

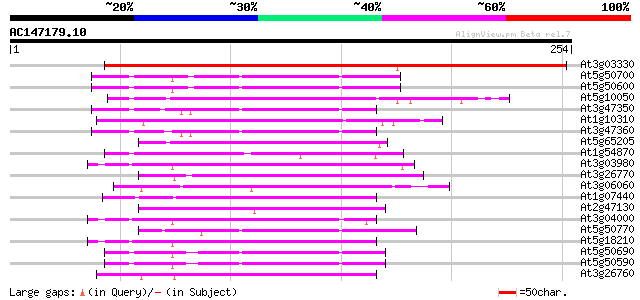
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147179.10 - phase: 1 /pseudo
(254 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g03330 unknown protein 307 4e-84
At5g50700 11-beta-hydroxysteroid dehydrogenase-like 68 5e-12
At5g50600 11-beta-hydroxysteroid dehydrogenase-like 68 5e-12
At5g10050 unknown protein 65 5e-11
At3g47350 putative protein 62 4e-10
At1g10310 unknown protein 61 7e-10
At3g47360 putative protein 60 9e-10
At5g65205 unknown protein 60 1e-09
At1g54870 putative protein (At1g54870) 59 3e-09
At3g03980 putative short-chain type dehydrogenase/reductase 59 3e-09
At3g26770 short chain alcohol dehydrogenase like protein 57 1e-08
At3g06060 unknown protein 57 1e-08
At1g07440 tropinone reductase-I, putative 57 1e-08
At2g47130 putative alcohol dehydrogenase 55 3e-08
At3g04000 short-chain type dehydrogenase/reductase like protein 54 6e-08
At5g50770 putative protein 54 8e-08
At5g18210 Brn1-like protein 54 8e-08
At5g50690 11-beta-hydroxysteroid dehydrogenase-like 54 1e-07
At5g50590 11-beta-hydroxysteroid dehydrogenase-like 54 1e-07
At3g26760 putative short chain alcohol dehydrogenase 53 2e-07
>At3g03330 unknown protein
Length = 345
Score = 307 bits (786), Expect = 4e-84
Identities = 150/231 (64%), Positives = 182/231 (77%), Gaps = 22/231 (9%)
Query: 44 RKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTP 103
+ VV++A SLFP +GVDY++HNAA+ERPKS D +EE+LK TFDVNVFGTI+LT+L+ P
Sbjct: 111 KHVVEQAVSLFPGAGVDYLVHNAAFERPKSKASDASEETLKTTFDVNVFGTISLTKLVAP 170
Query: 104 FMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGP 163
ML++G GHFVV+SSAAGK P+PGQA+YSASK+AL+GYFHSLRSE CQKGI+VTVVCPGP
Sbjct: 171 HMLKQGGGHFVVISSAAGKVPSPGQAIYSASKHALHGYFHSLRSEFCQKGIKVTVVCPGP 230
Query: 164 IETANNSGSQV----------------------PSEKRVSAEKCVELTIIAATHGLKEAW 201
IET N +G+ ++KRVS+E+C ELTIIAA+H LKEAW
Sbjct: 231 IETWNGTGTSTSEDSKSPELLAQLDFSQASILFATQKRVSSERCAELTIIAASHNLKEAW 290
Query: 202 ISYQPVLAVMYLVQYMPTIGYWLMDKVGKNRVEAAKEKGNAYSLSLLFGKK 252
ISYQPVL VMYLVQYMP++G+WLMDKVG RVE A++KGN YS LLFGKK
Sbjct: 291 ISYQPVLLVMYLVQYMPSLGFWLMDKVGGKRVEVAEKKGNTYSWDLLFGKK 341
>At5g50700 11-beta-hydroxysteroid dehydrogenase-like
Length = 349
Score = 67.8 bits (164), Expect = 5e-12
Identities = 47/142 (33%), Positives = 75/142 (52%), Gaps = 8/142 (5%)
Query: 38 SKVTAKRKVVDEAESLFPDSGVDYMIHNAAYERPK--SSVLDVTEESLKATFDVNVFGTI 95
SK R++VD+ + F +D++++NA + ++ D+T KA D N +G++
Sbjct: 107 SKPDDCRRIVDDTITHF--GRLDHLVNNAGMTQISMFENIEDITRT--KAVLDTNFWGSV 162
