

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147179.8 + phase: 0
(307 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
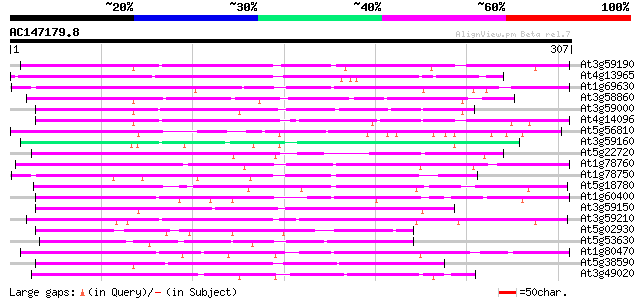
Score E
Sequences producing significant alignments: (bits) Value
At3g59190 unknown protein 76 3e-14
At4g13965 putative protein 75 4e-14
At1g69630 hypothetical protein 75 6e-14
At3g58860 putative protein 72 4e-13
At3g59000 unknown protein 72 5e-13
At4g14096 F-box protein - like 71 6e-13
At5g56810 putative protein 71 8e-13
At3g59160 putative protein 70 1e-12
At5g22720 unknown protein 70 2e-12
At1g78760 hypothetical protein 69 4e-12
At1g78750 hypothetical protein 67 9e-12
At5g18780 putative protein 66 2e-11
At1g60400 66 3e-11
At3g59150 unknown protein 65 5e-11
At3g59210 unknown protein 64 8e-11
At5g02930 putative protein 64 1e-10
At5g53630 heat shock transcription factor HSF30-like protein 63 2e-10
At1g80470 hypothetical protein 63 2e-10
At5g38590 unknown protein 62 4e-10
At3g49020 putative protein 62 4e-10
>At3g59190 unknown protein
Length = 388
Score = 75.9 bits (185), Expect = 3e-14
Identities = 84/332 (25%), Positives = 146/332 (43%), Gaps = 44/332 (13%)
Query: 7 KHNDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMR 66
K D +D +S+LP+ +L +LS L EA T ++ RWR L +P L L + + R
Sbjct: 4 KRMDSGSKDIISNLPDALLCHVLSFLPTTEAASTSVLAKRWRFLLAFVPNLDLDNMIYDR 63
Query: 67 -----------IKSFNRFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNV 115
KSF FV R++ L+ + PL C+ DP + V + V
Sbjct: 64 PKMGRRKRLELRKSFKLFVDRVMALQG-NAPLKKFSLRCKIGSDPSRVNGWVLKVLDRGV 122
Query: 116 EKLRVHACCDFQH-FSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESF 174
E+L ++ ++++ TL SL + TDE LP L TL L +
Sbjct: 123 EELDLYIASEYEYPLPPKVLMTKTLVSLKV----SGTDEFTIDVGEFFLPKLKTLHLSAI 178
Query: 175 AFCVGDDG---CAEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPVYYS-- 229
+F GD+G A+ SA ++L+ L+++ E +SS +L ++I + + +
Sbjct: 179 SF--GDEGGPPFAKLISACHALEELVMIKMMWDYWEFCSVSSPSLKRVSIDCENIDENPK 236
Query: 230 TVELSTPSLFTFDFVCYEGIPVLKLCRSKINLSSVKHVNIEVRMWTDYADASLVLLN--- 286
+V TP+L +F + + K+N S+ +I +RM D + L+N
Sbjct: 237 SVSFDTPNLVYLEFT-----DTVAVKYPKVNFDSLVEASIGLRMTPDQVFDARDLVNRHH 291
Query: 287 ------------WLAELANIKSLTLNHTALKV 306
++ + N+K++ L+ AL+V
Sbjct: 292 GYKRCKGANAADFMMGVCNVKTMYLSSEALEV 323
>At4g13965 putative protein
Length = 294
Score = 75.1 bits (183), Expect = 4e-14
Identities = 85/295 (28%), Positives = 133/295 (44%), Gaps = 37/295 (12%)
Query: 1 MKRGRKKHNDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILC 60
MKR RK+ V DR+S LP ++++ILSLL + AV T +S +W+ LWK LP L
Sbjct: 1 MKR-RKRGRGIVNADRISQLPEALIIQILSLLPTEVAVTTSVLSKQWQFLWKMLPKLNFD 59
Query: 61 S-SHFMRIKSFNRFVSRILCLRDVSTPLHAVDFLCR-GVVDPRLLKKVVTYAVSHNVEKL 118
S K+F++ V R L L + LH++ + + + K++ A + N+ KL
Sbjct: 60 SLDQRHEFKTFSKNVKRAL-LSHKAPVLHSLHLIVHLHLCNSMNTAKLIGIAFACNLRKL 118
Query: 119 RVHACCDFQHFSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFAFCV 178
+ ++C TL +L+L + + P S+ L +L TL L F
Sbjct: 119 VLEVDGGRFSIPESLYNCETLDTLEL-----KYSILMDVPSSICLKSLRTLHLHYVDFKD 173
