

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147178.4 - phase: 0
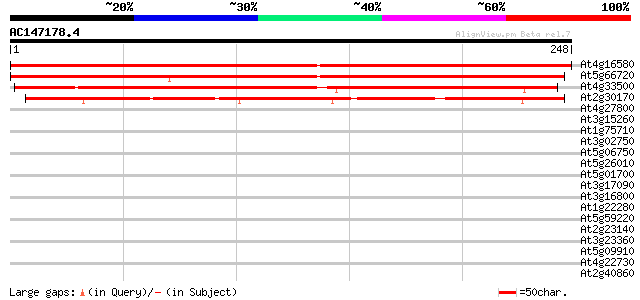
(248 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g16580 unknown protein 357 4e-99
At5g66720 unknown protein 342 1e-94
At4g33500 unknown protein 177 4e-45
At2g30170 unknown protein 154 3e-38
At4g27800 protein phosphatase homolog (PPH1) 34 0.083
At3g15260 putative protein phosphatase type 2C 34 0.083
At1g75710 unknown protein 33 0.11
At3g02750 protein phosphatase-2C (PP2C) like protein 33 0.19
At5g06750 protein phosphatase 2C-like 32 0.41
At5g26010 protein phosphatase - like 31 0.54
At5g01700 putative protein 31 0.70
At3g17090 protein phosphatase-2c, putative 30 1.2
At3g16800 protein phosphatase, putative 30 1.2
At1g22280 protein phosphatase type 2C like protein 29 2.7
At5g59220 protein phosphatase 2C - like 28 3.5
At2g23140 hypothetical protein 28 3.5
At3g23360 hypothetical protein 28 4.6
At5g09910 unknown protein 28 6.0
At4g22730 leucine rich repeat receptor kinase-like protein 27 7.8
At2g40860 putative protein phosphatase 2C 27 7.8
>At4g16580 unknown protein
Length = 467
Score = 357 bits (915), Expect = 4e-99
Identities = 170/248 (68%), Positives = 209/248 (83%), Gaps = 1/248 (0%)
Query: 1 MLSGSCYLPHPDKVATGGEDAHFICEDEQAIGVADGVGGWADVGVNAGLYAQELVANSAR 60
++SGSCYLPHPDK ATGGEDAHFIC +EQA+GVADGVGGWA++G++AG Y++EL++NS
Sbjct: 218 LVSGSCYLPHPDKEATGGEDAHFICAEEQALGVADGVGGWAELGIDAGYYSRELMSNSVN 277
Query: 61 AIREEPKGSFNPVRVLEKAHSKTKAMGSSTVCIIALIDEALNAINLGDSGFIVIRDGSVI 120
AI++EPKGS +P RVLEKAH+ TK+ GSST CIIAL ++ L+AINLGDSGF+V+R+G +
Sbjct: 278 AIQDEPKGSIDPARVLEKAHTCTKSQGSSTACIIALTNQGLHAINLGDSGFMVVREGHTV 337
Query: 121 FKSPVQQRGFNFPYQLARSGTEGDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIVG 180
F+SPVQQ FNF YQL SG GDLPSSG+VFTV VAPGD+I+AGTDGLFDN+YNN+I
Sbjct: 338 FRSPVQQHDFNFTYQL-ESGRNGDLPSSGQVFTVAVAPGDVIIAGTDGLFDNLYNNEITA 396
Query: 181 VVVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLTVVVS 240
+VV A RA + PQ TAQKIAALARQRA D RQ+PFS AA + G+R+ GGKLDD+TVVVS
Sbjct: 397 IVVHAVRANIDPQVTAQKIAALARQRAQDKNRQTPFSTAAQDAGFRYYGGKLDDITVVVS 456
Query: 241 YISNSVNE 248
Y++ S E
Sbjct: 457 YVAASKEE 464
>At5g66720 unknown protein
Length = 414
Score = 342 bits (877), Expect = 1e-94
Identities = 167/246 (67%), Positives = 207/246 (83%), Gaps = 2/246 (0%)
