

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147363.5 - phase: 0
(433 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
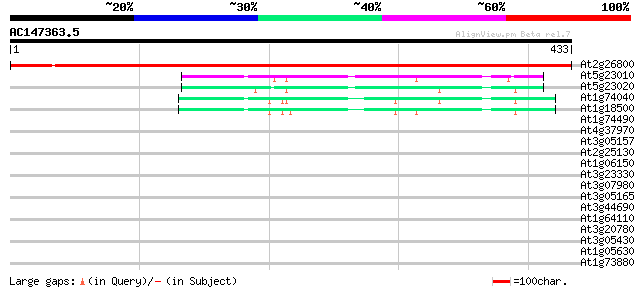
Score E
Sequences producing significant alignments: (bits) Value
At2g26800 hydroxymethylglutaryl-CoA lyase like protein 629 e-180
At5g23010 2-isopropylmalate synthase-like; homocitrate synthase-... 62 6e-10
At5g23020 2-isopropylmalate synthase-like protein 54 2e-07
At1g74040 2-isopropylmalate synthase like protein 49 7e-06
At1g18500 2-isopropylmalate synthase, putative 42 7e-04
At1g74490 putative protein kinase 34 0.14
At4g37970 cinnamyl alcohol dehydrogenase -like protein, LCADa 32 0.68
At3g05157 sugar transporter like protein 30 2.0
At2g25130 unknown protein 30 3.4
At1g06150 unknown protein 30 3.4
At3g23330 hypothetical protein 29 4.4
At3g07980 putative MAP3K epsilon protein kinase 29 4.4
At3g05165 sugar transporter like protein 29 4.4
At3g44690 putative protein 29 5.8
At1g64110 unknown protein 29 5.8
At3g20780 unknown protein 28 7.5
At3g05430 hypothetical protein 28 7.5
At1g05630 putative inositol 1,4,5-trisphosphate 5-phosphatase 28 7.5
At1g73880 putative glucosyltransferase (At1g73880) 28 9.8
>At2g26800 hydroxymethylglutaryl-CoA lyase like protein
Length = 468
Score = 629 bits (1621), Expect = e-180
Identities = 316/434 (72%), Positives = 363/434 (82%), Gaps = 2/434 (0%)
Query: 1 MSSLEEPLGHDKLPSMSTMDRVQRFSSGCCRPQVDNLGMGNCFIEGRSCSTSNSCNEDNE 60
MSSLEEPL DKLPSMSTMDR+QRFSSG CRP+ D++GMG+ +IEGR C+TSNSC +D++
Sbjct: 36 MSSLEEPLSFDKLPSMSTMDRIQRFSSGACRPR-DDVGMGHRWIEGRDCTTSNSCIDDDK 94
Query: 61 DYTAETYPWKRQTRDMSRGDSFSPRTMTTGRNTLKSGIVDNSFYTSDYQYSQKRNNKDMQ 120
+ E++PW+R TR +S G+ GR + SG + S + +YS N
Sbjct: 95 SFAKESFPWRRHTRKLSEGEHMFRNISFAGRTSTVSGTLRESKSFKEQKYSTFSNENGTS 154
Query: 121 DMAYKFMKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSP 180
++ K KG+P+FVKIVEVGPRDGLQNEKNIVPT VK+ELI RL S+GL V+EATSFVSP
Sbjct: 155 HISNKISKGIPKFVKIVEVGPRDGLQNEKNIVPTSVKVELIQRLVSSGLPVVEATSFVSP 214
Query: 181 KWVPQLADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNI 240
KWVPQLADAKDVM AV+ L G RLPVLTPNLKGF+AAV+AGA+EVA+FASASESFS SNI
Sbjct: 215 KWVPQLADAKDVMDAVNTLDGARLPVLTPNLKGFQAAVSAGAKEVAIFASASESFSLSNI 274
Query: 241 NCSIEESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFE 300
NC+IEESL RYR V AAKE S+PVRGYVSCVVGCPVEGPV PSKVAYV K LYDMGCFE
Sbjct: 275 NCTIEESLLRYRVVATAAKEHSVPVRGYVSCVVGCPVEGPVLPSKVAYVVKELYDMGCFE 334
Query: 301 ISLGDTIGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSS 360