Query: 179 GD------DGCAE---------PFS-------ALNSLKNLMILYCNVLDAES-LCISSVT 215
+ GC PFS A++SLK L I + +D + I+S +
Sbjct: 174 NESALNLLSGCPNLENLVVHRYPFSSVKTYTIAVSSLKRLTIYTSSTVDPRAGYVINSPS 233
Query: 216 LTNLTIVGDPVYYSTVELSTPSLFTFDFVCYEGIPVLKLCRSKINLSSVKHVNIE 270
LT L IVG + + +E + P L + I L S L+SVK + +E
Sbjct: 234 LTYLKIVGQ-IGFCLIE-NVPELVEASMIVSSQIINKNLLES---LTSVKRLFLE 283
>At1g69630 hypothetical protein
Length = 451
Score = 74.7 bits (182), Expect = 6e-14
Identities = 88/325 (27%), Positives = 135/325 (41%), Gaps = 36/325 (11%)
Query: 2 KRGRKKHNDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCS 61
K R K +D V D +S LP+C+L +L L + VKT +S RWR+LWKH+P L L +
Sbjct: 7 KHVRAKGSDEV--DWISKLPDCLLCEVLLNLPTKDVVKTSVLSRRWRNLWKHVPGLDLDN 64
Query: 62 SHFMRIKSFNRFVSRILCLRDVS-TPLHAVDFLCRGVVDP------RLLKKVVTYAVSHN 114
+ F +F FV L S + + C DP R + +VT V H
Sbjct: 65 TDFQEFNTFLSFVDSFLDFNSESFLQKFILKYDCDDEYDPDIFLIGRWINTIVTRKVQHI 124
Query: 115 VEKLRVHACCDFQHFSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESF 174
+ + Q S ++C +L SL LC L+ P+ ++LP+L + L
Sbjct: 125 DVLDDSYGSWEVQ-LPSSIYTCESLVSLKLC------GLTLASPEFVSLPSLKVMDLIIT 177
Query: 175 AFCVGDDGCAEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDP----VYYST 230
F D G + L++L I + E L + S +L T V D V
Sbjct: 178 KF-ADDMGLETLITKCPVLESLTIERSFCDEIEVLRVRSQSLLRFTHVADSDEGVVEDLV 236
Query: 231 VELSTPSLFTFDFVCYEGIPVL-----KLCRSKI----NLSSVKHVNIEVRMWTDYADAS 281
V + P L + + KL ++ I NLSSV N D
Sbjct: 237 VSIDAPKLEYLRLSDHRVASFILNKPGKLVKADIDIVFNLSSVNKFN------PDDLPKR 290
Query: 282 LVLLNWLAELANIKSLTLNHTALKV 306
++ N+L ++ IK + + + L+V
Sbjct: 291 TMIRNFLLGISTIKDMIIFSSTLEV 315
>At3g58860 putative protein
Length = 457
Score = 72.0 bits (175), Expect = 4e-13
Identities = 78/283 (27%), Positives = 120/283 (41%), Gaps = 33/283 (11%)
Query: 10 DYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMR--- 66
D K D S LP+ V+ ILS L EA T ++ +WR+L+ +P+L S F+
Sbjct: 2 DSEKMDLFSKLPDEVISHILSSLPTKEAASTSVLAKKWRYLFAFVPSLDFNDSDFLHPQE 61
Query: 67 --------IKSFNRFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKL 118
++SF FV R+L L+ S P+ + VDP + + + + V L
Sbjct: 62 GKREKDGILRSFMDFVDRVLALQGAS-PIKKFSLNVKTGVDPDRVDRWICNVLQRGVSHL 120
Query: 119 RVHACCDFQHFSSCFFS---CHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFA 175
+ DF+ S + TL L G + + + LP L TL LES
Sbjct: 121 ALF--MDFEEEYSLPYEISVSKTLVELKTGYGV----DLYLWDDDMFLPMLKTLVLESVE 174
Query: 176 FCVGDDGCAEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPVYYSTVELST 235
F G P A L+ LM+L D ++ +SS +L NL I + T+ T
Sbjct: 175 FGRGQFQTLLP--ACPVLEELMLLNMEWKD-RNVILSSSSLKNLKITSEDGCLGTLSFDT 231
Query: 236 PSLFTFDFVCY--EGIPVLKLCRSKINLSSVKHVNIEVRMWTD 276
P+L D+ Y E P+ +NL ++ V I + + D
Sbjct: 232 PNLVFLDYYDYVAEDYPI-------VNLKNLVEVGINLVLTAD 267
>At3g59000 unknown protein
Length = 491
Score = 71.6 bits (174), Expect = 5e-13
Identities = 77/258 (29%), Positives = 116/258 (44%), Gaps = 27/258 (10%)
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMR-------- 66
DR+ LP+ +L ILS L EA T +S RWR+L +P L F+