Query: 1 MLSGSCYLPHPDKVATGGEDAHFICEDEQAIGVADGVGGWADVGVNAGLYAQELVANSAR 60
++SGSCYLPHP+K ATGGEDAHFIC++EQAIGVADGVGGWA+VGVNAGL+++EL++ S
Sbjct: 170 LVSGSCYLPHPEKEATGGEDAHFICDEEQAIGVADGVGGWAEVGVNAGLFSRELMSYSVS 229
Query: 61 AIREEPKGS-FNPVRVLEKAHSKTKAMGSSTVCIIALIDEALNAINLGDSGFIVIRDGSV 119
AI+E+ KGS +P+ VLEKAHS+TKA GSST CII L D+ L+AINLGDSGF V+R+G+
Sbjct: 230 AIQEQHKGSSIDPLVVLEKAHSQTKAKGSSTACIIVLKDKGLHAINLGDSGFTVVREGTT 289
Query: 120 IFKSPVQQRGFNFPYQLARSGTEGDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIV 179
+F+SPVQQ GFNF YQL SG D+PSSG+VFT+ V GD+IVAGTDG++DN+YN +I
Sbjct: 290 VFQSPVQQHGFNFTYQL-ESGNSADVPSSGQVFTIDVQSGDVIVAGTDGVYDNLYNEEIT 348
Query: 180 GVVVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLTVVV 239
GVVV + RA L P+ TAQKIA LARQRA+D KRQSPF+ AA E GYR+ GGKLDD+T VV
Sbjct: 349 GVVVSSVRAGLDPKGTAQKIAELARQRAVDKKRQSPFATAAQEAGYRYYGGKLDDITAVV 408
Query: 240 SYISNS 245
SY+++S
Sbjct: 409 SYVTSS 414
>At4g33500 unknown protein
Length = 724
Score = 177 bits (450), Expect = 4e-45
Identities = 101/242 (41%), Positives = 147/242 (60%), Gaps = 7/242 (2%)
Query: 3 SGSCYLPHPDKVATGGEDAHFICEDEQAIGVADGVGGWADVGVNAGLYAQELVANSARAI 62
SG L P K G EDA+FI IG+ADGV W+ G+N G+YAQEL++N + I
Sbjct: 483 SGFASLQSPFKALAGREDAYFISHHNW-IGIADGVSQWSFEGINKGMYAQELMSNCEKII 541
Query: 63 REEPKGSFNPVRVLEKAHSKTKAMGSSTVCIIALIDEALNAINLGDSGFIVIRDGSVIFK 122
E +PV+VL ++ ++TK+ GSST I L + L+ N+GDSGF+VIRDG+V+
Sbjct: 542 SNETAKISDPVQVLHRSVNETKSSGSSTALIAHLDNNELHIANIGDSGFMVIRDGTVLQN 601
Query: 123 SPVQQRGFNFPYQLARSGTEG-DLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIVGV 181
S F FP + T+G D+ EV+ V + GD+++A TDGLFDN+Y +IV +
Sbjct: 602 SSPMFHHFCFPLHI----TQGCDVLKLAEVYHVNLEEGDVVIAATDGLFDNLYEKEIVSI 657
Query: 182 VVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAAALEYGYR-FDGGKLDDLTVVVS 240
V G+ + L PQ A+ +AA A++ ++PF+ AA E GY GGKLD +TV++S
Sbjct: 658 VCGSLKQSLEPQKIAELVAAKAQEVGRSKTERTPFADAAKEEGYNGHKGGKLDAVTVIIS 717
Query: 241 YI 242
++
Sbjct: 718 FV 719
>At2g30170 unknown protein
Length = 298
Score = 154 bits (390), Expect = 3e-38
Identities = 91/253 (35%), Positives = 157/253 (61%), Gaps = 23/253 (9%)
Query: 8 LPHPDKVATGGEDAHFICEDEQAI-GVADGVGGWADVGVNAGLYAQELVANSARAIREEP 66
+PHPDKV GGEDA F+ + VADGV GWA+ V+ L+++EL+AN++R + ++
