

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147178.8 + phase: 0
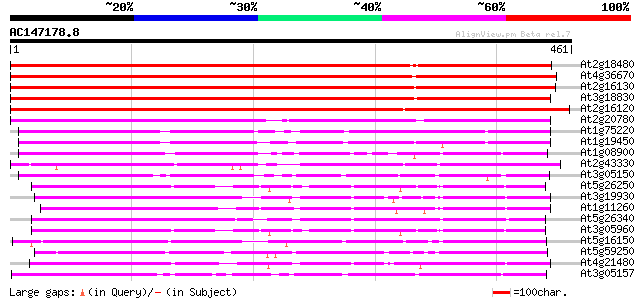
(461 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g18480 putative sugar transporter 481 e-136
At4g36670 sugar transporter like protein 467 e-132
At2g16130 putative sugar transporter 434 e-122
At3g18830 sugar transporter like protein 431 e-121
At2g16120 putative sugar transporter 431 e-121
At2g20780 putative sugar transporter 322 3e-88
At1g75220 integral membrane protein, putative 182 3e-46
At1g19450 integral membrane protein, putative 177 1e-44
At1g08900 Beta integral like membrane protein 174 7e-44
At2g43330 membrane transporter like protein 173 2e-43
At3g05150 putative sugar transporter 171 8e-43
At5g26250 hexose transporter - like protein 170 2e-42
At3g19930 monosaccharide transport protein, STP4 169 2e-42
At1g11260 glucose transporter 169 2e-42
At5g26340 hexose transporter - like protein 167 1e-41
At3g05960 putative hexose transporter 165 4e-41
At5g16150 sugar transporter like protein 163 2e-40
At5g59250 D-xylose-H+ symporter - like protein 163 2e-40
At4g21480 glucose transporter 160 1e-39
At3g05157 sugar transporter like protein 160 2e-39
>At2g18480 putative sugar transporter
Length = 508
Score = 481 bits (1239), Expect = e-136
Identities = 234/445 (52%), Positives = 324/445 (72%), Gaps = 3/445 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA +FI+++L+I+D Q+++LAGILN CAL + AG+ SD IGRRYTI LS++ F +G
Sbjct: 43 MSGAQIFIRDDLKINDTQIEVLAGILNLCALVGSLTAGKTSDVIGRRYTIALSAVIFLVG 102
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
S+LMGYG ++P+LM+GRCIAG GVGFAL+I VYSAEISS S+RGFLTSLP+L I++G L
Sbjct: 103 SVLMGYGPNYPVLMVGRCIAGVGVGFALMIAPVYSAEISSASHRGFLTSLPELCISLGIL 162
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SNY GKL+L+LGWR+ML I + PS+ L + ++ ESPRWLVMQGRL +AKK+++
Sbjct: 163 LGYVSNYCFGKLTLKLGWRLMLGIAAFPSLILAFGITRMPESPRWLVMQGRLEEAKKIMV 222
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
L+SN+++EAE+R ++I A +D + + KK G +E+ KP P V ILI
Sbjct: 223 LVSNTEEEAEERFRDILTAAEVDVTEIKEVGGGVKKKNHGKSVWRELVIKPRPAVRLILI 282
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
AA+G+H F++ G+E + LYSPRIF + G+ K LLLATVG+G+++ F +++ FLLDK
Sbjct: 283 AAVGIHFFEHATGIEAVVLYSPRIFKKAGVVSKDKLLLATVGVGLTKAFFIIIATFLLDK 342
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
+GRR LLL S+GG++F++ L V +V+ + G WA+ +I+ Y F +IG+G
Sbjct: 343 VGRRKLLLTSTGGMVFALTSLAVSLTMVQ--RFG-RLAWALSLSIVSTYAFVAFFSIGLG 399
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+TWVYS+EIFPLRLRAQG + V +NRI N V SF+S+ K IT GG FF+F G V
Sbjct: 400 PITWVYSSEIFPLRLRAQGASIGVAVNRIMNATVSMSFLSMTKAITTGGVFFVFAGIAVA 459
Query: 421 GWWFYYSFLPETKGRSLEDMETIFG 445
WWF++ LPETKG LE+ME +FG
Sbjct: 460 AWWFFFFMLPETKGLPLEEMEKLFG 484
>At4g36670 sugar transporter like protein
Length = 493
Score = 467 bits (1201), Expect = e-132
Identities = 221/449 (49%), Positives = 318/449 (70%), Gaps = 2/449 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA++FI+E+L+ +D+Q+++L GILN CAL ++AGR SD IGRRYTI+L+SI F LG
Sbjct: 38 MSGAMVFIEEDLKTNDVQIEVLTGILNLCALVGSLLAGRTSDIIGRRYTIVLASILFMLG 97
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
SILMG+G ++P+L+ GRC AG GVGFAL++ VYSAEI++ S+RG L SLP L I+IG L
Sbjct: 98 SILMGWGPNYPVLLSGRCTAGLGVGFALMVAPVYSAEIATASHRGLLASLPHLCISIGIL 157
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+ NYF KL + +GWR+ML I ++PS+ L +L++ ESPRWL+MQGRL + K++L
Sbjct: 158 LGYIVNYFFSKLPMHIGWRLMLGIAAVPSLVLAFGILKMPESPRWLIMQGRLKEGKEILE 217
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
L+SNS +EAE R ++IK A GID C ++V + K G G KE+ +P+P V R+L+
Sbjct: 218 LVSNSPEEAELRFQDIKAAAGIDPKCVDDVVKMEGKKTHGEGVWKELILRPTPAVRRVLL 277
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
A+G+H FQ+ G+E + LY PRIF + GIT K L L T+G+GI +T F + LLDK
Sbjct: 278 TALGIHFFQHASGIEAVLLYGPRIFKKAGITTKDKLFLVTIGVGIMKTTFIFTATLLLDK 337
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
+GRR LLL S GG++ ++ L + +N+ G + WA++ +I+ Y F +IG+G
Sbjct: 338 VGRRKLLLTSVGGMVIALTMLGFGLTMAQNA--GGKLAWALVLSIVAAYSFVAFFSIGLG 395
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+TWVYS+E+FPL+LRAQG + V +NR+ N V SF+S+ IT GG FF+F G +
Sbjct: 396 PITWVYSSEVFPLKLRAQGASLGVAVNRVMNATVSMSFLSLTSAITTGGAFFMFAGVAAV 455
Query: 421 GWWFYYSFLPETKGRSLEDMETIFGKNSN 449
W F++ LPETKG+SLE++E +F ++ +