Sbjct: 2 DRVGSLPDELLSHILSFLTTKEAALTSLLSKRWRYLIAFVPNLAFDDIVFLHPEEGKPER 61
Query: 67 ---IKSFNRFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKLRVHAC 123
+SF FV R+L L+ +P+ CR VD + ++ ++ V +L +
Sbjct: 62 DEIRQSFMDFVDRVLALQ-AESPIKKFSLKCRIGVDSDRVDGWISNVLNRGVSELDLLII 120
Query: 124 C-----DFQHFSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFAFCV 178
D S F+ TL L++ G + S+ LP L TL L+S F V
Sbjct: 121 LGMTMEDSYRLSPKGFASKTLVKLEIGCG----IDISWVAGSIFLPMLKTLVLDSVWFYV 176
Query: 179 GDDGCAEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPVYYSTVELSTPSL 238
D AL +L+ L+++ N LD++ + IS+ +L LTI D + T TPSL
Sbjct: 177 --DKFETLLLALPALEELVLVDVNWLDSD-VTISNASLKTLTIDSDG-HLGTFSFDTPSL 232
Query: 239 FTFDFVCY--EGIPVLKL 254
F + Y E PV+K+
Sbjct: 233 VYFCYSDYAAEDYPVVKM 250
>At4g14096 F-box protein - like
Length = 468
Score = 71.2 bits (173), Expect = 6e-13
Identities = 82/309 (26%), Positives = 137/309 (43%), Gaps = 31/309 (10%)
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMR-------I 67
D +S LP + ILS L EA T +S +WR+L+ +P L L S ++
Sbjct: 8 DIISSLPEAISCHILSFLPTKEAASTSVLSKKWRYLFAFVPNLDLDESVYLNPENETEVS 67
Query: 68 KSFNRFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKLRVHACCDFQ 127
SF FV R+L L+ ++PLH V+P + + + V L +H + +
Sbjct: 68 SSFMDFVDRVLALQG-NSPLHKFSLKIGDGVEPDRIIPWINNVLERGVSDLDLHVYMETE 126
Query: 128 H-FSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFAFCVGDDGCAEP 186
F S F TL L L + P L F + + LP L TL ++S F G +
Sbjct: 127 FVFPSEMFLSKTLVRLKLMLYP-----LLEF-EDVYLPKLKTLYIDSCYFEKYGIGLTKL 180
Query: 187 FSALNSLKNLM---ILYCNVLDAESLCISSV-TLTNLTIVGDPVYYSTVELSTPSLFTFD 242
S L++L+ I +C D S+ + ++ LT T V D + +V + TP+L
Sbjct: 181 LSGCPILEDLVLDDIPWC-TWDFASVSVPTLKRLTFSTQVRDE-FPKSVSIDTPNLVYLK 238
Query: 243 FVCYEGIPVLKLCRSKINLSSVKHVNIEVRM---WTDYADASLV--LLNWLAELANIKSL 297
F + K+N S+ +I++R+ Y + +V +++ + N+K+L
Sbjct: 239 FT-----DTVAGKYPKVNFDSLVEAHIDLRLLQGHQGYGENDMVGNATDFIMRICNVKTL 293
Query: 298 TLNHTALKV 306
L+ L+V
Sbjct: 294 YLSSNTLQV 302
>At5g56810 putative protein
Length = 435
Score = 70.9 bits (172), Expect = 8e-13
Identities = 98/372 (26%), Positives = 152/372 (40%), Gaps = 100/372 (26%)
Query: 1 MKRGRKKHNDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILC 60
M+ R D DR+S LPN +L RILSL+ +A+ T +S RW+ +WK LPTL+
Sbjct: 1 MEAARSGIEDVTSPDRISQLPNDLLFRILSLIPVSDAMSTSLLSKRWKSVWKMLPTLVYN 60
Query: 61 SSHFMRIKS--FNRFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKL 118
+ I S F++F R L L + L K +T + + L
Sbjct: 61 ENSCSNIGSLGFDQFCGRSLQLHEAP------------------LLKTLTLELRKQTDSL 102
Query: 119 RVHACCDFQHFSSCFFSCHTLTSLDLCV---GPPRTDERLSFPKSLNL-PALTTLSLESF 174
SS F + H+ T L+ + G P +SFP +L++ L L L+
Sbjct: 103 D----------SSIFPNIHS-TLLEFSIKSTGYPVYYSTISFPNNLDVFQTLVVLKLQG- 150
Query: 175 AFCVGDDGCAEPFSALNSLK------------NLMILYCNVLD---AESLC--------I 211
C+ F +L SL + ++ C VL+ + LC I
Sbjct: 151 NICLDVVDSPVCFQSLKSLYLTCVNFENEESFSKLLSACPVLEDLFLQRLCSVGRFLFSI 210
Query: 212 SSVTLTNLTIVGDPVYYST----VELSTPS---LFTFD----FVCYEGIPVLKLCRSKIN 260
S +L LT + YYS +E++ PS L FD F E +P L ++