Sbjct: 54 IPHPDKVEKGGEDAFFVSSYRGGVMAVADGVSGWAEQDVDPSLFSKELMANASRLV-DDQ 112
Query: 67 KGSFNPVRVLEKAHSKTKAMGSSTVCIIALIDEA--LNAINLGDSGFIVIRDGSVIFKSP 124
+ ++P +++KAH+ T + GS+T+ I+A+++E L N+GD G ++R+G +IF +
Sbjct: 113 EVRYDPGFLIDKAHTATTSRGSATI-ILAMLEEVGILKIGNVGDCGLKLLREGQIIFATA 171
Query: 125 VQQRGFNFPYQLARSGT-EGDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIVGVVV 183
Q+ F+ PYQL+ G+ + L +S + V V GD+IV G+DGLFDN+++++IV +V
Sbjct: 172 PQEHYFDCPYQLSSEGSAQTYLDASFSI--VEVQKGDVIVMGSDGLFDNVFDHEIVSIVT 229
Query: 184 GATRARLGPQATAQKIAALARQRALDTKRQSPFSAAALEYGY-----------RFDGGKL 232
T +++ +A +A + DT+ +SP++ A G+ + GGKL
Sbjct: 230 KHTDV----AESSRLLAEVASSHSRDTEFESPYALEARAKGFDVPLWKKVLGKKLTGGKL 285
Query: 233 DDLTVVVSYISNS 245
DD+TV+V+ + +S
Sbjct: 286 DDVTVIVAKVVSS 298
>At4g27800 protein phosphatase homolog (PPH1)
Length = 388
Score = 33.9 bits (76), Expect = 0.083
Identities = 25/91 (27%), Positives = 45/91 (48%), Gaps = 6/91 (6%)
Query: 136 LARSGTEGDLP-SSGEVFTVPVAPG-DIIVAGTDGLFDNMYNNDIVGVVVGATRARLGPQ 193
++R +GD+ ++ ++F VP+ + I+ +DGL+D M ++D+V V R Q
Sbjct: 263 VSRIEFKGDMVVATPDIFQVPLTSDVEFIILASDGLWDYMKSSDVVSYVRDQLRKHGNVQ 322
Query: 194 ATAQKIAALARQRALDTKRQSPFSAAALEYG 224
+ +A Q ALD + Q S + G
Sbjct: 323 LACESLA----QVALDRRSQDNISIIIADLG 349
>At3g15260 putative protein phosphatase type 2C
Length = 289
Score = 33.9 bits (76), Expect = 0.083
Identities = 43/162 (26%), Positives = 71/162 (43%), Gaps = 30/162 (18%)
Query: 75 VLEKAHSKTKAMGSSTVCIIALIDEALNAINLGDSGFIVIRDGSV----------IFKSP 124
+L+KA K GS+ V I + + L N+GDS ++ ++G + K
Sbjct: 122 ILDKADDLGKG-GSTAVTAILINCQKLVVANVGDSRAVICQNGVAKPLSVDHEPNMEKDE 180
Query: 125 VQQRG---FNFP-------YQLARSGTEGDLP-----SSGEVFTVPVAPGD--IIVAGTD 167
++ RG NFP QLA + GD SS TV + D ++ +D
Sbjct: 181 IENRGGFVSNFPGDVPRVDGQLAVARAFGDKSLKMHLSSEPYVTVEIIDDDAEFLILASD 240
Query: 168 GLFDNMYNNDIVGVVVGATRARLGPQATAQKIAALARQRALD 209
GL+ M N + V + G A+ + A++ A+AR+ + D
Sbjct: 241 GLWKVMSNQEAVDSIKGIKDAKAAAKHLAEE--AVARKSSDD 280
>At1g75710 unknown protein
Length = 462
Score = 33.5 bits (75), Expect = 0.11
Identities = 22/57 (38%), Positives = 31/57 (53%), Gaps = 4/57 (7%)
Query: 8 LPHPDKVATGGEDAHFICEDEQAIGVADGVGGWADVGVNAGLYA--QELVANSARAI 62
LP D AT + + ED +GV+ G + V VNAG+Y+ +ELV + RAI
Sbjct: 395 LPAADASATAEKKS--TVEDNSVVGVSSSGGTFDSVAVNAGVYSNLEELVVYNPRAI 449
