 133
Query: 113 KSKVFWGTLITIF-DALMVALLFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVA 171
+ L I D +M+ L A+ AI L + GNS W VC F +C +
Sbjct: 134 RPLAVPPRLFLILCDTVMMGLTLMAASASAAIVYLAHNGNSSSNWLPVCQQFGDFCQGTS 193
Query: 172 ASIILSQLGSLVFLLLVVL 190
+++ S + + + + LV+L
Sbjct: 194 GAVVASFIAATLLMFLVIL 212
>At4g25040 unknown protein
Length = 170
Score = 53.9 bits (128), Expect = 6e-08
Identities = 35/118 (29%), Positives = 51/118 (42%), Gaps = 4/118 (3%)
Query: 74 VAVSAKWHYLSAFVYFLVANAIACTYGAISMLLTLLN-RGKSKVFWGTLITIFDALMVAL 132
+A AK+ Y SAF Y + A C +++ L R + VF + FD L
Sbjct: 48 IAFEAKYSYSSAFRYLVYAQIAVCAATLFTLVWACLAVRRRGLVF---ALFFFDLLTTLT 104
Query: 133 LFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVL 190
S AA A G +G GN W +C YC +V S+ +S ++ +L VL
Sbjct: 105 AISAFSAAFAEGYVGKYGNKQAGWLPICGYVHGYCSRVTISLAMSFASFILLFILTVL 162
>At4g25825 unknown protein
Length = 175
Score = 48.9 bits (115), Expect = 2e-06
Identities = 35/156 (22%), Positives = 68/156 (43%), Gaps = 14/156 (8%)
Query: 34 ELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVAN 93
E++LR L + ++G D QT + + + + YL A L +
Sbjct: 8 EVILRLCIVFFLLLTSCLIGLDSQTKEIAY----------IHKNVSFRYLLALEAELYID 57
Query: 94 AIACTYGAISMLLTLLN--RGKSKVFWGTLITIFDALMVALLFSGNGAATAIGVLGYQGN 151
+ Y + + L N + S W + + D ++F+G AA +L G+
Sbjct: 58 VVVAAYNLVQLGLGWYNVEQKTSNPKWFSYL--LDQTAAYVVFAGTSAAAQHSLLVVTGS 115
Query: 152 SHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLL 187
++W K C F ++C Q+ ++IIL+ + + + +LL
Sbjct: 116 RELQWMKWCYKFTRFCFQMGSAIILNYIAAALMVLL 151
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.141 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,185,136
Number of Sequences: 26719
Number of extensions: 155047
Number of successful extensions: 526
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 479
Number of HSP's gapped (non-prelim): 31
length of query: 200
length of database: 11,318,596
effective HSP length: 94
effective length of query: 106
effective length of database: 8,807,010
effective search space: 933543060
effective search space used: 933543060
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC147364.1