ISLGDTIG+GTPG+VVPML AVMA+VP +KLAVHFHDTYGQ+L NILVSLQMGIS VDSS
Sbjct: 335 ISLGDTIGIGTPGSVVPMLEAVMAVVPADKLAVHFHDTYGQALANILVSLQMGISIVDSS 394
Query: 361 VAGLGGCPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTA 420
+AGLGGCPYAKGASGNVATEDVVYMLNGLG+ TNVD+GKL++AGDFI K LGRP+GSK A
Sbjct: 395 IAGLGGCPYAKGASGNVATEDVVYMLNGLGVHTNVDLGKLIAAGDFISKHLGRPNGSKAA 454
Query: 421 IALN-RVTADASKI 433
+ALN R+TADASKI
Sbjct: 455 VALNRRITADASKI 468
>At5g23010 2-isopropylmalate synthase-like; homocitrate
synthase-like
Length = 506
Score = 62.0 bits (149), Expect = 6e-10
Identities = 68/302 (22%), Positives = 125/302 (40%), Gaps = 37/302 (12%)
Query: 133 FVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADAKDV 192
+V++ + RDG Q+ + K+E+ +LA + ++E S + +L K +
Sbjct: 84 YVRVFDTTLRDGEQSPGGSLTPPQKLEIARQLAKLRVDIMEVGFPGSSE--EELETIKTI 141
Query: 193 MQAVHNLRGIR---LPVLTPNLK--------GFEAAVAAGAREVAVFASASESFSKSNIN 241
+ V N +PV+ + +EA A + VF S S+ K +
Sbjct: 142 AKTVGNEVDEETGYVPVICAIARCKHRDIEATWEALKYAKRPRILVFTSTSDIHMKYKLK 201
Query: 242 CSIEESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEI 301
+ EE + + R AK L G+ GC G + + G +
Sbjct: 202 KTQEEVIEMAVSSIRFAKSL-----GFNDIQFGCEDGGRSDKDFLCKILGEAIKAGVTVV 256
Query: 302 SLGDTIGVGTP---GTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVD 358
++GDT+G+ P G +V L A + +AVH H+ G + N + ++ G V+
Sbjct: 257 TIGDTVGINMPHEYGELVTYLKANTPGIDDVVVAVHCHNDLGLATANSIAGIRAGARQVE 316
Query: 359 SSVAGLGGCPYAKGASGNVATEDVV--------YMLNGLGIKTNVDIGKLMSAGDFIGKQ 410
++ G+G SGN + E+VV Y++N G+ T +D ++M+ + +
Sbjct: 317 VTINGIG------ERSGNASLEEVVMALKCRGAYVIN--GVYTKIDTRQIMATSKMVQEY 368
Query: 411 LG 412
G
Sbjct: 369 TG 370
>At5g23020 2-isopropylmalate synthase-like protein
Length = 503
Score = 53.5 bits (127), Expect = 2e-07
Identities = 65/299 (21%), Positives = 115/299 (37%), Gaps = 31/299 (10%)
Query: 133 FVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLAD--AK 190
+V++++ RDG Q+ + K+E+ +LA + ++E VS + + AK
Sbjct: 84 YVRVLDTTLRDGEQSPGAALTPPQKLEIARQLAKLRVDIMEVGFPVSSEEEFEAIKTIAK 143
Query: 191 DVMQAVHNLRGIRLPVLTPNLK--------GFEAAVAAGAREVAVFASASESFSKSNINC 242
V V G +PV+ + +EA A V +F S SE K +
Sbjct: 144 TVGNEVDEETGY-VPVICGIARCKKRDIEATWEALKYAKRPRVMLFTSTSEIHMKYKLKK 202
Query: 243 SIEESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEIS 302
+ EE + + AK L G+ GC G + + G +
Sbjct: 203 TKEEVIEMAVNSVKYAKSL-----GFKDIQFGCEDGGRTEKDFICKILGESIKAGATTVG 257
Query: 303 LGDTIGVGTPGTVVPMLLAVMAIVPTEK---LAVHFHDTYGQSLPNILVSLQMGISAVDS 359