Sbjct: 456 AWNFFFFLLPETKGKSLEEIEALFQRDGD 484
>At2g16130 putative sugar transporter
Length = 511
Score = 434 bits (1115), Expect = e-122
Identities = 210/448 (46%), Positives = 309/448 (68%), Gaps = 1/448 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA +FIK++L++SD+Q+++L GILN +L AGR SD+IGRRYTI+L+ FFF G
Sbjct: 47 MSGAAIFIKDDLKLSDVQLEILMGILNIYSLIGSGAAGRTSDWIGRRYTIVLAGFFFFCG 106
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
++LMG+ +++P +M+GR +AG GVG+A++I VY+ E++ S RGFL+S P++ INIG L
Sbjct: 107 ALLMGFATNYPFIMVGRFVAGIGVGYAMMIAPVYTTEVAPASSRGFLSSFPEIFINIGIL 166
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SNYF KL +GWR ML I ++PS+ L I +L + ESPRWLVMQGRLGDA KVL
Sbjct: 167 LGYVSNYFFAKLPEHIGWRFMLGIGAVPSVFLAIGVLAMPESPRWLVMQGRLGDAFKVLD 226
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
SN+K+EA R+ +IK AVGI ++ T +++ V K +G G K++ +P+P V ILI
Sbjct: 227 KTSNTKEEAISRLNDIKRAVGIPDDMTDDVIVVPNKKSAGKGVWKDLLVRPTPSVRHILI 286
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
A +G+H Q G++ + LYSP IF R G+ K LLATV +G+ +TLF ++ L+D+
Sbjct: 287 ACLGIHFSQQASGIDAVVLYSPTIFSRAGLKSKNDQLLATVAVGVVKTLFIVVGTCLVDR 346
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
GRR LLL S GG+ FS+ L +++ + G+ WAI + + ++G G
Sbjct: 347 FGRRALLLTSMGGMFFSLTALGTSLTVIDRNP-GQTLKWAIGLAVTTVMTFVATFSLGAG 405
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
VTWVY++EIFP+RLRAQG + V++NR+ + + +F+S+ K +T+GG F LF G V
Sbjct: 406 PVTWVYASEIFPVRLRAQGASLGVMLNRLMSGIIGMTFLSLSKGLTIGGAFLLFAGVAVA 465
Query: 421 GWWFYYSFLPETKGRSLEDMETIFGKNS 448
W F+++FLPET+G LE++E++FG S
Sbjct: 466 AWVFFFTFLPETRGVPLEEIESLFGSYS 493
>At3g18830 sugar transporter like protein
Length = 539
Score = 431 bits (1108), Expect = e-121
Identities = 213/444 (47%), Positives = 307/444 (68%), Gaps = 2/444 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA+++IK +L+I+D+Q+ +LAG LN +L AGR SD+IGRRYTI+L+ FF G
Sbjct: 57 MSGAMIYIKRDLKINDLQIGILAGSLNIYSLIGSCAAGRTSDWIGRRYTIVLAGAIFFAG 116
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
+ILMG ++ LM GR IAG GVG+AL+I VY+AE+S S RGFL S P++ IN G +
Sbjct: 117 AILMGLSPNYAFLMFGRFIAGIGVGYALMIAPVYTAEVSPASSRGFLNSFPEVFINAGIM 176
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SN L L++GWR+ML I ++PS+ L I +L + ESPRWLVMQGRLGDAK+VL
Sbjct: 177 LGYVSNLAFSNLPLKVGWRLMLGIGAVPSVILAIGVLAMPESPRWLVMQGRLGDAKRVLD 236
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
S+S EA R+++IK+A GI +C ++V VS++ G G +E+ +P+P V R++I
Sbjct: 237 KTSDSPTEATLRLEDIKHAAGIPADCHDDVVQVSRRNSHGEGVWRELLIRPTPAVRRVMI 296
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
AAIG+H FQ G++ + L+SPRIF G+ LLATV +G+ +T F L++ FLLD+
Sbjct: 297 AAIGIHFFQQASGIDAVVLFSPRIFKTAGLKTDHQQLLATVAVGVVKTSFILVATFLLDR 356
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
IGRR LLL S GG++ S+ L I++ S+ ++ +WA++ I + +IG G
Sbjct: 357 IGRRPLLLTSVGGMVLSLAALGTSLTIIDQSE--KKVMWAVVVAIATVMTYVATFSIGAG 414
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
+TWVYS+EIFPLRLR+QG + V++NR+T+ + SF+ + K +T GG F+LF G +
Sbjct: 415 PITWVYSSEIFPLRLRSQGSSMGVVVNRVTSGVISISFLPMSKAMTTGGAFYLFGGIATV 474
Query: 421 GWWFYYSFLPETKGRSLEDMETIF 444
W F+Y+FLPET+GR LEDM+ +F
Sbjct: 475 AWVFFYTFLPETQGRMLEDMDELF 498
>At2g16120 putative sugar transporter
Length = 511
Score = 431 bits (1108), Expect = e-121
Identities = 212/460 (46%), Positives = 310/460 (67%), Gaps = 1/460 (0%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA +FIK++L++SD+Q+++L GILN +L AGR SD++GRRYTI+L+ FFF G
Sbjct: 47 MSGASIFIKDDLKLSDVQLEILMGILNIYSLVGSGAAGRTSDWLGRRYTIVLAGAFFFCG 106
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
++LMG+ +++P +M+GR +AG GVG+A++I VY+AE++ S RGFLTS P++ INIG L
Sbjct: 107 ALLMGFATNYPFIMVGRFVAGIGVGYAMMIAPVYTAEVAPASSRGFLTSFPEIFINIGIL 166
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SNYF KL LGWR ML + ++PS+ L I +L + ESPRWLV+QGRLGDA KVL
Sbjct: 167 LGYVSNYFFSKLPEHLGWRFMLGVGAVPSVFLAIGVLAMPESPRWLVLQGRLGDAFKVLD 226
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
SN+K+EA R+ +IK AVGI ++ T +++ V K +G G K++ +P+P V ILI
Sbjct: 227 KTSNTKEEAISRLDDIKRAVGIPDDMTDDVIVVPNKKSAGKGVWKDLLVRPTPSVRHILI 286
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
A +G+H Q G++ + LYSP IF + G+ K LLATV +G+ +TLF ++ ++D+
Sbjct: 287 ACLGIHFAQQASGIDAVVLYSPTIFSKAGLKSKNDQLLATVAVGVVKTLFIVVGTCVVDR 346
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
GRR LLL S GG+ S+ L S V N G+ WAI + + +IG G