Query: 96 TLTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQ 155
TR P+ LR+ G V MSS+A AP + Y+ASK AL +F ++R EL +
Sbjct: 163 YTTRAALPY-LRQSNGKIVAMSSSAAWLTAPRMSFYNASKAALLSFFETMRIEL-GGDVH 220
Query: 156 VTVVCPGPIETANNSGSQVPSE 177
+T+V PG IE+ G E
Sbjct: 221 ITIVTPGYIESELTQGKYFSGE 242
>At5g50600 11-beta-hydroxysteroid dehydrogenase-like
Length = 349
Score = 67.8 bits (164), Expect = 5e-12
Identities = 47/142 (33%), Positives = 75/142 (52%), Gaps = 8/142 (5%)
Query: 38 SKVTAKRKVVDEAESLFPDSGVDYMIHNAAYERPK--SSVLDVTEESLKATFDVNVFGTI 95
SK R++VD+ + F +D++++NA + ++ D+T KA D N +G++
Sbjct: 107 SKPDDCRRIVDDTITHF--GRLDHLVNNAGMTQISMFENIEDITRT--KAVLDTNFWGSV 162
Query: 96 TLTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQ 155
TR P+ LR+ G V MSS+A AP + Y+ASK AL +F ++R EL +
Sbjct: 163 YTTRAALPY-LRQSNGKIVAMSSSAAWLTAPRMSFYNASKAALLSFFETMRIEL-GGDVH 220
Query: 156 VTVVCPGPIETANNSGSQVPSE 177
+T+V PG IE+ G E
Sbjct: 221 ITIVTPGYIESELTQGKYFSGE 242
>At5g10050 unknown protein
Length = 279
Score = 64.7 bits (156), Expect = 5e-11
Identities = 61/211 (28%), Positives = 93/211 (43%), Gaps = 39/211 (18%)
Query: 45 KVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPF 104
KV+ E F +D +++NA + + + +++ TF+ NVFG++ +T+ + P
Sbjct: 69 KVLSEVIDKF--GKIDVLVNNAGVQCV-GPLAETPISAMENTFNTNVFGSMRMTQAVVPH 125
Query: 105 MLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPI 164
M+ + KG V + S P P VY+A+K A++ +LR EL GI V V PG I
Sbjct: 126 MVSKKKGKIVNVGSITVMAPGPWAGVYTATKAAIHALTDTLRLELRPFGIDVINVVPGGI 185
Query: 165 ETANNSGSQV-------------PSEKRV-------------SAEKCVELTIIAATHGLK 198
T N + S V P E+ + AE T+ A
Sbjct: 186 RT-NIANSAVATFNKMPELKLYKPYEEAIRERAFISQRMNPTPAETFARDTVAAVLKKNP 244
Query: 199 EAWIS---YQPVLAVMYLVQYMPTIGYWLMD 226
AW S Y ++AVMY +MP WL D
Sbjct: 245 PAWFSSGRYSTLMAVMY---HMP---LWLKD 269
>At3g47350 putative protein
Length = 308
Score = 61.6 bits (148), Expect = 4e-10
Identities = 46/131 (35%), Positives = 71/131 (54%), Gaps = 8/131 (6%)
Query: 38 SKVTAKRKVVDEAESLFPDSGVDYMIHNAAYERPKSSVL-DVTE-ESLKATFDVNVFGTI 95
S V +K +DE F +D++I+NA P++ + D T+ + + D+N +G+
Sbjct: 106 SNVEDCKKFIDETIHHF--GKLDHLINNAGV--PQTVIFEDFTQIQDANSIMDINFWGST 161
Query: 96 TLTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQ 155
+T P LR+ KG VV+SSA P +VYSASK AL +F +LR E+ I+
Sbjct: 162 YITYFAIPH-LRKSKGKIVVISSATAIIPLQAASVYSASKAALVKFFETLRVEI-SPDIK 219
Query: 156 VTVVCPGPIET 166
+T+ PG I T
Sbjct: 220 ITIALPGFIST 230
>At1g10310 unknown protein
Length = 242
Score = 60.8 bits (146), Expect = 7e-10