Sbjct: 211 SVPSLQRLTYTKEQAYYSNDEAILEITAPSLKHLNIFDRVGVFSFIEDMPKLVEASVRVK 270
Query: 261 LS----------SVKHVNIE------------------VRMWTDYAD--ASLVLLNWLAE 290
LS SV+H++++ + + D D S +LL+ L +
Sbjct: 271 LSKNEKLPKVLTSVEHLSLDLYPSMVFHLDDRFISKQLLHLKLDIYDNFQSNLLLSLLKD 330
Query: 291 LANIKSLTLNHT 302
L N++SL LNH+
Sbjct: 331 LPNLQSLKLNHS 342
>At3g59160 putative protein
Length = 464
Score = 70.1 bits (170), Expect = 1e-12
Identities = 83/312 (26%), Positives = 124/312 (39%), Gaps = 48/312 (15%)
Query: 7 KHNDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFM- 65
K D +D ++DLP+ +L +LS L EA T +S RWR+L +P L S ++
Sbjct: 5 KRMDSGSKDMINDLPDALLCHVLSYLTTKEAASTSLLSRRWRYLLAFVPNLEFDDSAYLH 64
Query: 66 ---RIK------------------------SFNRFVSRILCLRDVSTPLHAVDFLC---R 95
R+K SF FV RIL L+ ++PL
Sbjct: 65 RDKRVKNPLHEKGLVGFVLTVDDKRKKLSTSFPDFVDRILDLQG-NSPLDKFSLKMVDDH 123
Query: 96 GVVDPRLLKKVVTYAVSHNVEKLRVHACCDFQHFSSC---FFSCHTLTSLDLCV--GPPR 150
VDP + + + V L H D ++S F TL L L + GPP
Sbjct: 124 DPVDPDCVAPWIHKVLVRGVSDL--HLVIDMNEWTSLPAKIFLTETLVKLTLKIRDGPPI 181
Query: 151 TDERLSFPKSLNLPALTTLSLESFAFCVGDDGCAEPFSALNSLKNLMILYCNVLDAESLC 210
K ++LP L TL LES F D G ++ S L+ L++ + S
Sbjct: 182 D------VKHVHLPKLKTLHLESVMFDEEDIGFSKLLSGCPELEELVLHHIWSCVWTSCS 235
Query: 211 ISSVTLTNLTIVGDPVYYSTVELSTPSLFTFD---FVCYEGIPVLKLCRSKINLSSVKHV 267
+S TL LT + + + + P+ +FD V E V+ K+N S+
Sbjct: 236 VSVATLKRLTFCCNNMKFCGMHEENPNNVSFDTPNLVYLEYAEVIANNYPKVNFDSLVEA 295
Query: 268 NIEVRMWTDYAD 279
I++ M D D
Sbjct: 296 KIDIWMTNDQLD 307
>At5g22720 unknown protein
Length = 422
Score = 69.7 bits (169), Expect = 2e-12
Identities = 73/270 (27%), Positives = 116/270 (42%), Gaps = 47/270 (17%)
Query: 13 KEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIKSFNR 72
KED +S LP+ ++ +IL L +AV T +S RWR LW P L+L S+ F +F
Sbjct: 21 KEDLISQLPDSLITQILFYLQTKKAVTTSVLSKRWRSLWLSTPGLVLISNDFTDYNAFVS 80
Query: 73 FVSRILCL-RDVSTPLHAVDFLCR-GVVDPRLLKKVVTYAVSHNVEKLRVH----ACCDF 126
FV + L R+ LH + R G D + + + + + ++ L V C F
Sbjct: 81 FVDKFLGFSREQKLCLHKLKLSIRKGENDQDCVTRWIDFVATPKLKHLDVEIGPTRCECF 140
Query: 127 QHFSSCFFSCHTLTSLDL---CVGPPRTDERLSFPKSLNLPALTTLSLESFAFCVGDDGC 183
+ +SC +L L L C+G +S++LP L T+SLE
Sbjct: 141 EVIPLSLYSCESLLYLRLNHVCLGK---------FESVSLPCLKTMSLE----------- 180
Query: 184 AEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPVYYSTVELSTPSLFTFDF 243
+Y N D ESL + L +L+ V Y V + T
Sbjct: 181 -------------QNIYANEADLESLISTCPVLEDLSFVSGA--YDKVNVLRVQSQTLTS 225
Query: 244 VCYEG-IPVLKLCRSK--INLSSVKHVNIE 270
+ EG + L L +S+ I+ + +K++N+E
Sbjct: 226 LNIEGCVEYLDLDKSEVLIDATRLKYLNLE 255
>At1g78760 hypothetical protein
Length = 452
Score = 68.6 bits (166), Expect = 4e-12
Identities = 76/318 (23%), Positives = 143/318 (44%), Gaps = 28/318 (8%)
Query: 4 GRKKHNDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSH 63
G +K +EDRLS+LP ++ +I+ + + VK+ +S RWR+LW+++P L + +
Sbjct: 5 GDEKRARRSEEDRLSNLPESLICQIMLNIPTKDVVKSSVLSKRWRNLWRYVPGLNVEYNQ 64
Query: 64 FMRIKSFNRFVSRILCL-RDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKLRVHA 122
F+ +F FV R L L R+ + + C + R + V + +KL+V