Sbjct: 347 FGRRALLLTSMGGMFLSLTALGT-SLTVINRNPGQTLKWAIGLAVTTVMTFVATFSIGAG 405
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
VTWVY +EIFP+RLRAQG + V++NR+ + + +F+S+ K +T+GG F LF G
Sbjct: 406 PVTWVYCSEIFPVRLRAQGASLGVMLNRLMSGIIGMTFLSLSKGLTIGGAFLLFAGVAAA 465
Query: 421 GWWFYYSFLPETKGRSLEDMETIFGKNSNSEVQMKYGSNN 460
W F+++FLPET+G LE+MET+FG + ++ +N
Sbjct: 466 AWVFFFTFLPETRGIPLEEMETLFGSYTANKKNNSMSKDN 505
>At2g20780 putative sugar transporter
Length = 547
Score = 322 bits (825), Expect = 3e-88
Identities = 176/444 (39%), Positives = 266/444 (59%), Gaps = 19/444 (4%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLG 60
M+GA+LFI+++L+I+++Q ++L G L+ +L + GR SD IGR++T+ L+++ F G
Sbjct: 97 MSGAVLFIQQDLKITEVQTEVLIGSLSIISLFGSLAGGRTSDSIGRKWTMALAALVFQTG 156
Query: 61 SILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFL 120
+ +M SF +LMIGR +AG G+G ++I VY AEIS RGF TS P++ IN+G L
Sbjct: 157 AAVMAVAPSFEVLMIGRTLAGIGIGLGVMIAPVYIAEISPTVARGFFTSFPEIFINLGIL 216
Query: 121 LGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLL 180
LGY+SNY LS+ + WRIMLA+ +PS+ + + + ESPRWLVM+GR+ A++VL+
Sbjct: 217 LGYVSNYAFSGLSVHISWRIMLAVGILPSVFIGFALCVIPESPRWLVMKGRVDSAREVLM 276
Query: 181 LISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILI 240
+ EAE+R+ EI+ A E V +E+ PSP V ++LI
Sbjct: 277 KTNERDDEAEERLAEIQLAAAHTEGSEDRPV------------WRELL-SPSPVVRKMLI 323
Query: 241 AAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDK 300
G+ FQ I G++ YSP I GI D+ LL ATV +G+++T+F L + FL+D
Sbjct: 324 VGFGIQCFQQITGIDATVYYSPEILKEAGIQDETKLLAATVAVGVTKTVFILFATFLIDS 383
Query: 301 IGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIG 360
+GR+ LL VS+ G+ + L + LG I ++ + F +IG+G
Sbjct: 384 VGRKPLLYVSTIGMTLCLFCLSFTLTFLGQGTLG------ITLALLFVCGNVAFFSIGMG 437
Query: 361 AVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVL 420
V WV ++EIFPLRLRAQ + + NR+ + V SF+S+ + IT+GGTFF+F + L
Sbjct: 438 PVCWVLTSEIFPLRLRAQASALGAVGNRVCSGLVAMSFLSVSRAITVGGTFFVFSLVSAL 497
Query: 421 GWWFYYSFLPETKGRSLEDMETIF 444
F Y +PET G+SLE +E +F
Sbjct: 498 SVIFVYVLVPETSGKSLEQIELMF 521
>At1g75220 integral membrane protein, putative
Length = 487
Score = 182 bits (463), Expect = 3e-46
Identities = 120/438 (27%), Positives = 220/438 (49%), Gaps = 28/438 (6%)
Query: 8 IKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYG 67
I ++L ++ + + + N A+ + +G++++YIGR+ ++M+++I +G + + +
Sbjct: 76 ITKDLGLTVSEYSVFGSLSNVGAMVGAIASGQIAEYIGRKGSLMIAAIPNIIGWLCISFA 135
Query: 68 SSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNY 127
L +GR + GFGVG V VY AEI+ + RG L S+ LS+ IG +L YL
Sbjct: 136 KDTSFLYMGRLLEGFGVGIISYTVPVYIAEIAPQNMRGGLGSVNQLSVTIGIMLAYL--- 192
Query: 128 FLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQ 187
L L + WRI+ + +P L+ + + ESPRWL G + + L ++ +
Sbjct: 193 ----LGLFVPWRILAVLGILPCTLLIPGLFFIPESPRWLAKMGMTDEFETSLQVLRGFET 248
Query: 188 EAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHI 247
+ + EIK +V + +N V R +Y P L+ IG+ +
Sbjct: 249 DITVEVNEIKRSVA--SSTKRNTVRFVDLKR-------RRYYFP-------LMVGIGLLV 292
Query: 248 FQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILL 307
Q + G+ G+ YS IF G+T AT G+G Q + T +S +L+DK GRR+LL
Sbjct: 293 LQQLGGINGVLFYSSTIFESAGVTSSNA---ATFGVGAIQVVATAISTWLVDKAGRRLLL 349
Query: 308 LVSSGGVIFSMLGLCVCSAIVE-NSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVY 366
+SS G+ S++ + + E S + W I +++ + M F ++G+G + W+
Sbjct: 350 TISSVGMTISLVIVAAAFYLKEFVSPDSDMYSWLSILSVVGVVAMVVFFSLGMGPIPWLI 409
Query: 367 STEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYY 426
+EI P+ ++ + + N + ++T ++ + GGTF L+ F
Sbjct: 410 MSEILPVNIKGLAGSIATLANWFFS-WLITMTANLLLAWSSGGTFTLYGLVCAFTVVFVT 468
Query: 427 SFLPETKGRSLEDMETIF 444
++PETKG++LE+++++F
Sbjct: 469 LWVPETKGKTLEELQSLF 486
>At1g19450 integral membrane protein, putative
Length = 488
Score = 177 bits (448), Expect = 1e-44
Identities = 120/439 (27%), Positives = 224/439 (50%), Gaps = 30/439 (6%)
Query: 8 IKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYG 67
I ++L ++ + + + N A+ + +G++++Y+GR+ ++M+++I +G + + +
Sbjct: 77 ITKDLGLTVSEYSVFGSLSNVGAMVGAIASGQIAEYVGRKGSLMIAAIPNIIGWLSISFA 136
Query: 68 SSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNY 127
L +GR + GFGVG V VY AEI+ + RG L S+ LS+ IG +L YL
Sbjct: 137 KDTSFLYMGRLLEGFGVGIISYTVPVYIAEIAPQTMRGALGSVNQLSVTIGIMLAYL--- 193