Identities = 51/166 (30%), Positives = 76/166 (45%), Gaps = 11/166 (6%)
Query: 40 VTAKRKVVDEAESLFPDSGV-DYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLT 98
V + V + A ++ GV D +++NA S + +V+ E D NV G +
Sbjct: 74 VKSNSSVEEMAHTIVEKKGVPDIIVNNAGTINKNSKIWEVSAEDFDNVMDTNVKGVANVL 133
Query: 99 RLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTV 158
R P ML R +G V MSS G++ A A Y ASK+A+ G ++ E+ + G+ V
Sbjct: 134 RHFIPLMLPRKQGIIVNMSSGWGRSGAALVAPYCASKWAIEGLSRAVAKEVVE-GMAVVA 192
Query: 159 VCPGPIETA------NNSGS--QVPSEKRVSAEKCVELTIIAATHG 196
+ PG I T NS S Q P V A + L + A +G
Sbjct: 193 LNPGVINTELLTSCFGNSASLYQAPDAWAVKAATMI-LNLTAGDNG 237
>At3g47360 putative protein
Length = 309
Score = 60.5 bits (145), Expect = 9e-10
Identities = 45/132 (34%), Positives = 71/132 (53%), Gaps = 10/132 (7%)
Query: 38 SKVTAKRKVVDEAESLFPDSGVDYMIHNAAYERPKSSVL--DVTE-ESLKATFDVNVFGT 94
S V +K +DE F +D++I+NA + +VL D T+ + D+N +GT
Sbjct: 107 SNVEDCKKFIDETIRHF--GKLDHLINNAGVFQ---TVLFEDFTQIQDANPIMDINFWGT 161
Query: 95 ITLTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGI 154
+T P LR+ KG V ++S + P P ++Y+ASK AL +F +LR EL I
Sbjct: 162 TYITYFAIPH-LRKSKGKIVAITSGSANIPLPLASIYAASKAALLRFFETLRIEL-SPDI 219
Query: 155 QVTVVCPGPIET 166
++T+V PG + T
Sbjct: 220 KITIVLPGVVST 231
>At5g65205 unknown protein
Length = 285
Score = 60.1 bits (144), Expect = 1e-09
Identities = 37/114 (32%), Positives = 62/114 (53%), Gaps = 2/114 (1%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +++NA + + ++ ++ TF+ NV G++ +T+ + P M + KG V + S
Sbjct: 82 IDVLVNNAGVQCI-GPLAEIPISAMDYTFNTNVLGSMRMTQAVVPHMASKKKGKIVNIGS 140
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET-ANNSG 171
+ P P VY+ASK AL+ +LR EL GI V + PG I++ NSG
Sbjct: 141 ISIMAPGPWAGVYTASKAALHALTDTLRLELKPFGIDVINIVPGGIQSNIANSG 194
>At1g54870 putative protein (At1g54870)
Length = 288
Score = 58.9 bits (141), Expect = 3e-09
Identities = 42/147 (28%), Positives = 72/147 (48%), Gaps = 17/147 (11%)
Query: 44 RKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTP 103
++VVDE + F +D +I+NAA + S++ ++ E L+ F N+F LTR
Sbjct: 109 KRVVDEVVNAF--GRIDVLINNAAEQYESSTIEEIDEPRLERVFRTNIFSYFFLTRHALK 166
Query: 104 FMLRRGKGHFVVMSSAAGKTPAPGQAV-YSASKYALNGYFHSLRSELCQKGIQVTVVCPG 162
M +G ++ +++ + Y+A+K A+ + L +L +KGI+V V PG
Sbjct: 167 HMK---EGSSIINTTSVNAYKGNASLLDYTATKGAIVAFTRGLALQLAEKGIRVNGVAPG 223
Query: 163 PI-----------ETANNSGSQVPSEK 178
PI E N GS+VP ++
Sbjct: 224 PIWTPLIPASFNEEKIKNFGSEVPMKR 250
>At3g03980 putative short-chain type dehydrogenase/reductase
Length = 270
Score = 58.5 bits (140), Expect = 3e-09