Sbjct: 65 FLDYNAFVSFVDRFLALDRESCFQSFRLRYDCD--EEERTISNVKRWINIVVDQKLKVLD 122
Query: 123 CCDFQ------HFSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFAF 176
D+ ++C +L SL LC + L PK ++LP + + L+ F
Sbjct: 123 VLDYTWGNEEVQIPPSVYTCESLVSLKLC------NVILPNPKVISLPLVKVIELDIVKF 176
Query: 177 CVGDDGCAEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVG---DPVYYSTVEL 233
+ S+ ++L++L+I +V D L +SS +L + +G D V +
Sbjct: 177 S-NALVLEKIISSCSALESLIISRSSVDDINVLRVSSRSLLSFKHIGNCSDGWDELEVAI 235
Query: 234 STPSLFTFDFVCYEGIPVL-----KLCRSKINLSSVKHVNIEVRMWTDYADASLVLLNWL 288
P L + + L +KIN+ N+E + ++ ++L
Sbjct: 236 DAPKLEYLNISDHSTAKFKMKNSGSLVEAKINII----FNMEELPHPNDRPKRKMIQDFL 291
Query: 289 AELANIKSLTLNHTALKV 306
AE++++K L ++ L+V
Sbjct: 292 AEISSVKKLFISSHTLEV 309
>At1g78750 hypothetical protein
Length = 458
Score = 67.4 bits (163), Expect = 9e-12
Identities = 78/269 (28%), Positives = 114/269 (41%), Gaps = 34/269 (12%)
Query: 2 KRGRKKHNDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLP--TLIL 59
KR R K + V D +S+LP +L ++L L + VK+ +SSRWR+LWK++P L
Sbjct: 7 KRVRAKGSREV--DWISNLPETLLCQVLFYLPTKDVVKSSVLSSRWRNLWKYVPGFNLSY 64
Query: 60 CSSHFMRIKSFN-----RFVSRILCLRDVS-TPLHAVDFLCRGVVDPR--LLKKVVTYAV 111
C H S++ RFV + S +++ G +P+ L+++ + V
Sbjct: 65 CDFHVRNTFSYDHNTFLRFVDSFMGFNSQSCLQSFRLEYDSSGYGEPKLALIRRWINSVV 124
Query: 112 SHNVEKLRV-HACCDFQHFS--SCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTT 168
S V+ L V CD F ++C TL L L L+ PK ++LP+L
Sbjct: 125 SRKVKYLGVLDDSCDNYEFEMPPTLYTCETLVYLTL------DGLSLASPKFVSLPSLKE 178
Query: 169 LSLESFAFCVGDDGCAEP-FSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPVY 227
L L F D E S L+NL I D E C+ S +L + T V D
Sbjct: 179 LHLSIVKF--ADHMALETLISQCPVLENLNINRSFCDDFEFTCVRSQSLLSFTHVAD--- 233
Query: 228 YSTVELSTPSLFTFDFVCYEGIPVLKLCR 256
T + D V P LK R
Sbjct: 234 -------TDEMLNEDLVVAIDAPKLKYLR 255
>At5g18780 putative protein
Length = 441
Score = 66.2 bits (160), Expect = 2e-11
Identities = 84/312 (26%), Positives = 134/312 (42%), Gaps = 42/312 (13%)
Query: 14 EDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIKSFNRF 73
EDR+S LP +L ILS L ++V+T +SSRWR LW +P L L S F F
Sbjct: 10 EDRISILPEPLLCHILSFLRTKDSVRTSVLSSRWRDLWLWVPRLDLDKSDFSDDNPSASF 69
Query: 74 VSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKLRVHACCDFQHF--SS 131
V + L R S RG +L Y +S L C QHF +
Sbjct: 70 VDKFLNFRGES--------YLRGF---KLNTDHDVYDISTLDACLMRLDKCKIQHFEIEN 118
Query: 132 CFFSCHTLTSLDLCVGPPRTDERLSFP-----KSLNLPALTTLSLESFAFCVGDDGCAEP 186
CF C L L + + +LSF SL+LP L + E F D AE
Sbjct: 119 CFGFCILLMPLIIPMCHTLVSLKLSFVILSKFGSLSLPCLEIMHFEKVIF--PSDKSAEV 176
Query: 187 FSALNS-LKNLMILYCNVLDAESLCISSVTLTNLTI-------VGDPVYYSTVELSTPSL 238
A + L++L I E L + S +L + T+ VG+ Y TV + TP L
Sbjct: 177 LIACSPVLRDLRISQSGDDAVEVLRVCSASLKSFTLKRTDHDYVGNGEY--TVVIDTPRL 234
Query: 239 FTFDFVCYEGIPVLKLCRSKINLSSVKHVNIEVRMWTDYADASL-----VLLNWLAELAN 293
+ Y+ C+ +S ++V + V + + ++ ++ ++L+ ++N
Sbjct: 235 EYLNLKDYQ-------CQGFKIVSMSEYVRVHVDVVFEVIGGTVLSKRNIICDFLSCVSN 287
Query: 294 IKSLTLNHTALK 305
++ +T++ +L+
Sbjct: 288 VRYMTISRRSLE 299