DT+G+ P ++ V+ P A+H H+ G + N + + G V+
Sbjct: 258 FADTVGINMPQEFGELVAYVIENTPGADDIVFAIHCHNDLGVATANTISGICAGARQVEV 317
Query: 360 SVAGLGGCPYAKGASGNVATEDVVYMLNGL------GIKTNVDIGKLMSAGDFIGKQLG 412
++ G+G SGN E+VV L G+ T +D ++M+ + + G
Sbjct: 318 TINGIG------ERSGNAPLEEVVMALKCRGESLMDGVYTKIDSRQIMATSKMVQEHTG 370
>At1g74040 2-isopropylmalate synthase like protein
Length = 631
Score = 48.5 bits (114), Expect = 7e-06
Identities = 67/317 (21%), Positives = 122/317 (38%), Gaps = 46/317 (14%)
Query: 131 PEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADAK 190
P +V+I + RDG Q+ + + K+++ +LA G+ +IEA + K K
Sbjct: 84 PNYVRIFDTTLRDGEQSPGATLTSKEKLDIARQLAKLGVDIIEAGFPAASK--DDFEAVK 141
Query: 191 DVMQAVHNL--RGIRLPVLTP----NLK----GFEAAVAAGAREVAVFASASESFSKSNI 240
+ + V N +PV+ N K +EA A + F + S+ K +
Sbjct: 142 TIAETVGNTVDENGYVPVICGLSRCNKKDIETAWEAVKYAKRPRIHTFIATSDIHLKYKL 201
Query: 241 NCSIEESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDM---- 296
S EE + R + R A+ L GC P + LY++
Sbjct: 202 KKSKEEVIEIARNMVRFARSL------------GCEDVEFSPEDAGRSEREYLYEILGEV 249
Query: 297 ---GCFEISLGDTIGVGTPGTVVPMLLAVMAIVPTEK---LAVHFHDTYGQSLPNILVSL 350
G +++ DT+G+ P ++ + A P + ++ H + G S N L
Sbjct: 250 IKAGATTLNIPDTVGITLPSEFGQLIADIKANTPGIQNVIISTHCQNDLGLSTANTLSGA 309
Query: 351 QMGISAVDSSVAGLGGCPYAKGASGNVATEDVVYMLNGL------GIKTNVDIGKLMSAG 404
G V+ ++ G+G +GN + E+VV + G+ T +D ++
Sbjct: 310 HSGARQVEVTINGIG------ERAGNASLEEVVMAIKCRGDHVLGGLFTGIDTRHIVMTS 363
Query: 405 DFIGKQLGRPSGSKTAI 421
+ + G + AI
Sbjct: 364 KMVEEYTGMQTQPHKAI 380
>At1g18500 2-isopropylmalate synthase, putative
Length = 660
Score = 42.0 bits (97), Expect = 7e-04
Identities = 63/317 (19%), Positives = 121/317 (37%), Gaps = 46/317 (14%)
Query: 131 PEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADAK 190
P +V++ + RDG Q+ + + K+++ +LA G+ +IEA + K K
Sbjct: 86 PNYVRVFDTTLRDGEQSPGATLTSKEKLDIARQLAKLGVDIIEAGFPAASK--DDFEAVK 143
Query: 191 DVMQAVHNL--RGIRLPVLTP----NLKGFE----AAVAAGAREVAVFASASESFSKSNI 240
+ + V N +PV+ N K E A A + F + S+ + +
Sbjct: 144 TIAETVGNTVDENGYVPVICGLSRCNKKDIERAWDAVKYAKRPRIHTFIATSDIHLEYKL 203
Query: 241 NCSIEESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDM---- 296
+ E + R++ R A+ L GC P + LY++
Sbjct: 204 KKTKAEVIEIARSMVRFARSL------------GCEDVEFSPEDAGRSEREYLYEILGEV 251
Query: 297 ---GCFEISLGDTIGVGTP---GTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSL 350
G +++ DT+G+ P G ++ L A + ++ H + G S N L
Sbjct: 252 IKAGATTLNIPDTVGITLPSEFGQLITDLKANTPGIENVVISTHCQNDLGLSTANTLSGA 311