Identities = 43/150 (28%), Positives = 74/150 (48%), Gaps = 6/150 (4%)
Query: 36 KPSKVTAKRKVVDEAESLFPDSGVDYMIHNAAYERPK-SSVLDVTEESLKATFDVNVFGT 94
+PS+V + + D AES F ++ V ++++A PK ++ D + E TF VN G
Sbjct: 86 EPSQV---KSMFDAAESAF-EAPVHILVNSAGILDPKYPTIADTSVEDFDHTFSVNTKGA 141
Query: 95 ITLTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGI 154
++ + + G G ++++S+ ++ PG Y+ASK A+ L EL GI
Sbjct: 142 FLCSKEAANRLKQGGGGRIILLTSSQTRSLKPGFGAYAASKAAVETMVKILAKELKGTGI 201
Query: 155 QVTVVCPGPIETANNSGSQVPS-EKRVSAE 183
V PGPI T + P ++++AE
Sbjct: 202 TANCVAPGPIATEMFFDGKTPELVEKIAAE 231
>At3g26770 short chain alcohol dehydrogenase like protein
Length = 306
Score = 57.0 bits (136), Expect = 1e-08
Identities = 40/133 (30%), Positives = 60/133 (45%), Gaps = 6/133 (4%)
Query: 59 VDYMIHNAAYERPKS----SVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFV 114
+D M +NA P + S LD+TE + +NVFG ++ + FM+ G +
Sbjct: 118 LDVMYNNAGIVGPMTPASISQLDMTE--FERVMRINVFGVVSGIKHAAKFMIPARSGCIL 175
Query: 115 VMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIETANNSGSQV 174
SS AG T Y+ SK+ G S SELC+ G+++ + PG + T
Sbjct: 176 CTSSVAGVTGGLAPHSYTISKFTTPGIVKSAASELCEHGVRINCISPGTVATPLTLSYLQ 235
Query: 175 PSEKRVSAEKCVE 187
+VS EK E
Sbjct: 236 KVFPKVSEEKLRE 248
>At3g06060 unknown protein
Length = 326
Score = 56.6 bits (135), Expect = 1e-08
Identities = 49/156 (31%), Positives = 69/156 (43%), Gaps = 13/156 (8%)
Query: 48 DEAESLFPDSG-VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFML 106
D +SG +D +I N K V + E +K T DVN+ G+ + + P M
Sbjct: 102 DAVSKAIDESGPIDVLIVNQGVFTAKELVKH-SPEDVKFTIDVNLVGSFNVIKAALPAMK 160
Query: 107 RR---GKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGP 163
R G ++SS AG+ G A YSASK+ L G +L+ E+ I VT++ P
Sbjct: 161 ARKDRGPASISLVSSQAGQVGVYGYAAYSASKFGLQGLAQALQQEVISDDIHVTLIFPPD 220
Query: 164 IETANNSGSQVPSEKRVSAEKCVELTIIAATHGLKE 199
T Q S V+A IIAA+ G E
Sbjct: 221 TNTPGFEEEQ-KSRPEVTA-------IIAASSGSME 248
>At1g07440 tropinone reductase-I, putative
Length = 266
Score = 56.6 bits (135), Expect = 1e-08
Identities = 39/124 (31%), Positives = 58/124 (46%), Gaps = 2/124 (1%)
Query: 43 KRKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLT 102
+ K++ S+F +D +I+N R K + LD T E N+ L++L
Sbjct: 78 REKLMQTVSSMFGGK-LDILINNLGAIRSKPT-LDYTAEDFSFHISTNLESAYHLSQLAH 135
Query: 103 PFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPG 162
P + G G+ + MSS AG A ++YSA+K ALN +L E GI+ V P
Sbjct: 136 PLLKASGCGNIIFMSSIAGVVSASVGSIYSATKGALNQLARNLACEWASDGIRANAVAPA 195
Query: 163 PIET 166
I T