Query: 128 FLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQ 187
L L + WRI+ + +P L+ + + ESPRWL G D + L ++ +
Sbjct: 194 ----LGLFVPWRILAVLGVLPCTLLIPGLFFIPESPRWLAKMGLTDDFETSLQVLRGFET 249
Query: 188 EAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHI 247
+ + EIK +V SK++ LK Y Y L+ IG+
Sbjct: 250 DITVEVNEIKRSVASS----------SKRSAVRFVDLKRRRY------YFPLMVGIGLLA 293
Query: 248 FQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILL 307
Q + G+ G+ YS IF G+T +AT G+G+ Q + T ++ +L+DK GRR+LL
Sbjct: 294 LQQLGGINGVLFYSSTIFESAGVTSSN---VATFGVGVVQVVATGIATWLVDKAGRRLLL 350
Query: 308 LVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGF--NAIGIGAVTWV 365
++SS G+ S++ + V + E + ++ I+ + V+ ++A ++G+G + W+
Sbjct: 351 MISSIGMTISLVIVAVAFYLKEFVS-PDSNMYNILSMVSVVGVVAMVISCSLGMGPIPWL 409
Query: 366 YSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFY 425
+EI P+ ++ + ++N + +VT ++ + GGTF L+ F
Sbjct: 410 IMSEILPVNIKGLAGSIATLLNWFVS-WLVTMTANMLLAWSSGGTFTLYALVCGFTVVFV 468
Query: 426 YSFLPETKGRSLEDMETIF 444
++PETKG++LE+++ +F
Sbjct: 469 SLWVPETKGKTLEEIQALF 487
>At1g08900 Beta integral like membrane protein
Length = 462
Score = 174 bits (442), Expect = 7e-44
Identities = 125/441 (28%), Positives = 223/441 (50%), Gaps = 43/441 (9%)
Query: 8 IKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYG 67
I EEL +S ++ + + +G++S +GRR T+ +S + G + + +
Sbjct: 53 IMEELGLSVADYSFFTSVMTLGGMITAVFSGKISALVGRRQTMWISDVCCIFGWLAVAFA 112
Query: 68 SSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNY 127
+L GR GFGVG +V VY AEI+ ++RG + L +G L + +
Sbjct: 113 HDIIMLNTGRLFLGFGVGLISYVVPVYIAEITPKTFRGGFSYSNQLLQCLGISLMFFTGN 172
Query: 128 FLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQ 187
F WR + + +IPS VI + + ESPRWL M G+ + + L +
Sbjct: 173 F-------FHWRTLALLSAIPSAFQVICLFFIPESPRWLAMYGQDQELEVSLKKLRGENS 225
Query: 188 EAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHI 247
+ + EI+ V I K+++SG ++++F+ + H LI +G+ +
Sbjct: 226 DILKEAAEIRETVEISR----------KESQSG---IRDLFHIGNAH---SLIIGLGLML 269
Query: 248 FQNICGVEGIFLYSPRIFGRMGI-TDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRIL 306
Q CG I Y+ RIF + G +D GT +LA I I Q++ +L+ +D+ GRR L
Sbjct: 270 LQQFCGSAAISAYAARIFDKAGFPSDIGTTILAV--ILIPQSIVVMLT---VDRWGRRPL 324
Query: 307 LLVSSGGVIFSMLGLCVCSAIVENS----KLGE-EPLWAIIFTIIVIYIMAGFNAIGIGA 361
L++SS +G+C+CS + S K GE + L +++ + ++ ++ F IG+G
Sbjct: 325 LMISS-------IGMCICSFFIGLSYYLQKNGEFQKLCSVMLIVGLVGYVSSF-GIGLGG 376
Query: 362 VTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLG 421
+ WV +EIFP+ ++ + + N N ++ SF + + + GT+F+F G +++
Sbjct: 377 LPWVIMSEIFPVNVKITAGSLVTMSNWFFNWIIIYSFNFMIQW-SASGTYFIFSGVSLVT 435
Query: 422 WWFYYSFLPETKGRSLEDMET 442
F ++ +PETKGR+LE+++T
Sbjct: 436 IVFIWTLVPETKGRTLEEIQT 456
>At2g43330 membrane transporter like protein
Length = 509
Score = 173 bits (439), Expect = 2e-43
Identities = 139/466 (29%), Positives = 219/466 (46%), Gaps = 44/466 (9%)
Query: 1 MAGALLFIKEELEISDMQVQLLAGILNACALPACMIA----GRLSDYIGRRYTIMLSSIF 56
++GALL+IK++ E+ Q L + + AL MI G ++DY GR+ + + +
Sbjct: 52 ISGALLYIKDDFEVVK-QSSFLQETIVSMALVGAMIGAAAGGWINDYYGRKKATLFADVV 110
Query: 57 FFLGSILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSIN 116
F G+I+M +L+ GR + G GVG A + VY AE S RG L S L I
Sbjct: 111 FAAGAIVMAAAPDPYVLISGRLLVGLGVGVASVTAPVYIAEASPSEVRGGLVSTNVLMIT 170
Query: 117 IGFLLGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAK 176
G L YL N ++ WR ML + +P++ ILML + ESPRWL M+ R +A
Sbjct: 171 GGQFLSYLVNSAFTQVPGT--WRWMLGVSGVPAVIQFILMLFMPESPRWLFMKNRKAEAI 228
Query: 177 KVLLL---ISNSKQE------AEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEM 227
+VL IS + E AE+ K+ K VG + V SK+ R
Sbjct: 229 QVLARTYDISRLEDEIDHLSAAEEEEKQRKRTVGYLD------VFRSKELRLA------- 275
Query: 228 FYKPSPHVYRILIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQ 287
+A G+ FQ G+ + YSP I G L ++ +
Sbjct: 276 -----------FLAGAGLQAFQQFTGINTVMYYSPTIVQMAGFHSNQLALFLSLIVAAMN 324
Query: 288 TLFTLLSCFLLDKIGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTI-I 346
T++ + +D GR+ L L S GVI S+L L V S ++ + L+ + + +
Sbjct: 325 AAGTVVGIYFIDHCGRKKLALSSLFGVIISLLILSV-SFFKQSETSSDGGLYGWLAVLGL 383
Query: 347 VIYIMAGFNAIGIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTIT 406
+YI+ F A G+G V W ++EI+P + R G+ +N I+N+ V +F++I +
Sbjct: 384 ALYIV--FFAPGMGPVPWTVNSEIYPQQYRGICGGMSATVNWISNLIVAQTFLTIAEAAG 441
Query: 407 LGGTFFLFVGTNVLGWWFYYSFLPETKGRSLEDMETIFGKNSNSEV 452