>At3g02750 protein phosphatase-2C (PP2C) like protein
Length = 492
Score = 32.7 bits (73), Expect = 0.19
Identities = 20/86 (23%), Positives = 35/86 (40%), Gaps = 20/86 (23%)
Query: 160 DIIVAGTDGLFDNMYNNDIVGVVVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAA 219
+ IV TDG++D + N D+V +V A +A + +A
Sbjct: 331 EFIVLATDGIWDVLSNEDVVAIVASAPSRSSAARALVE--------------------SA 370
Query: 220 ALEYGYRFDGGKLDDLTVVVSYISNS 245
+ Y++ K+DD V Y+ +S
Sbjct: 371 VRAWRYKYPTSKVDDCAAVCLYLDSS 396
>At5g06750 protein phosphatase 2C-like
Length = 393
Score = 31.6 bits (70), Expect = 0.41
Identities = 30/132 (22%), Positives = 53/132 (39%), Gaps = 9/132 (6%)
Query: 117 GSVIFKSPVQQRGFNFP-YQLARSGTEGDLPSSGEVFTVPVAPGD-IIVAGTDGLFDNMY 174
G K P +FP + LA L + V+T + D ++ +DGL++ M
Sbjct: 239 GDAYLKRPEFSLDPSFPRFHLAEELQRPVLSAEPCVYTRVLQTSDKFVIFASDGLWEQMT 298
Query: 175 NNDIVGVVVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDD 234
N V +V R + + + I A++R ++ + + DD
Sbjct: 299 NQQAVEIVNKHPRPGIARRLVRRAITIAAKKREMNYDDLKKVERGVRRFFH-------DD 351
Query: 235 LTVVVSYISNSV 246
+TVVV +I N +
Sbjct: 352 ITVVVIFIDNEL 363
>At5g26010 protein phosphatase - like
Length = 337
Score = 31.2 bits (69), Expect = 0.54
Identities = 22/83 (26%), Positives = 43/83 (51%), Gaps = 19/83 (22%)
Query: 162 IVAGTDGLFDNMYNNDIVGVVVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAAAL 221
+V TDG++D + N+++V ++ + + QA+A K+ A A + A + L
Sbjct: 269 LVLATDGVWDMLSNDEVVSLIWSSGKK----QASAAKMVAEAAEAAWKKR---------L 315
Query: 222 EYGYRFDGGKLDDLTVVVSYISN 244
+Y K+DD+TV+ ++ N
Sbjct: 316 KYT------KVDDITVICLFLQN 332
>At5g01700 putative protein
Length = 320
Score = 30.8 bits (68), Expect = 0.70
Identities = 25/85 (29%), Positives = 39/85 (45%), Gaps = 20/85 (23%)
Query: 160 DIIVAGTDGLFDNMYNNDIVGVVVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAA 219
+ +V TDG++D + N ++V VV G+ + R A + QRA T R
Sbjct: 243 EFVVLATDGIWDVLSNEEVVKVV-GSCKDR-------SVAAEMLVQRAARTWRT------ 288
Query: 220 ALEYGYRFDGGKLDDLTVVVSYISN 244
+F K DD VVV Y+++
Sbjct: 289 ------KFPASKADDCAVVVLYLNH 307
>At3g17090 protein phosphatase-2c, putative
Length = 384
Score = 30.0 bits (66), Expect = 1.2
Identities = 26/126 (20%), Positives = 57/126 (44%), Gaps = 13/126 (10%)
Query: 122 KSPVQQRGFNFPYQLARSGTEGDLPSSGEVFTVPVAPGD-IIVAGTDGLFDNMYNNDIVG 180
K P+ Q+ +++A + ++ + + P+ P D ++ +DGL++++ N V