Sbjct: 196 VIAT 199
>At2g47130 putative alcohol dehydrogenase
Length = 257
Score = 55.5 bits (132), Expect = 3e-08
Identities = 38/113 (33%), Positives = 52/113 (45%), Gaps = 1/113 (0%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRG-KGHFVVMS 117
+D + NA S LD+ E T VNV G + M+ +G +G V +
Sbjct: 84 LDVLFSNAGVMEQPGSFLDLNLEQFDRTMAVNVRGAAAFIKHAARAMVEKGTRGSIVCTT 143
Query: 118 SAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIETANNS 170
S A + PG Y+ASK+AL G S L + GI+V V P + TA NS
Sbjct: 144 SVASEIGGPGPHAYTASKHALLGLVKSACGGLGKYGIRVNGVAPYAVATAINS 196
>At3g04000 short-chain type dehydrogenase/reductase like protein
Length = 272
Score = 54.3 bits (129), Expect = 6e-08
Identities = 42/134 (31%), Positives = 68/134 (50%), Gaps = 9/134 (6%)
Query: 36 KPSKVTAKRKVVDEAESLFPDSGVDYMIHNAAYERPK-SSVLDVTEESLKATFDVNVFGT 94
+PS+V + + DEAE +F +S V ++++AA P S++ D++ E VN G
Sbjct: 87 EPSQV---KSLFDEAERVF-ESPVHILVNSAAIADPNHSTISDMSVELFDRIISVNTRGA 142
Query: 95 ITLTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGI 154
R + R G G +++S++ +T Y+ASK A+ L EL KG
Sbjct: 143 FICAREAANRLKRGGGGRIILLSTSLVQTLNTNYGSYTASKAAVEAMAKILAKEL--KGT 200
Query: 155 QVTVVC--PGPIET 166
++TV C PGP+ T
Sbjct: 201 EITVNCVSPGPVAT 214
>At5g50770 putative protein
Length = 342
Score = 53.9 bits (128), Expect = 8e-08
Identities = 34/127 (26%), Positives = 69/127 (53%), Gaps = 4/127 (3%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKA-TFDVNVFGTITLTRLLTPFMLRRGKGHFVVMS 117
+D+++ NA P D+ + S + D+N +G++ T +P+ L++ +G VV++
Sbjct: 126 LDHLVTNAGVA-PLYFFADIEDVSKASPAMDINFWGSVYCTFFASPY-LKKFRGRIVVIA 183
Query: 118 SAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIETANNSGSQVPSE 177
S G +P + Y ASK A+ ++ +LR+E I VT+V PG +++ + G + +
Sbjct: 184 SGCGYIASPRLSFYCASKAAVIAFYETLRTEF-GSDIGVTIVAPGIVDSEMSRGKFMTKD 242
Query: 178 KRVSAEK 184
++ +K
Sbjct: 243 GKLVVDK 249
>At5g18210 Brn1-like protein
Length = 261
Score = 53.9 bits (128), Expect = 8e-08
Identities = 37/132 (28%), Positives = 62/132 (46%), Gaps = 5/132 (3%)
Query: 36 KPSKVTAKRKVVDEAESLFPDSGVDYMIHNAAYERPK-SSVLDVTEESLKATFDVNVFGT 94
+PS++ + + D AE F +S V ++++A P ++ + E F VN G+
Sbjct: 76 EPSQI---KSLFDAAEKAF-NSPVHILVNSAGILNPNYPTIANTPIEEFDRIFKVNTRGS 131
Query: 95 ITLTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGI 154
+ + R G G ++++S+ + PGQ Y+ASK A+ L EL GI
Sbjct: 132 FLCCKEAAKRLKRGGGGRIILLTSSLTEALIPGQGAYTASKAAVEAMVKILAKELKGLGI 191
Query: 155 QVTVVCPGPIET 166
V PGP+ T
Sbjct: 192 TANCVSPGPVAT 203
>At5g50690 11-beta-hydroxysteroid dehydrogenase-like
Length = 303