G TF + G VL F F+PET+G + ++E I+ + + +
Sbjct: 442 TGMTFLILAGIAVLAVIFVIVFVPETQGLTFSEVEQIWKERAYGNI 487
>At3g05150 putative sugar transporter
Length = 463
Score = 171 bits (433), Expect = 8e-43
Identities = 129/439 (29%), Positives = 222/439 (50%), Gaps = 36/439 (8%)
Query: 8 IKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYG 67
I EEL +S Q + ILN A+ + +G++SD+IGR+ + LSS+ +G +++
Sbjct: 56 IMEELNLSYSQFSVFGSILNMGAVLGAITSGKISDFIGRKGAMRLSSVISAIGWLIIYLA 115
Query: 68 SSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNY 127
L GR + G+G G +V V+ AEIS RG L +L L I IG L++
Sbjct: 116 KGDVPLDFGRFLTGYGCGTLSFVVPVFIAEISPRKLRGALATLNQLFIVIG-----LASM 170
Query: 128 FLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQ 187
FL + + WR + P + L + ESPRWL M GR D + L + +
Sbjct: 171 FL--IGAVVNWRTLALTGVAPCVVLFFGTWFIPESPRWLEMVGRHSDFEIALQKLRGPQA 228
Query: 188 EAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHI 247
+ EI+ + ++ H+ K T L ++ K + R +I +G+
Sbjct: 229 NITREAGEIQEYLA-------SLAHLPKAT------LMDLIDKKN---IRFVIVGVGLMF 272
Query: 248 FQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLL-SCFLLDKIGRRIL 306
FQ G+ G+ Y+ +IF G + L ++ I Q + T L + L+D++GRR L
Sbjct: 273 FQQFVGINGVIFYAQQIFVSAGASP----TLGSILYSIEQVVLTALGATLLIDRLGRRPL 328
Query: 307 LLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVY 366
L+ S+ G++ L L S +++ L + + A+ + +++YI G +IG+GA+ WV
Sbjct: 329 LMASAVGMLIGCL-LIGNSFLLKAHGLALDIIPALAVSGVLVYI--GSFSIGMGAIPWVI 385
Query: 367 STEIFPLRLRAQGLGVCVIMNRITN--VAVVTSFISIYKTITLGGTFFLFVGTNVLGWWF 424
+EIFP+ L+ G+ ++N +++ V+ +F+ I+ GTF+++ G VL F
Sbjct: 386 MSEIFPINLKGTAGGLVTVVNWLSSWLVSFTFNFLMIWSP---HGTFYVYGGVCVLAIIF 442
Query: 425 YYSFLPETKGRSLEDMETI 443
+PETKGR+LE+++ +
Sbjct: 443 IAKLVPETKGRTLEEIQAM 461
>At5g26250 hexose transporter - like protein
Length = 507
Score = 170 bits (430), Expect = 2e-42
Identities = 131/427 (30%), Positives = 205/427 (47%), Gaps = 32/427 (7%)
Query: 19 VQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYGSSFPILMIGRC 78
+QL L AL A A +GRR T+ L+SIFF +G L + +L+IGR
Sbjct: 80 LQLFTSSLYLAALVASFFASATCSKLGRRPTMQLASIFFLIGVGLAAGAVNIYMLIIGRI 139
Query: 79 IAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKLSLRLGW 138
+ GFGVGF V ++ +EI+ RG L + L + IG L+ + NYF + GW
Sbjct: 140 LLGFGVGFGNQAVPLFLSEIAPARLRGGLNIVFQLMVTIGILIANIVNYFTSSIH-PYGW 198
Query: 139 RIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKN 198
RI L IP++ L+ L + E+P L+ + N +E ++ +K+I+
Sbjct: 199 RIALGGAGIPALILLFGSLLICETPTSLIER--------------NKTKEGKETLKKIRG 244
Query: 199 AVGIDENCTQNIVH---VSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVE 255
+DE ++IVH ++++ + L + +P P V +L+ FQ G+
Sbjct: 245 VEDVDEE-YESIVHACDIARQVKDPYTKLMKPASRP-PFVIGMLL-----QFFQQFTGIN 297
Query: 256 GIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVI 315
I Y+P +F +G + LL A V G L T + FL+DK GRR LLL SS ++
Sbjct: 298 AIMFYAPVLFQTVGFGNDAALLSAVV-TGTINVLSTFVGIFLVDKTGRRFLLLQSSVHML 356
Query: 316 FSML--GLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPL 373
L G+ + + L P ++ + +Y+M GF A G + W+ +E FPL
Sbjct: 357 ICQLVIGIILAKDLDVTGTLA-RPQALVVVIFVCVYVM-GF-AWSWGPLGWLIPSETFPL 413
Query: 374 RLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSFLPETK 433
R +G + V N + +F+S+ + G FF F G V+ F F+PETK
Sbjct: 414 ETRTEGFALAVSCNMFFTFVIAQAFLSMLCAMK-SGIFFFFSGWIVVMGLFALFFVPETK 472
Query: 434 GRSLEDM 440
G S++DM
Sbjct: 473 GVSIDDM 479
>At3g19930 monosaccharide transport protein, STP4
Length = 514
Score = 169 bits (429), Expect = 2e-42
Identities = 126/435 (28%), Positives = 214/435 (48%), Gaps = 41/435 (9%)
Query: 21 LLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYGSSFPILMIGRCIA 80
L L AL + + A ++ GR++++ L FF+GS G+ + +L+IGR +
Sbjct: 83 LFTSSLYVAALVSSLFASTITRVFGRKWSMFLGGFTFFIGSAFNGFAQNIAMLLIGRILL 142
Query: 81 GFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKLSLRLGWRI 140
GFGVGFA V VY +E++ P+ RG + ++I G ++ + NYF ++ +GWRI
Sbjct: 143 GFGVGFANQSVPVYLSEMAPPNLRGAFNNGFQVAIIFGIVVATIINYFTAQMKGNIGWRI 202
Query: 141 MLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKNAV 200
L + +P++ ++I L L ++P L+ +G +AK++L I + + E+
Sbjct: 203 SLGLACVPAVMIMIGALILPDTPNSLIERGYTEEAKEMLQSIRGTNEVDEE--------- 253
Query: 201 GIDENCTQNIVHVSKKTRSGGGALKEMF---YKPSPHVYRILIAAIGVHIFQNICGVEGI 257
Q+++ S++++ K + Y+P LI + FQ + G+ I
