

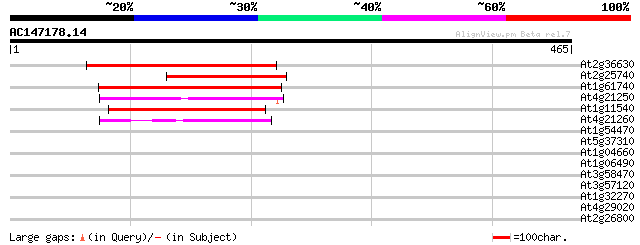
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147178.14 + phase: 0
(465 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g36630 unknown protein 219 3e-57
At2g25740 unknown protein 150 2e-36
At1g61740 unknown protein 120 1e-27
At4g21250 hypothetical protein 116 3e-26
At1g11540 hypothetical protein; similar to EST gb|AI998648.1 116 3e-26
At4g21260 hypothetical protein 77 2e-14
At1g54470 hypothetical protein 40 0.002
At5g37310 multispanning membrane protein - like 33 0.33
At1g04660 unknown protein 30 2.8
At1g06490 glucan synthase, putative 30 3.7
At3g58470 unknown protein 29 4.8
At3g57120 unknown protein 29 6.3
At1g32270 syntaxin, putative 29 6.3
At4g29020 glycine-rich protein like 28 8.2
At2g26800 hydroxymethylglutaryl-CoA lyase like protein 28 8.2
>At2g36630 unknown protein
Length = 459
Score = 219 bits (558), Expect = 3e-57
Identities = 97/158 (61%), Positives = 135/158 (85%)
Query: 64 SGKSSYEPVWPEMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSST 123
S S+ E +WP++KF WK+++ ++I FLG+A G+VGGVGGGGIFVPML LI+GFD KS+
Sbjct: 42 SSLSATEKIWPDLKFSWKLVLATVIAFLGSACGTVGGVGGGGIFVPMLTLILGFDTKSAA 101
Query: 124 AISKCMIMGAALSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADW 183
AISKCMIMGA+ S+V+YN+R+R+PT ++P++DYDLALLFQPML+LGI++GV +V+F W
Sbjct: 102 AISKCMIMGASASSVWYNVRVRHPTKEVPILDYDLALLFQPMLLLGITVGVSLSVVFPYW 161
Query: 184 MVTVLLIILFIGTSTKALIKGINTWKKETMLKKETAKQ 221
++TVL+IILF+GTS+++ KGI WK+ET+LK E A+Q
Sbjct: 162 LITVLIIILFVGTSSRSFFKGIEMWKEETLLKNEMAQQ 199
Score = 32.7 bits (73), Expect = 0.44
Identities = 23/77 (29%), Positives = 40/77 (51%), Gaps = 5/77 (6%)
Query: 75 EMKFDW---KIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISK-CMI 130
E +W +I + G +G +G + G GGG + P+L L IG P+ ++A + M+
Sbjct: 307 EATIEWTPLSLIFCGLCGLIGGIVGGLLGSGGGFVLGPLL-LEIGVIPQVASATATFVMM 365
Query: 131 MGAALSTVYYNMRLRNP 147
++LS V + + R P
Sbjct: 366 FSSSLSVVEFYLLKRFP 382
>At2g25740 unknown protein
Length = 922
Score = 150 bits (378), Expect = 2e-36
Identities = 72/99 (72%), Positives = 85/99 (85%)
Query: 131 MGAALSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLI 190
MGA++STVYYN+RLR+PTLDMP+IDYDLALL QPMLMLGISIGV NV+F DW+VTVLLI
Sbjct: 1 MGASVSTVYYNLRLRHPTLDMPIIDYDLALLIQPMLMLGISIGVAFNVIFPDWLVTVLLI 60
Query: 191 ILFIGTSTKALIKGINTWKKETMLKKETAKQLEEEPKTG 229
+LF+GTSTKA +KG TW KET+ KKE AK+LE +G
Sbjct: 61 VLFLGTSTKAFLKGSETWNKETIEKKEAAKRLESNGVSG 99
>At1g61740 unknown protein
Length = 458
Score = 120 bits (302), Expect = 1e-27