Score = 53.5 bits (127), Expect = 1e-07
Identities = 39/132 (29%), Positives = 66/132 (49%), Gaps = 14/132 (10%)
Query: 44 RKVVDEAESLFPDSGVDYMIHNAAYERPK-----SSVLDVTEESLKATFDVNVFGTITLT 98
++ V E S F +D++++NA K S + DV + N +G + T
Sbjct: 113 KRFVQETISRF--GRLDHLVNNAGIAEAKFFEDYSEISDVLP-----IVNTNFWGPVYAT 165
Query: 99 RLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTV 158
P L++ KG + ++S AG + P ++Y+ASK A+ ++ +LR EL + VT+
Sbjct: 166 HFAIPH-LKKTKGKIIAVASPAGWSGVPRMSIYAASKAAMINFYETLRIEL-HPEVGVTI 223
Query: 159 VCPGPIETANNS 170
V PG IE N +
Sbjct: 224 VFPGLIENGNTN 235
>At5g50590 11-beta-hydroxysteroid dehydrogenase-like
Length = 299
Score = 53.5 bits (127), Expect = 1e-07
Identities = 39/132 (29%), Positives = 66/132 (49%), Gaps = 14/132 (10%)
Query: 44 RKVVDEAESLFPDSGVDYMIHNAAYERPK-----SSVLDVTEESLKATFDVNVFGTITLT 98
++ V E S F +D++++NA K S + DV + N +G + T
Sbjct: 113 KRFVQETISRF--GRLDHLVNNAGIAEAKFFEDYSEISDVLP-----IVNTNFWGPVYAT 165
Query: 99 RLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTV 158
P L++ KG + ++S AG + P ++Y+ASK A+ ++ +LR EL + VT+
Sbjct: 166 HFAIPH-LKKTKGKIIAVASPAGWSGVPRMSIYAASKAAMINFYETLRIEL-HPEVGVTI 223
Query: 159 VCPGPIETANNS 170
V PG IE N +
Sbjct: 224 VFPGLIENGNTN 235
>At3g26760 putative short chain alcohol dehydrogenase
Length = 269
Score = 52.8 bits (125), Expect = 2e-07
Identities = 34/130 (26%), Positives = 62/130 (47%), Gaps = 3/130 (2%)
Query: 40 VTAKRKVVDEAESLFPDSG-VDYMIHNAAYERPKS--SVLDVTEESLKATFDVNVFGTIT 96
VT + ++ E+ G +D M+++A S S+ D+ ++ +NV GT+
Sbjct: 62 VTEEEQIAKAVETAVTRHGKLDVMLNSAGISCSISPPSIADLDMDTYDKVMRLNVRGTVL 121
Query: 97 LTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQV 156
+ M+ G G + +SS +G G YS SK+ + G ++ SELC+ G+++
Sbjct: 122 GIKHAARAMIPAGSGSILCLSSISGLMGGLGPHAYSISKFTIPGVVKTVASELCKHGLRI 181
Query: 157 TVVCPGPIET 166
+ P I T
Sbjct: 182 NCISPAGIPT 191
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.134 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,526,056
Number of Sequences: 26719
Number of extensions: 210588
Number of successful extensions: 553
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 54
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 478
Number of HSP's gapped (non-prelim): 66
length of query: 254
length of database: 11,318,596
effective HSP length: 97
effective length of query: 157
effective length of database: 8,726,853
effective search space: 1370115921
effective search space used: 1370115921
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC147179.10