Sbjct: 254 ------FQDLIDASEESKQVKHPWKNIMLPRYRPQ------LIMTCFIPFFQQLTGINVI 301
Query: 258 FLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGV--- 314
Y+P +F +G K +LL A V GI + L T +S F +D+ GRRIL L GG+
Sbjct: 302 TFYAPVLFQTLGFGSKASLLSAMV-TGIIELLCTFVSVFTVDRFGRRILFL--QGGIQML 358
Query: 315 -----IFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTE 369
I +M+G V + +G+ +I +I IY+ AGF A G + W+ +E
Sbjct: 359 VSQIAIGAMIG--VKFGVAGTGNIGKSDA-NLIVALICIYV-AGF-AWSWGPLGWLVPSE 413
Query: 370 IFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSFL 429
I PL +R+ + V +N V F+++ + G FF F V+ F Y L
Sbjct: 414 ISPLEIRSAAQAINVSVNMFFTFLVAQLFLTMLCHMKF-GLFFFFAFFVVIMTIFIYLML 472
Query: 430 PETKGRSLEDMETIF 444
PETK +E+M ++
Sbjct: 473 PETKNVPIEEMNRVW 487
>At1g11260 glucose transporter
Length = 522
Score = 169 bits (429), Expect = 2e-42
Identities = 121/424 (28%), Positives = 213/424 (49%), Gaps = 28/424 (6%)
Query: 26 LNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYGSSFPILMIGRCIAGFGVG 85
L AL + ++A ++ GRR +++ I F G+++ G+ +L++GR + GFG+G
Sbjct: 89 LYLAALISSLVASTVTRKFGRRLSMLFGGILFCAGALINGFAKHVWMLIVGRILLGFGIG 148
Query: 86 FALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKLSLRLGWRIMLAIP 145
FA V +Y +E++ YRG L LSI IG L+ + NYF K+ GWR+ L
Sbjct: 149 FANQAVPLYLSEMAPYKYRGALNIGFQLSITIGILVAEVLNYFFAKIKGGWGWRLSLGGA 208
Query: 146 SIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKNAVGIDEN 205
+P++ + I L L ++P ++ +G+ +EA+ +++ I+ + +
Sbjct: 209 VVPALIITIGSLVLPDTPNSMIERGQ--------------HEEAKTKLRRIRGVDDVSQE 254
Query: 206 CTQNIVHVSKKTRSGGGALKEMF-YKPSPHVYRILIAAIGVHIFQNICGVEGIFLYSPRI 264
++V SK+++S + + K PH L A+ + FQ + G+ I Y+P +
Sbjct: 255 -FDDLVAASKESQSIEHPWRNLLRRKYRPH----LTMAVMIPFFQQLTGINVIMFYAPVL 309
Query: 265 FGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVIF--SMLGLC 322
F +G T +L+ A V G TL+S + +D+ GRR L L ++ +++ C
Sbjct: 310 FNTIGFTTDASLMSAVV-TGSVNVAATLVSIYGVDRWGRRFLFLEGGTQMLICQAVVAAC 368
Query: 323 VCSAIVENSKLGEEPLW--AIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRLRAQGL 380
+ + + GE P W ++ T I IY+ AGF A G + W+ +EIFPL +R+
Sbjct: 369 IGAKFGVDGTPGELPKWYAIVVVTFICIYV-AGF-AWSWGPLGWLVPSEIFPLEIRSAAQ 426
Query: 381 GVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSFLPETKGRSLEDM 440
+ V +N I + F+++ + G F +F V+ F Y FLPETKG +E+M
Sbjct: 427 SITVSVNMIFTFIIAQIFLTMLCHLKF-GLFLVFAFFVVVMSIFVYIFLPETKGIPIEEM 485
Query: 441 ETIF 444
++
Sbjct: 486 GQVW 489
>At5g26340 hexose transporter - like protein
Length = 526
Score = 167 bits (423), Expect = 1e-41
Identities = 123/424 (29%), Positives = 201/424 (47%), Gaps = 24/424 (5%)
Query: 19 VQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYGSSFPILMIGRC 78
+QL L L A A + +GRR T++++ +FF +G L +L+ GR
Sbjct: 83 LQLFTSSLYLAGLTATFFASYTTRTLGRRLTMLIAGVFFIIGVALNAGAQDLAMLIAGRI 142
Query: 79 IAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKLSLRLGW 138
+ G GVGFA V ++ +EI+ RG L L L++ IG L L NY K+ GW
Sbjct: 143 LLGCGVGFANQAVPLFLSEIAPTRIRGGLNILFQLNVTIGILFANLVNYGTAKIKGGWGW 202
Query: 139 RIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKN 198
R+ L + IP++ L + L + E+P LV +GRL + K VL I + E ++
Sbjct: 203 RLSLGLAGIPALLLTVGALLVTETPNSLVERGRLDEGKAVLRRIRGT-DNVEPEFADLLE 261
Query: 199 AVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVEGIF 258
A ++K+ + L + +P L+ A+ + IFQ G+ I
Sbjct: 262 AS-----------RLAKEVKHPFRNLLQRRNRPQ------LVIAVALQIFQQCTGINAIM 304
Query: 259 LYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVIFSM 318
Y+P +F +G +L A V G L TL+S + +DK+GRR+LLL + + FS
Sbjct: 305 FYAPVLFSTLGFGSDASLYSAVV-TGAVNVLSTLVSIYSVDKVGRRVLLLEAGVQMFFSQ 363
Query: 319 LGLCVCSAI-VENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLRLRA 377
+ + + + V ++ +AI+ +++ +A F A G + W+ +E FPL R+
Sbjct: 364 VVIAIILGVKVTDTSTNLSKGFAILVVVMICTYVAAF-AWSWGPLGWLIPSETFPLETRS 422
Query: 378 QGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSF-LPETKGRS 436
G V V +N + + +F+S+ G F+F VL + F LPETK
Sbjct: 423 AGQSVTVCVNLLFTFIIAQAFLSMLCHFKFG--IFIFFSAWVLIMSVFVMFLLPETKNIP 480
Query: 437 LEDM 440
+E+M
Sbjct: 481 IEEM 484
>At3g05960 putative hexose transporter
Length = 507
Score = 165 bits (418), Expect = 4e-41
Identities = 124/427 (29%), Positives = 206/427 (48%), Gaps = 32/427 (7%)
Query: 19 VQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYGSSFPILMIGRC 78
+QL L AL A +A +GRR T+ +SIFF +G L + +L+IGR