>At1g60400
Length = 403
Score = 65.9 bits (159), Expect = 3e-11
Identities = 76/318 (23%), Positives = 133/318 (40%), Gaps = 54/318 (16%)
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIKSFNRFV 74
DRLS LP +L RILS L ++V+T +S WR+LW H+P L L +S F F F+
Sbjct: 14 DRLSALPEHLLCRILSELSTKDSVRTSVLSKHWRNLWLHVPVLELETSDFPDNLVFREFI 73
Query: 75 SRILCLRDVSTPLHAVD--------------FLCRGVVDPRLLKKVVT---YAVSHNVEK 117
R + D L + D ++ VV R+ +VT Y V+ + K
Sbjct: 74 DRFVGF-DKEIDLKSFDIFYDVNVLWYDDFLWMIDDVVKRRVCDLMVTNNPYVVNEKLVK 132
Query: 118 LRV--HACCDFQHFSSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFA 175
+ + ++C + + F + + L S +C LP + TL L
Sbjct: 133 MPISLYSCATLVNLNLSFVAMNNLPSESVC-----------------LPRVKTLYLHGVK 175
Query: 176 FCVGDDGCAEPFSALNSLKNLMIL-----YCNVLDAESLCISSVTLTNLTIVGDPVYYST 230
GD S+ + L++L ++ Y V+ S + S T+ + DP
Sbjct: 176 L-DGDSILGTLVSSCSVLEDLTVVTHPGDYEKVVCFRSQSVKSFTIESQCEYKDP----N 230
Query: 231 VELSTPSLFTFDFVCYEGIPVLKLCRSKINLSSVKHVNIEVRMWTDYAD--ASLVLLNWL 288
VE+ P L +++C I V++++ +Y D A ++ N+L
Sbjct: 231 VEIDCPRL---EYMCIREYQSESFVVHSI--GPYAKVDVDIFFEVEYEDPLAISMIRNFL 285
Query: 289 AELANIKSLTLNHTALKV 306
++ ++ +T++ L+V
Sbjct: 286 TGISKVREMTISSRTLEV 303
>At3g59150 unknown protein
Length = 475
Score = 65.1 bits (157), Expect = 5e-11
Identities = 75/243 (30%), Positives = 107/243 (43%), Gaps = 18/243 (7%)
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIKS----- 69
D +S+LPN +L RILS L EA T +S RW +L +P L S ++ +
Sbjct: 13 DVISNLPNDLLCRILSYLSTKEAALTSILSKRWSNLLLSIPILDFDDSVLLKPQKGQRKN 72
Query: 70 --FNRFVSRILCLR-DVSTPLHAVDFLCR-GVVDPRLLKKVVTYAVSHNVEKLRVHACCD 125
F FV R+L R + S+ + V CR G V+P + K + V ++ L + C D
Sbjct: 73 VFFKAFVDRLLSQRVETSSHVQRVSLKCRQGGVEPDCVIKWILTTV-RDLGVLDLSLCID 131
Query: 126 FQHFSSCFFSCHTLTSLDLCVGPPRTDERLS-FPKSLNLPALTTLSLESFAFCVGDD-GC 183
F F F + T + L +G T RLS FP + P L +L L+ F GD
Sbjct: 132 FGIFHLPFNVFRSKTLVKLRIG---TMIRLSQFPSDVVSPTLNSLVLDLVEFRDGDKVEF 188
Query: 184 AEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGD---PVYYSTVELSTPSLFT 240
+ A SL++L + N + SS TL +L D V TPSL
Sbjct: 189 RQILLAFPSLQSLRVHESNKWKFWNGSASSRTLKSLVYRSDDDSSAPKPCVSFDTPSLVY 248
Query: 241 FDF 243
D+
Sbjct: 249 LDY 251
>At3g59210 unknown protein
Length = 484
Score = 64.3 bits (155), Expect = 8e-11
Identities = 80/316 (25%), Positives = 136/316 (42%), Gaps = 25/316 (7%)
Query: 10 DYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTL---ILCSSH--- 63
D +D ++ LP+ +L +ILS L EA T +S RWR+L+ +P L +L S H
Sbjct: 2 DDCSKDIINCLPDNLLCQILSNLSTKEAALTSLLSKRWRYLFALVPNLDFDVLPSLHPEV 61
Query: 64 ---FMRIKSFNRFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKLRV 120
SF FV R+L LR ++ C ++ + + + H V L +
Sbjct: 62 AMQDQDQTSFIDFVDRVLKLRG-KDHINKFSLKCGDGIEDEDVFPWILNTLRHGVSDLSL 120
Query: 121 HACCDFQHF-SSCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFAFCVG 179
H ++ S F+ TL L + P+ R+ +++ LP L TL+L+S F G
Sbjct: 121 HVSPSLVYWLPSKVFASKTLVRLKI---GPKDGPRVKL-RNVCLPKLKTLNLDSVVFEEG 176
Query: 180 DDGCAEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTI--VGDPVYYSTVELSTPS 237