Query: 351 QMGISAVDSSVAGLGGCPYAKGASGNVATEDVVYMLNGL------GIKTNVDIGKLMSAG 404
G ++ ++ G+G +GN + E+VV + G+ T +D ++
Sbjct: 312 HAGARQMEVTINGIG------ERAGNASLEEVVMAIKCRGDHVLGGLFTGIDTRHIVMTS 365
Query: 405 DFIGKQLGRPSGSKTAI 421
+ + G + AI
Sbjct: 366 KMVEEYTGMQTQPHKAI 382
>At1g74490 putative protein kinase
Length = 399
Score = 34.3 bits (77), Expect = 0.14
Identities = 17/45 (37%), Positives = 22/45 (48%), Gaps = 3/45 (6%)
Query: 39 MGNCFIEGRSCSTSNSCNEDNEDYTAETYPWKRQTRDMSRGDSFS 83
MGNCF SCS N +DN D + + P+ R D R + S
Sbjct: 1 MGNCF---ESCSKRNDNEDDNLDSSVNSKPFSRANSDTGRSSNLS 42
>At4g37970 cinnamyl alcohol dehydrogenase -like protein, LCADa
Length = 363
Score = 32.0 bits (71), Expect = 0.68
Identities = 19/55 (34%), Positives = 32/55 (57%), Gaps = 10/55 (18%)
Query: 9 GHDKLPSMSTM-DRVQRFSSGCCRPQVDNLGMGNCFIEGRSCSTSNSCNEDNEDY 62
GH+ + +S + ++V +F+ G D +G+G C ++ SC T SC ED E+Y
Sbjct: 72 GHEIIGEVSEIGNKVSKFNLG------DKVGVG-CIVD--SCRTCESCREDQENY 117
>At3g05157 sugar transporter like protein
Length = 458
Score = 30.4 bits (67), Expect = 2.0
Identities = 18/61 (29%), Positives = 29/61 (47%), Gaps = 12/61 (19%)
Query: 179 SPKWVPQLADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKS 238
SP+W+ ++ K+V ++H LR G +A V+ A E+ V E SKS
Sbjct: 196 SPRWLAKIGSVKEVENSLHRLR------------GKDADVSDEAAEIQVMTKMLEEDSKS 243
Query: 239 N 239
+
Sbjct: 244 S 244
>At2g25130 unknown protein
Length = 468
Score = 29.6 bits (65), Expect = 3.4
Identities = 51/210 (24%), Positives = 80/210 (37%), Gaps = 35/210 (16%)
Query: 207 LTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEESLSRYRAVTRAAKELSIPVR 266
L LK FE ++ ARE A+ A + S N++ +E L + T E+S +
Sbjct: 244 LVKTLKNFEETSSSQAREDALRALYNLSIYHQNVSFILETDLIPFLLNTLGDMEVSERIL 303
Query: 267 GYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDTIGVGTPGTVVPMLLAVMAIV 326
++ VV P EG +V L D+ ++ D+I + ML+A
Sbjct: 304 AILTNVVSVP-EGRKAIGEVVEAFPILVDV----LNWNDSIKCQEKAVYILMLMA----- 353
Query: 327 PTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGCPYAKGASGNVATEDVVYML 386
H YG N ++ + S ++ + L G P A+ + V
Sbjct: 354 ---------HKGYGDR--NAMIEAGIESSLLELT---LVGSPLAQKRASRV--------- 390
Query: 387 NGLGIKTNVDIGKLMSAGDFIGKQLGRPSG 416
L VD GK +SA + LGR G
Sbjct: 391 --LECLRVVDKGKQVSAPIYGTSSLGRERG 418
>At1g06150 unknown protein
Length = 1288
Score = 29.6 bits (65), Expect = 3.4
Identities = 19/82 (23%), Positives = 36/82 (43%), Gaps = 4/82 (4%)
Query: 213 GFEAAVAAGAREVAVFASASESFSKSNINCSIEESLSRYRAVTRAAKELSIPVRGYVS-- 270