Sbjct: 79 LQLFTSSLYLAALVASFVASATCSKLGRRPTMQFASIFFLIGVGLTAGAVNLVMLIIGRL 138
Query: 79 IAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKLSLRLGW 138
GFGVGF V ++ +EI+ RG L + L + IG L+ + NYF + GW
Sbjct: 139 FLGFGVGFGNQAVPLFLSEIAPAQLRGGLNIVFQLMVTIGILIANIVNYFTATVH-PYGW 197
Query: 139 RIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKN 198
RI L IP++ L+ L ++E+P L+ + N +E ++ +++I+
Sbjct: 198 RIALGGAGIPAVILLFGSLLIIETPTSLIER--------------NKNEEGKEALRKIRG 243
Query: 199 AVGIDENCTQNIVH---VSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGVE 255
I++ ++IVH ++ + + L + +P P + +L+ +FQ G+
Sbjct: 244 VDDINDE-YESIVHACDIASQVKDPYRKLLKPASRP-PFIIGMLL-----QLFQQFTGIN 296
Query: 256 GIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGVI 315
I Y+P +F +G LL A + G L T + +L+D+ GRR LLL SS ++
Sbjct: 297 AIMFYAPVLFQTVGFGSDAALLSAVI-TGSINVLATFVGIYLVDRTGRRFLLLQSSVHML 355
Query: 316 FSML--GLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPL 373
L G+ + + LG P ++ + +Y+M GF A G + W+ +E FPL
Sbjct: 356 ICQLIIGIILAKDLGVTGTLG-RPQALVVVIFVCVYVM-GF-AWSWGPLGWLIPSETFPL 412
Query: 374 RLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSFLPETK 433
R+ G V V N + +F+S+ + G FF F G ++ F + F+PETK
Sbjct: 413 ETRSAGFAVAVSCNMFFTFVIAQAFLSMLCGMR-SGIFFFFSGWIIVMGLFAFFFIPETK 471
Query: 434 GRSLEDM 440
G +++DM
Sbjct: 472 GIAIDDM 478
>At5g16150 sugar transporter like protein
Length = 546
Score = 163 bits (413), Expect = 2e-40
Identities = 128/445 (28%), Positives = 215/445 (47%), Gaps = 39/445 (8%)
Query: 3 GALLFIKEELEISD---MQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFL 59
GAL ++ ++L I++ +Q +++ +L A A G L+D GR T L +I +
Sbjct: 128 GALEYLAKDLGIAENTVLQGWIVSSLL-AGATVGSFTGGALADKFGRTRTFQLDAIPLAI 186
Query: 60 GSILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGF 119
G+ L S +++GR +AG G+G + IV +Y +EIS RG L S+ L I IG
Sbjct: 187 GAFLCATAQSVQTMIVGRLLAGIGIGISSAIVPLYISEISPTEIRGALGSVNQLFICIGI 246
Query: 120 LLGYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVL 179
L ++ L + L WR M + IPS+ L I M ESPRWLV QG++ +A+K +
Sbjct: 247 LAALIAGLPLA--ANPLWWRTMFGVAVIPSVLLAIGMAFSPESPRWLVQQGKVSEAEKAI 304
Query: 180 LLISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKE---MFYKPSPHVY 236
+ ++ E V + SG G+ + F S +
Sbjct: 305 KTLYGKERVVEL---------------------VRDLSASGQGSSEPEAGWFDLFSSRYW 343
Query: 237 RILIAAIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCF 296
+++ + +FQ + G+ + YS +F GI + A+ +G S T ++
Sbjct: 344 KVVSVGAALFLFQQLAGINAVVYYSTSVFRSAGIQSD---VAASALVGASNVFGTAVASS 400
Query: 297 LLDKIGRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNA 356
L+DK+GR+ LLL S GG+ SML L + + + A++ T V+Y+++ +
Sbjct: 401 LMDKMGRKSLLLTSFGGMALSMLLLSL--SFTWKALAAYSGTLAVVGT--VLYVLSF--S 454
Query: 357 IGIGAVTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVG 416
+G G V + EIF R+RA+ + + + M+ I+N + F+S+ + + F G
Sbjct: 455 LGAGPVPALLLPEIFASRIRAKAVALSLGMHWISNFVIGLYFLSVVTKFGISSVYLGFAG 514
Query: 417 TNVLGWWFYYSFLPETKGRSLEDME 441
VL + + ETKGRSLE++E
Sbjct: 515 VCVLAVLYIAGNVVETKGRSLEEIE 539
>At5g59250 D-xylose-H+ symporter - like protein
Length = 558
Score = 163 bits (412), Expect = 2e-40
Identities = 123/428 (28%), Positives = 216/428 (49%), Gaps = 26/428 (6%)
Query: 21 LLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYGSSFPILMIGRCIA 80
+++G L L + + G ++D++GRR ++++++ + LGS++ G IL++GR +
Sbjct: 147 VVSGSLYGALLGSISVYG-VADFLGRRRELIIAAVLYLLGSLITGCAPDLNILLVGRLLY 205
Query: 81 GFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKLSLRLGWRI 140
GFG+G A+ +Y AE RG L SL +L I +G LLG+ F ++ + GWR
Sbjct: 206 GFGIGLAMHGAPLYIAETCPSQIRGTLISLKELFIVLGILLGFSVGSF--QIDVVGGWRY 263
Query: 141 MLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEIKNAV 200
M + ++ + + M L SPRWL+++ G + + K++A + +++
Sbjct: 264 MYGFGTPVALLMGLGMWSLPASPRWLLLRAVQGKGQ-----LQEYKEKAMLALSKLRGRP 318
Query: 201 GIDENCTQNI--VHVSKKT----RSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGV 254
D+ + + ++S KT GG E+F P+ + L G+ +FQ I G
Sbjct: 319 PGDKISEKLVDDAYLSVKTAYEDEKSGGNFLEVFQGPN---LKALTIGGGLVLFQQITGQ 375
Query: 255 EGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGV 314
+ Y+ I G + +V IG+ + L T ++ +D +GRR LL+ GGV
Sbjct: 376 PSVLYYAGSILQTAGFSAAADATRVSVIIGVFKLLMTWVAVAKVDDLGRRPLLI---GGV 432
Query: 315 IFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGAVTWVYSTEIFPLR 374
L L + SA + LG PL A+ +++Y+ G I G ++W+ +EIFPLR