Identities = 54/154 (35%), Positives = 100/154 (64%), Gaps = 2/154 (1%)
Query: 74 PEMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMGA 133
P ++ I+ ++ FL +++ S GG+GGGG++VP++ ++ G D K++++ S M+ G
Sbjct: 51 PRIELTTSTIIAGLLSFLASSISSAGGIGGGGLYVPIMTIVAGLDLKTASSFSAFMVTGG 110
Query: 134 ALSTVYYNMRLRNPTLD-MPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIIL 192
+++ V N+ +RNP LID+DLALL +P ++LG+SIGVICN++F +W++T L +
Sbjct: 111 SIANVGCNLFVRNPKSGGKTLIDFDLALLLEPCMLLGVSIGVICNLVFPNWLITSLFAVF 170
Query: 193 FIGTSTKALIKGINTWKKET-MLKKETAKQLEEE 225
++ K G+ W+ E+ M+K + ++EE+
Sbjct: 171 LAWSTLKTFGNGLYYWRLESEMVKIRESNRIEED 204
Score = 32.7 bits (73), Expect = 0.44
Identities = 17/55 (30%), Positives = 30/55 (53%), Gaps = 2/55 (3%)
Query: 87 IIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIM-GAALSTVYY 140
++ L LG V G+GGG + P+L L +G P+ + A M++ + +S + Y
Sbjct: 324 VMALLAGVLGGVFGIGGGMLISPLL-LQVGIAPEVTAATCSFMVLFSSTMSAIQY 377
>At4g21250 hypothetical protein
Length = 449
Score = 116 bits (290), Expect = 3e-26
Identities = 57/161 (35%), Positives = 95/161 (58%), Gaps = 13/161 (8%)
Query: 75 EMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMGAA 134
E+K II+ ++ FL A + S GG+GGGG+F+P++ ++ G D K++++ S M+ G +
Sbjct: 51 ELKLSSAIIMAGVLCFLAALISSAGGIGGGGLFIPIMTIVAGVDLKTASSFSAFMVTGGS 110
Query: 135 LSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILFI 194
++ V N+ L+DYDLALL +P ++LG+SIGVICN + +W++TVL +
Sbjct: 111 IANVISNL-----FGGKALLDYDLALLLEPCMLLGVSIGVICNRVLPEWLITVLFAVFLA 165
Query: 195 GTSTKALIKGINTWKKETMLKKETAK--------QLEEEPK 227
+ K G+ WK E+ + +E+ Q+EEE K
Sbjct: 166 WSILKTCRSGVKFWKLESEIARESGHGRPERGQGQIEEETK 206
Score = 34.7 bits (78), Expect = 0.11
Identities = 28/110 (25%), Positives = 49/110 (44%), Gaps = 10/110 (9%)
Query: 88 IGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMI-MGAALSTVYYNMRLRN 146
+ FL LG + G+GGG + P+L L G P+ + A + M+ A +S V Y
Sbjct: 316 MSFLAGLLGGIFGIGGGMLISPLL-LQSGIPPQITAATTSFMVFFSATMSAVQY------ 368
Query: 147 PTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILFIGT 196
L + + + D A +F + L +G++ +I+ +GT
Sbjct: 369 --LLLGMQNTDTAYVFSFICFLASLLGLVLVQKAVAQFGRASIIVFSVGT 416
>At1g11540 hypothetical protein; similar to EST gb|AI998648.1
Length = 450
Score = 116 bits (290), Expect = 3e-26
Identities = 53/131 (40%), Positives = 86/131 (65%), Gaps = 1/131 (0%)
Query: 83 IVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMGAALSTVYYNM 142
I+ +++ F A++ S GG+GGGG+F+ ++ +I G + K++++ S M+ G + + V N+
Sbjct: 56 IIAAVLSFFAASISSAGGIGGGGLFLSIMTIIAGLEMKTASSFSAFMVTGVSFANVGCNL 115