Sbjct: 249 KEPISQK-----FRIAEPMKRPLMSATPTILSHPLHPNDSFLIFASDGLWEHLTNEKAVE 303
Query: 181 VVVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLTVVVS 240
+V RA + + AR+R + R S + F DD+TV+V
Sbjct: 304 IVHNHPRAGSAKRLIKAALHEAARKREM---RYSDLRKIDKKVRRHFH----DDITVIVV 356
Query: 241 YISNSV 246
++++ +
Sbjct: 357 FLNHDL 362
>At3g16800 protein phosphatase, putative
Length = 351
Score = 30.0 bits (66), Expect = 1.2
Identities = 13/45 (28%), Positives = 24/45 (52%)
Query: 162 IVAGTDGLFDNMYNNDIVGVVVGATRARLGPQATAQKIAALARQR 206
++ TDG++D M NN+ V +V G R + ++ L R++
Sbjct: 288 LILATDGMWDVMTNNEAVEIVRGVKERRKSAKRLVERAVTLWRRK 332
>At1g22280 protein phosphatase type 2C like protein
Length = 281
Score = 28.9 bits (63), Expect = 2.7
Identities = 42/174 (24%), Positives = 71/174 (40%), Gaps = 42/174 (24%)
Query: 71 NPVRVLEKAHSKTKAM-----------GSSTVCIIALIDEALNAINLGDSGFIVIRDGSV 119
+P R + KA+ KT GS+ V I + L N+GDS ++ G++
Sbjct: 98 DPRRSIAKAYEKTDQAILSNSSDLGRGGSTAVTAILINGRKLWIANVGDSRAVLSHGGAI 157
Query: 120 ----------IFKSPVQQRG---FNFP-------YQLARSGTEGD------LPSSGEVFT 153
+S ++ RG N P QLA S GD L S ++
Sbjct: 158 TQMSTDHEPRTERSSIEDRGGFVSNLPGDVPRVNGQLAVSRAFGDKGLKTHLSSEPDIKE 217
Query: 154 VPV-APGDIIVAGTDGLFDNMYNNDIVGVVVGATRARLGPQATAQKIAALARQR 206
V + D+++ +DG++ M N + + + R PQ A+++ A A +R
Sbjct: 218 ATVDSQTDVLLLASDGIWKVMTNEEAMEI----ARRVKDPQKAAKELTAEALRR 267
>At5g59220 protein phosphatase 2C - like
Length = 413
Score = 28.5 bits (62), Expect = 3.5
Identities = 35/135 (25%), Positives = 51/135 (36%), Gaps = 27/135 (20%)
Query: 89 STVCIIALIDEALNAINLGDSGFIVIRDGSVIFKSP------------VQQRGFNFPY-- 134
ST + L E + N GDS ++ R+G I S +Q G Y
Sbjct: 222 STAVVSVLTPEKIIVANCGDSRAVLCRNGKAIALSSDHKPDRPDELDRIQAAGGRVIYWD 281
Query: 135 ------QLARSGTEGD------LPSSGEVFTVPVAPG-DIIVAGTDGLFDNMYNNDIVGV 181
LA S GD + S EV A G D ++ +DGL+D + N V
Sbjct: 282 GPRVLGVLAMSRAIGDNYLKPYVISRPEVTVTDRANGDDFLILASDGLWDVVSNETACSV 341
Query: 182 VVGATRARLGPQATA 196
V R ++ Q ++
Sbjct: 342 VRMCLRGKVNGQVSS 356
>At2g23140 hypothetical protein
Length = 924
Score = 28.5 bits (62), Expect = 3.5
Identities = 19/57 (33%), Positives = 33/57 (57%), Gaps = 5/57 (8%)
Query: 57 NSARAIREEPKGSFNP-VRVLEKAHSKTKAMGSSTVCIIALIDEALNAINLGDSGFI 112
N+ +AI + G+ P + VLE S+ K ++T+ +++I+E N I +G SG I