G A+ S L+ L +L +S +SS L LT+ +V TP+
Sbjct: 177 KIGFAKLLSGCPVLEELSLLNLAWDRWDSCSVSSKILKRLTLYCAHSSRNPKSVSFDTPN 236
Query: 238 LFTFDFV-----CYEGIPVLKLCRSKINLSSVKHVNIEVRMWTDYADASLVLLN---WLA 289
+ F++ Y + L + I + K R +D + + ++ N L
Sbjct: 237 VVYFEYSDNIANKYPKVNFDSLVEASIGIRMTKVQKANARYVSDVDEETEMVGNATDLLM 296
Query: 290 ELANIKSLTLNHTALK 305
+ N+K+L L++ L+
Sbjct: 297 GICNVKTLYLSYDTLE 312
>At5g02930 putative protein
Length = 469
Score = 63.5 bits (153), Expect = 1e-10
Identities = 59/216 (27%), Positives = 95/216 (43%), Gaps = 29/216 (13%)
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIKSFNRFV 74
D +SDLP+ VL I S + + A++T +S RWRH+W P L F +K + +
Sbjct: 28 DSISDLPDAVLQHIFSYIPTELAIRTSVLSKRWRHVWSETPHL-----SFEWLKVSPKLI 82
Query: 75 SRILCLRDVS--TPLHAVDFLCRGV---VDPRLLKKVVTYAVSHNVEKLRV--HACCDFQ 127
++ L S H LC D + + +A+SHNV+ L + C F
Sbjct: 83 NKTLASYTASKIKSFH----LCTRYSYEADTHHVNSSIEFAMSHNVDDLSLAFRRCSPFY 138
Query: 128 HFSSCFFSCHTLTSLDLCVGP--PRTDERLSFPKSLNLPALTTLSLESFAFCVGDDGCAE 185
+F CF++ +L ++L PR + K+L+L T + D+ E
Sbjct: 139 NFDDCFYTNSSLKRVELRYVDLMPRCMVSWTSLKNLSLTDCT----------MSDESFLE 188
Query: 186 PFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTI 221
S L++L + +C L +L S+ LT L I
Sbjct: 189 ILSGCPILESLSLKFCMSLKYLNLS-KSLRLTRLEI 223
>At5g53630 heat shock transcription factor HSF30-like protein
Length = 426
Score = 63.2 bits (152), Expect = 2e-10
Identities = 63/217 (29%), Positives = 94/217 (43%), Gaps = 26/217 (11%)
Query: 17 LSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMRIKSFNRFVS- 75
+S LP+ ++ ILS L + V T +S+RWR LW LP L L S +F FN FVS
Sbjct: 2 ISQLPDPLICHILSHLPIKDLVTTRVLSTRWRSLWLWLPCLELNSLYF---PDFNAFVSF 58
Query: 76 --------RILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKLRVHACCDFQ 127
R+ C+ ++ D GV DP L + AV ++ L V C +
Sbjct: 59 GDKFFDSNRVSCINKFKLYIYGYDV---GVDDPSYLTSWIDAAVKCKIQHLHVQ-CLPAK 114
Query: 128 HFSS---CFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFAFCVGDDGCA 184
+ + C TL L LC L + ++LP L T+ LE + + +
Sbjct: 115 YIYEMPLSLYICETLVYLKLC------RVMLDDAEFVSLPCLKTIHLE-YVWFPNEANLE 167
Query: 185 EPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTI 221
S L+ L I C +A +L + S +L L+I
Sbjct: 168 RFVSCCPVLEELKIYGCGNENAITLRLLSRSLKKLSI 204
>At1g80470 hypothetical protein
Length = 464
Score = 62.8 bits (151), Expect = 2e-10
Identities = 75/314 (23%), Positives = 133/314 (41%), Gaps = 31/314 (9%)
Query: 7 KHNDYVKEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMR 66
K + + D +S L + +LL+ILS + E+V T +S RWR+LW+H+P L L SS F
Sbjct: 7 KKSKLTESDWISGLADDLLLQILSKVPTRESVFTSRMSKRWRNLWRHVPALDLDSSKFPH 66
Query: 67 IKSFNRFVSRILCLRDVSTPLHAVDFL--CRGVVDPRLLKKVVTYAVSHNVEKL----RV 120
F L D + + ++ DP + + + + V ++ L +V
Sbjct: 67 ESDLEDFFDSFLQF-DGNLKIERFKWIYNVEEHCDPEFVSR-IDHVVKRGLKDLTILSKV 124
Query: 121 HACCDFQHFSSCFFSCHTLTSLDL---CVGPPRTDERLSFPKSLNLPALTTLSLESFAFC 177
+ D +SC TL +L L PR + ++LP L T+ LE+ F
Sbjct: 125 NIEDDSVRMPVSLYSCATLVNLTLYSVVFDAPRA-------QLVSLPCLKTMHLEAVKF- 176
Query: 178 VGDDGCAEPFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVG-----DPVYYSTVE 232