G+E+ ++AG + + + A S + C +EE + + L+ P+ + S
Sbjct: 77 GWESQISAGIKTILIVAVGSCGVVQLGSLCKVEEDPALVTHIRHLFLALTDPLADHASNL 136
Query: 271 --CVVGCPVEGPVPPSKVAYVA 290
C + P + P PSK + A
Sbjct: 137 MQCDINSPSDRPKIPSKCLHEA 158
>At3g23330 hypothetical protein
Length = 715
Score = 29.3 bits (64), Expect = 4.4
Identities = 22/103 (21%), Positives = 40/103 (38%), Gaps = 4/103 (3%)
Query: 37 LGMGNCFIEGRSCSTSNSCNEDNEDYTAETYPWKRQTRDMSRGDSFSPRTMTTGRNTLKS 96
+ +GN F E + N +ED AET + R PR NT+ +
Sbjct: 160 ISVGNVFDE----MPQRTSNSGDEDVKAETCIMPFGIDSVRRVFEVMPRKDVVSYNTIIA 215
Query: 97 GIVDNSFYTSDYQYSQKRNNKDMQDMAYKFMKGMPEFVKIVEV 139
G + Y + ++ D++ ++ +P F + V+V
Sbjct: 216 GYAQSGMYEDALRMVREMGTTDLKPDSFTLSSVLPIFSEYVDV 258
>At3g07980 putative MAP3K epsilon protein kinase
Length = 1367
Score = 29.3 bits (64), Expect = 4.4
Identities = 28/110 (25%), Positives = 45/110 (40%), Gaps = 25/110 (22%)
Query: 5 EEPLGHDKLPSMSTMDRVQRFSSGCCRPQVDNLGMGNCFIEGRSCSTSNSCNEDNEDYTA 64
E+ + D+ P++S D+ R S G C I + TS E++E Y
Sbjct: 359 EDDINSDQGPTLSMHDKSSRQS-------------GTCSISSDAKGTSQDVLENHEKYDR 405
Query: 65 ETYPWKRQTRDMSRGDSFSPRTMTTGRNTLKSGIVDNSF-YTSDYQYSQK 113
+ P +T + S G RNTL + +V + S + +SQK
Sbjct: 406 DEIPGNLET-EASEG----------RRNTLATKLVGKEYSIQSSHSFSQK 444
>At3g05165 sugar transporter like protein
Length = 467
Score = 29.3 bits (64), Expect = 4.4
Identities = 18/72 (25%), Positives = 33/72 (45%), Gaps = 12/72 (16%)
Query: 179 SPKWVPQLADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKS 238
SP+W+ ++ +K+V ++H LR G + V+ A E+ V E SKS
Sbjct: 205 SPRWLAKIRLSKEVESSLHRLR------------GKDTDVSGEAAEIQVMTKMLEEDSKS 252
Query: 239 NINCSIEESLSR 250
+ + ++ R
Sbjct: 253 SFSDMFQKKYRR 264
>At3g44690 putative protein
Length = 1176
Score = 28.9 bits (63), Expect = 5.8
Identities = 12/25 (48%), Positives = 15/25 (60%)
Query: 60 EDYTAETYPWKRQTRDMSRGDSFSP 84
+DY P +Q RD+SRGD SP
Sbjct: 245 DDYVGRNSPRDQQVRDLSRGDHDSP 269
>At1g64110 unknown protein
Length = 824
Score = 28.9 bits (63), Expect = 5.8
Identities = 12/42 (28%), Positives = 24/42 (56%)
Query: 385 MLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIALNRV 426
+L+ LG+ V +G +++G +GK G S S A+ +++
Sbjct: 2 LLSALGVGVGVGVGLGLASGQAVGKWAGGNSSSNNAVTADKM 43
>At3g20780 unknown protein
Length = 563
Score = 28.5 bits (62), Expect = 7.5
Identities = 29/127 (22%), Positives = 51/127 (39%), Gaps = 10/127 (7%)
Query: 60 EDYTAETYPWKRQTRDMSRGDSFSPRTMTTGRNTLKSGI---VDNSFYTSDYQYSQKRNN 116
+DY E KR ++ +R + + +G+ + G+ + S Y+ + K N