Sbjct: 433 SGIALSLFLLSAYYK--FLGGFPLVAV--GALLLYV--GCYQISFGPISWLMVSEIFPLR 486
Query: 375 LRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSFLPETKG 434
R +G+ + V+ N +N V +F + + + F LF G ++ F +PETKG
Sbjct: 487 TRGRGISLAVLTNFGSNAIVTFAFSPLKEFLGAENLFLLFGGIALVSLLFVILVVPETKG 546
Query: 435 RSLEDMET 442
SLE++E+
Sbjct: 547 LSLEEIES 554
>At4g21480 glucose transporter
Length = 508
Score = 160 bits (406), Expect = 1e-39
Identities = 122/436 (27%), Positives = 207/436 (46%), Gaps = 36/436 (8%)
Query: 17 MQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGSILMGYGSSFPILMIG 76
+ + L L AL + ++A ++ GR+ +++L + F G++L G+ ++ +L++G
Sbjct: 80 VSLTLFTSSLYLAALCSSLVASYVTRQFGRKISMLLGGVLFCAGALLNGFATAVWMLIVG 139
Query: 77 RCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLLGYLSNYFLGKLSLRL 136
R + GFG+GF V +Y +E++ YRG L LSI IG L+ + N+F K+S
Sbjct: 140 RLLLGFGIGFTNQSVPLYLSEMAPYKYRGALNIGFQLSITIGILVANVLNFFFSKIS--W 197
Query: 137 GWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLLISNSKQEAEQRMKEI 196
GWR+ L +P++ + + L L ++P ++ +G+ + AE ++++I
Sbjct: 198 GWRLSLGGAVVPALIITVGSLILPDTPNSMIERGQF--------------RLAEAKLRKI 243
Query: 197 KNAVGIDENCTQNIV--HVSKKTRSGGGALKEMFYKPSPHVYRILIAAIGVHIFQNICGV 254
+ ID+ I+ SK L + Y+P L AI + FQ + G+
Sbjct: 244 RGVDDIDDEINDLIIASEASKLVEHPWRNLLQRKYRPH------LTMAILIPAFQQLTGI 297
Query: 255 EGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKIGRRILLLVSSGGV 314
I Y+P +F +G L+ A V G+ T++S + +DK GRR L L GG
Sbjct: 298 NVIMFYAPVLFQTIGFGSDAALISAVV-TGLVNVGATVVSIYGVDKWGRRFLFL--EGG- 353
Query: 315 IFSMLGLCVCSAIVENSKLGEE------PLWAIIFTIIVIYIMAGFNAIGIGAVTWVYST 368
F ML V A +K G + P W I ++ I I A G + W+ +
Sbjct: 354 -FQMLISQVAVAAAIGAKFGVDGTPGVLPKWYAIVVVLFICIYVAAFAWSWGPLGWLVPS 412
Query: 369 EIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLGWWFYYSF 428
EIFPL +R+ + V +N I + F+ + + G F F V+ F Y F
Sbjct: 413 EIFPLEIRSAAQSITVSVNMIFTFLIAQVFLMMLCHLKF-GLFIFFAFFVVVMSIFVYLF 471
Query: 429 LPETKGRSLEDMETIF 444
LPET+G +E+M ++
Sbjct: 472 LPETRGVPIEEMNRVW 487
>At3g05157 sugar transporter like protein
Length = 458
Score = 160 bits (404), Expect = 2e-39
Identities = 124/440 (28%), Positives = 207/440 (46%), Gaps = 31/440 (7%)
Query: 2 AGALLFIKEELEISDMQVQLLAGILNACALPACMIAGRLSDYIGRRYTIMLSSIFFFLGS 61
+GA I +EL++S Q LN + +G+L+ +GRR T+ +F G
Sbjct: 43 SGAETAIMKELDLSMAQFSAFGSFLNLGGAVGALFSGQLAVILGRRRTLWACDLFCIFGW 102
Query: 62 ILMGYGSSFPILMIGRCIAGFGVGFALIIVSVYSAEISSPSYRGFLTSLPDLSINIGFLL 121
+ + + + L +GR G GVG +V VY AEI+ RG ++ L N G L
Sbjct: 103 LSIAFAKNVLWLDLGRISLGIGVGLTSYVVPVYIAEITPKHVRGAFSASTLLLQNSGISL 162
Query: 122 GYLSNYFLGKLSLRLGWRIMLAIPSIPSIGLVILMLQLVESPRWLVMQGRLGDAKKVLLL 181
YF G + + WR++ I ++P VI + + ESPRWL ++G K+V
Sbjct: 163 ----IYFFGTV---INWRVLAVIGALPCFIPVIGIYFIPESPRWL---AKIGSVKEV--- 209
Query: 182 ISNSKQEAEQRMKEIKNAVGIDENCTQNIVHVSKKTRSGGGALKEMFYKPSPHVYRILIA 241
NS + ++ + + T+ + SK + +MF K R L+
Sbjct: 210 -ENSLHRLRGKDADVSDEAAEIQVMTKMLEEDSK------SSFCDMFQK---KYRRTLVV 259
Query: 242 AIGVHIFQNICGVEGIFLYSPRIFGRMGITDKGTLLLATVGIGISQTLFTLLSCFLLDKI 301
IG+ + Q + G GI YS IF + G +++ L ++ G+ L+ L+D+
Sbjct: 260 GIGLMLIQQLSGASGITYYSNAIFRKAGFSER----LGSMIFGVFVIPKALVGLILVDRW 315
Query: 302 GRRILLLVSSGGVIFSMLGLCVCSAIVENSKLGEEPLWAIIFTIIVIYIMAGFNAIGIGA 361
GRR LLL S+ G+ L + V + E + P + +F I I + GF AIGIG
Sbjct: 316 GRRPLLLASAVGMSIGSLLIGVSFTLQEMNLF---PEFIPVFVFINILVYFGFFAIGIGG 372
Query: 362 VTWVYSTEIFPLRLRAQGLGVCVIMNRITNVAVVTSFISIYKTITLGGTFFLFVGTNVLG 421
+ W+ +EIFP+ ++ + + + T V F +++ + GTF++F L
Sbjct: 373 LPWIIMSEIFPINIKVSAGSIVALTSWTTGWFVSYGFNFMFEW-SAQGTFYIFAMVGGLS 431
Query: 422 WWFYYSFLPETKGRSLEDME 441
F + +PETKG+SLE+++
Sbjct: 432 LLFIWMLVPETKGQSLEELQ 451
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.144 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,438,673
Number of Sequences: 26719
Number of extensions: 389991
Number of successful extensions: 1628
Number of sequences better than 10.0: 89
Number of HSP's better than 10.0 without gapping: 67
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 1295
Number of HSP's gapped (non-prelim): 126
length of query: 461
length of database: 11,318,596
effective HSP length: 103
effective length of query: 358
effective length of database: 8,566,539
effective search space: 3066820962
effective search space used: 3066820962
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147178.8