G+ S+ + L L I+ + + + + S +L + D VE
Sbjct: 177 DGETILGTLISSCSFLDELTIITHDHDELGDVSVRSPSLRRFKLESMREDYDECEDPNVE 236
Query: 233 LSTPSLFTFDFVCYEGIPVLKLCRSKINLSSVKHVNIEVRMWTDYADASLVLLNWLAELA 292
+ TP L Y+ + N+S VNI+V D D V+ +++ ++
Sbjct: 237 VDTPGLEYMSITDYQSESFI-----IHNISPCAKVNIDVVF--DAEDEDSVIHDFITAIS 289
Query: 293 NIKSLTLNHTALKV 306
++ LT++ L++
Sbjct: 290 TVRELTISARTLEM 303
>At5g38590 unknown protein
Length = 410
Score = 62.0 bits (149), Expect = 4e-10
Identities = 62/229 (27%), Positives = 106/229 (46%), Gaps = 12/229 (5%)
Query: 15 DRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSSHFMR--IKSFNR 72
D+++ LP+ +L++ILS + D AV T +S RW LW LP L S +
Sbjct: 2 DKINGLPDDLLVKILSYVPTDIAVSTSILSKRWEFLWMWLPNLDYTSRWCRKPGDVGLRD 61
Query: 73 FVSRILCLRDVSTPLHAVDFLCRGV-VDPRLLKKVVTYAVSHNVEKLRV-HACCDFQHFS 130
F+ + L L + ++ F + P +++ + AVS +V L + H + F
Sbjct: 62 FIHKNLPLHRAPV-IESLRFHSNSPDIKPEDIRRWIEIAVSRHVHDLDIDHFSENENIFL 120
Query: 131 SCFFSCHTLTSLDLCVGPPRTDERLSFPKSLNLPALTTLSLESFAFCVGDDGCAEPFSAL 190
S FF+C +L +L L R+ P + LP+L TL L++ +F G E S
Sbjct: 121 SSFFACKSLVTLKL-----RSVTLRDIPSMVCLPSLKTLLLDNVSFVEG-KSLQELLSIC 174
Query: 191 NSLKNLMILYCNVLDAESLCISSVTLTNLTI-VGDPVYYSTVELSTPSL 238
L++L + + + + L I +L +L++ + D + TPSL
Sbjct: 175 PVLEDLSVYCDDYENTKELTIVVPSLLSLSLYIPDEWLLDGYWIDTPSL 223
>At3g49020 putative protein
Length = 447
Score = 62.0 bits (149), Expect = 4e-10
Identities = 74/254 (29%), Positives = 111/254 (43%), Gaps = 27/254 (10%)
Query: 13 KEDRLSDLPNCVLLRILSLLDADEAVKTCTISSRWRHLWKHLPTLILCSS-HFMRIKSFN 71
+EDR+S+LP +LL ILS L + + T +S +W+ LWK +P L S H+ S N
Sbjct: 20 EEDRISELPEALLLLILSSLPTETVIATSVLSKQWKSLWKLVPNLKFDSKYHYHYTFSDN 79
Query: 72 RFVSRILCLRDVSTPLHAVDFLCRGVVDPRLLKKVVTYAVSHNVEKLRVHACC---DFQH 128
F S I V LH +D +L + A S ++ K + F
Sbjct: 80 VFRSLISHKAPVLDSLHLKVEDKDDALDVGIL---IGIAFSRHMRKFVLKITLLEESFVR 136
Query: 129 FSSCFFSCH-TLTSLDL--CVGPPRTDERLSFPKSLNLPALTTLSLESFAFCVGDDGCAE 185
F S + SC+ TL L+L C+ L FP + L +L L L F + C
Sbjct: 137 FPSAWCSCNDTLEILELKYCI-------LLDFPSLVCLKSLRKLYLYHVRFNDEESVC-N 188
Query: 186 PFSALNSLKNLMILYCNVLDAESLCISSVTLTNLTIVGDPVYYST----VELSTPSLFTF 241
SL++L++ + D ES I+ +L LTI D YY ++ PSL
Sbjct: 189 LLCGCPSLEDLVVHRHSTTDVESYTIAVPSLQRLTIYDD--YYGEGVGGYVINAPSL--- 243
Query: 242 DFVCYEGIPVLKLC 255
++ +G L+ C
Sbjct: 244 KYLNIDGFNGLEFC 257
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.138 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,880,873
Number of Sequences: 26719
Number of extensions: 279641
Number of successful extensions: 1071
Number of sequences better than 10.0: 180
Number of HSP's better than 10.0 without gapping: 159
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 860
Number of HSP's gapped (non-prelim): 204
length of query: 307
length of database: 11,318,596
effective HSP length: 99
effective length of query: 208
effective length of database: 8,673,415
effective search space: 1804070320
effective search space used: 1804070320
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147179.8