Sbjct: 104 DDYETEKARGKRLAKE-ARASEIQAKNLASGKKNKEPGVSKVLKARGEASYYKVTCKDNG 162
Query: 117 KDMQDMAYKFMKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATS 176
K M +P V G + GL+ + K+ LI STGL + ++S
Sbjct: 163 KGMPH------DDIPNMFGRVLSGTKYGLKQTRGKFGLGAKMALIWSKMSTGLPIEISSS 216
Query: 177 FVSPKWV 183
S +V
Sbjct: 217 MKSQNYV 223
>At3g05430 hypothetical protein
Length = 965
Score = 28.5 bits (62), Expect = 7.5
Identities = 39/163 (23%), Positives = 66/163 (39%), Gaps = 24/163 (14%)
Query: 117 KDMQDMAYKFMKGMPEFVKIVEVGPRDGL------QNEKNIVPTDVKIELIHRLASTGLS 170
K Q + F K + E + EVG R L +N+ N P + + + +
Sbjct: 200 KSQQTSSDHFAKAVEEAMN--EVGRRSALGLTCKCRNQYNFRPINAQ-----GYFAVDVP 252
Query: 171 VIEATSFVSPKWVPQLADAKDVMQAVHNLRGIRLPVLTP------NLKGFEAAVAAGARE 224
E + S K Q+ A+D +V L ++ L P +LK F+ VA A
Sbjct: 253 DYEVQAIYSSK---QIQKARDSFSSVQTLAFVKRCALAPQECDTDSLKSFQKKVAVCAFR 309
Query: 225 VAVFASASESFSKSNINCSIEESLSRYRAVTRAAKELSIPVRG 267
AVF E++ ++ S+ + + + RA L +P+ G
Sbjct: 310 RAVFEEFDETYEQAFRARSVYCLMKTHEPLNRA--PLRVPLSG 350
>At1g05630 putative inositol 1,4,5-trisphosphate 5-phosphatase
Length = 1136
Score = 28.5 bits (62), Expect = 7.5
Identities = 26/117 (22%), Positives = 48/117 (40%), Gaps = 18/117 (15%)
Query: 168 GLSVIEATSFVSPK---WVPQLADAKDVMQAVHNLRGIRLPVLT-----------PNLKG 213
G++ EA F+S + W+ + + M+ +G+R ++T L G
Sbjct: 796 GITYDEARDFISQRSFDWLRERDQLRAEMKVGKVFQGMREALITFPPTYKFERNRSGLGG 855
Query: 214 FEAA----VAAGAREVAVFASASESFSKSNINCSIEESLSRYRAVTRAAKELSIPVR 266
+++ + A V + S FS+SN+ C + S+ Y A + PVR
Sbjct: 856 YDSGEKKRIPAWCDRVIYRDTQSSPFSESNLQCPVVSSVIMYEACMDVTESDHKPVR 912
>At1g73880 putative glucosyltransferase (At1g73880)
Length = 473
Score = 28.1 bits (61), Expect = 9.8
Identities = 16/48 (33%), Positives = 28/48 (58%), Gaps = 3/48 (6%)
Query: 158 IELIHRLASTGLSVIEATSFVSPKWVPQLADAKDVMQAVHNLRGIRLP 205
++ HRLA G + ++ T V+PK +P L+ ++ AV N+ + LP
Sbjct: 30 LDFTHRLALRGGAALKITVLVTPKNLPFLS---PLLSAVVNIEPLILP 74
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,185,835
Number of Sequences: 26719
Number of extensions: 384669
Number of successful extensions: 936
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 920
Number of HSP's gapped (non-prelim): 21
length of query: 433
length of database: 11,318,596
effective HSP length: 102
effective length of query: 331
effective length of database: 8,593,258
effective search space: 2844368398
effective search space used: 2844368398
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC147363.5