Sbjct: 694 NNKKAIADA--GAIEPLIHVLENGSSEAKENSAATLFSLSVIEE--NKIKIGQSGAI 746
>At3g23360 hypothetical protein
Length = 256
Score = 28.1 bits (61), Expect = 4.6
Identities = 30/156 (19%), Positives = 69/156 (44%), Gaps = 18/156 (11%)
Query: 76 LEKAHSKTKAMGSSTVCIIALIDEALNAINLGDSGFIVIRDGSVIFKSPVQQRGFNFPY- 134
+ +A+ + + G S ++ + E L ++GD +V +DG ++ R + +
Sbjct: 112 MRRAYVEEERTGGSAASVMVVNGEKLAIASIGDHRVVVCKDGEA---HQIRDRKASTKHW 168
Query: 135 -QLARSGTEGDL--PSSGEVFTVPV---APGDIIVAGTDGLFDNMYNNDIVGVVVGATRA 188
Q G E D P + E+ + + + I+ G+ G+++ M + + + ++ R
Sbjct: 169 SQFIFPGEEEDESDPRNSELVVITEKINSDTEFIIIGSPGIWEVMKSQEAINLI----RH 224
Query: 189 RLGPQATAQKIAALARQRALDTKRQSPFSAAALEYG 224
P+ A+ +A + AL+ +S S + +G
Sbjct: 225 IEDPKEAAKCLA----KEALNRISKSSISCVVIRFG 256
>At5g09910 unknown protein
Length = 431
Score = 27.7 bits (60), Expect = 6.0
Identities = 18/58 (31%), Positives = 28/58 (48%), Gaps = 2/58 (3%)
Query: 179 VGVVVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAAALEY--GYRFDGGKLDD 234
VG+++ A AR +A + L R+R + SP SA +L + R D G D+
Sbjct: 194 VGLIMAAKEARYDKEALTKIFHMLIRRRYFSDELPSPSSAWSLSHAPSQRLDEGTSDE 251
>At4g22730 leucine rich repeat receptor kinase-like protein
Length = 688
Score = 27.3 bits (59), Expect = 7.8
Identities = 15/42 (35%), Positives = 20/42 (46%)
Query: 41 ADVGVNAGLYAQELVANSARAIREEPKGSFNPVRVLEKAHSK 82
AD+G AGL +L NS + GS + VL H+K
Sbjct: 134 ADIGSMAGLQVMDLCCNSLTGKIPKNIGSLKKLNVLSLQHNK 175
>At2g40860 putative protein phosphatase 2C
Length = 404
Score = 27.3 bits (59), Expect = 7.8
Identities = 13/54 (24%), Positives = 29/54 (53%), Gaps = 3/54 (5%)
Query: 153 TVPVAPGDIIVAGTDGLFDNMYNNDIVGVVVGATRARLGPQATAQKIAALARQR 206
T+ A + +V +DGL+D M + +++G++ + P ++++A A R
Sbjct: 331 TILSADDEFLVMASDGLWDVMNDEEVIGIIRDTVKE---PSMCSKRLATEAAAR 381
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,413,887
Number of Sequences: 26719
Number of extensions: 220127
Number of successful extensions: 500
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 478
Number of HSP's gapped (non-prelim): 20
length of query: 248
length of database: 11,318,596
effective HSP length: 97
effective length of query: 151
effective length of database: 8,726,853
effective search space: 1317754803
effective search space used: 1317754803
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC147178.4