Query: 143 RLRNP-TLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILFIGTSTKAL 201
LRNP + D LID+DLAL QP L+LG+SIGVICN MF +W+V L + ++ K
Sbjct: 116 FLRNPKSRDKTLIDFDLALTIQPCLLLGVSIGVICNRMFPNWLVLFLFAVFLAWSTMKTC 175
Query: 202 IKGINTWKKET 212
KG++ W E+
Sbjct: 176 KKGVSYWNLES 186
Score = 32.3 bits (72), Expect = 0.57
Identities = 17/55 (30%), Positives = 31/55 (55%), Gaps = 2/55 (3%)
Query: 87 IIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIM-GAALSTVYY 140
++ L LG + G+GGG + P+L L IG P+ + A M++ +++S + Y
Sbjct: 316 VMALLAGVLGGLFGIGGGMLISPLL-LQIGIAPEVTAATCSFMVLFSSSMSAIQY 369
>At4g21260 hypothetical protein
Length = 393
Score = 77.0 bits (188), Expect = 2e-14
Identities = 40/143 (27%), Positives = 76/143 (52%), Gaps = 21/143 (14%)
Query: 75 EMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMGAA 134
E+K ++V ++ F A + S G+ D K++++ S M+ G +
Sbjct: 55 ELKLSPALVVAGVLCFTAALISSASGI----------------DLKAASSFSAFMVTGGS 98
Query: 135 LSTVYYNMRLRNPTLDMPLIDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILFI 194
++ + + N LIDYDLALL +P ++LG+S+GVICN +F +W++T L ++ +
Sbjct: 99 IANL-----INNHFGCKKLIDYDLALLLEPCMLLGVSVGVICNKVFPEWLITGLFVVFLM 153
Query: 195 GTSTKALIKGINTWKKETMLKKE 217
+S + G +WK +L+++
Sbjct: 154 WSSMETCENGHTSWKLSLILREK 176
Score = 31.6 bits (70), Expect = 0.97
Identities = 25/103 (24%), Positives = 46/103 (44%), Gaps = 10/103 (9%)
Query: 95 LGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMI-MGAALSTVYYNMRLRNPTLDMPL 153
LG + G+GGG I P+L L G P+ + A + M+ A +S V Y L + +
Sbjct: 267 LGGIFGIGGGMIISPLL-LRAGIPPQVTAATTSFMVFFSATMSGVQY--------LLLGM 317
Query: 154 IDYDLALLFQPMLMLGISIGVICNVMFADWMVTVLLIILFIGT 196
+ + A +F + ++G++ +I+ +GT
Sbjct: 318 QNTEAAYVFSVICFFASTLGLVFAQKVVPHFRRASIIVFLVGT 360
>At1g54470 hypothetical protein
Length = 112
Score = 40.4 bits (93), Expect = 0.002
Identities = 17/50 (34%), Positives = 29/50 (58%)
Query: 75 EMKFDWKIIVGSIIGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTA 124
E+K IV ++ FL A + S G+G GG+F+P+ L+ D K+ ++
Sbjct: 55 ELKLSLAAIVAGVLYFLAALISSACGIGSGGLFIPITTLVSRLDLKTGSS 104
>At5g37310 multispanning membrane protein - like
Length = 558
Score = 33.1 bits (74), Expect = 0.33
Identities = 45/185 (24%), Positives = 74/185 (39%), Gaps = 23/185 (12%)
Query: 70 EPVWPEMKFDWKIIVGSIIGF------LGAALGSVGGVGGGGIFVPMLALIIGFDPKSST 123
E V + + WK+I G + F L AALGS + +F+ MLAL+ F P +
Sbjct: 261 EAVDDQEETGWKLIHGDVFRFPKHKSLLAAALGSGTQLFTLAVFIFMLALVGVFYPYNRG 320
Query: 124 AISKCMIM---------GAALSTVYYNMR----LRNPTLDMPLIDYDLALLFQPMLMLGI 170
A+ +++ G ++ Y + +RN L L L + F + + I
Sbjct: 321 ALFTALVVIYALTSGIAGYTAASFYCQLEGTNWVRNVILTGSLFCGPLLITFSFLNTVAI 380
Query: 171 SIGVICNVMFADWMVTVLLIILFIGTSTKALIKGI--NTWKKETMLKKETAKQLEEEPKT 228
+ + F + V+ +I + TS ++ GI K E T K E P
Sbjct: 381 AYQATAALPFG--TIVVIFLIWALVTSPLLILGGIAGKNRKSEFQAPCRTTKYPREIPPM 438
Query: 229 GAYQR 233
Y+R
Sbjct: 439 RWYRR 443
>At1g04660 unknown protein
Length = 212
Score = 30.0 bits (66), Expect = 2.8
Identities = 13/21 (61%), Positives = 15/21 (70%)
Query: 84 VGSIIGFLGAALGSVGGVGGG 104
+G + G GA LG VGGVGGG
Sbjct: 139 LGGVGGLGGAGLGGVGGVGGG 159
>At1g06490 glucan synthase, putative
Length = 1933
Score = 29.6 bits (65), Expect = 3.7
Identities = 16/56 (28%), Positives = 28/56 (49%), Gaps = 6/56 (10%)
Query: 159 ALLFQPMLMLGISIGVICNVMFADWMVTVLLI------ILFIGTSTKALIKGINTW 208
ALLF L + + V+C + +D ++L IL IG + +++ KG+ W
Sbjct: 1812 ALLFLGFLSVMTVLFVVCGLTISDLFASILAFLPTGWAILLIGQALRSVFKGLGFW 1867
>At3g58470 unknown protein
Length = 248
Score = 29.3 bits (64), Expect = 4.8
Identities = 12/37 (32%), Positives = 22/37 (59%)
Query: 122 STAISKCMIMGAALSTVYYNMRLRNPTLDMPLIDYDL 158
S I C + A T+Y ++ R+P+L + L++YD+
Sbjct: 97 SQRIPGCRVACIACPTLYVYLKKRDPSLQVQLLEYDM 133
>At3g57120 unknown protein
Length = 456
Score = 28.9 bits (63), Expect = 6.3
Identities = 13/21 (61%), Positives = 15/21 (70%)
Query: 85 GSIIGFLGAALGSVGGVGGGG 105
GSII LGA++ G VGGGG
Sbjct: 168 GSIINLLGASVSGGGRVGGGG 188
>At1g32270 syntaxin, putative
Length = 416
Score = 28.9 bits (63), Expect = 6.3
Identities = 13/52 (25%), Positives = 28/52 (53%), Gaps = 1/52 (1%)
Query: 191 ILFIGTSTKALIKGINTWKKETMLKKETAKQLEEEPKTGAYQRVQVKYKMKW 242
+ ++G+ L++ + + +LK K+ E++ KTG Y R+ VK + +
Sbjct: 69 LFYLGSKNTMLLRALFEGQTLVLLKGNERKKFEDDQKTGVY-RIDVKLSINF 119
>At4g29020 glycine-rich protein like
Length = 158
Score = 28.5 bits (62), Expect = 8.2
Identities = 17/26 (65%), Positives = 19/26 (72%), Gaps = 3/26 (11%)
Query: 84 VGSIIGFLGAA--LGSVGGVGG-GGI 106
+GS IG LG A LG +GGVGG GGI
Sbjct: 120 IGSGIGGLGGAGGLGGIGGVGGLGGI 145
>At2g26800 hydroxymethylglutaryl-CoA lyase like protein
Length = 468
Score = 28.5 bits (62), Expect = 8.2
Identities = 15/57 (26%), Positives = 27/57 (47%)
Query: 88 IGFLGAALGSVGGVGGGGIFVPMLALIIGFDPKSSTAISKCMIMGAALSTVYYNMRL 144
+G +LG G+G G VPML ++ P A+ G AL+ + ++++
Sbjct: 330 MGCFEISLGDTIGIGTPGSVVPMLEAVMAVVPADKLAVHFHDTYGQALANILVSLQM 386
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.351 0.156 0.553
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,272,779
Number of Sequences: 26719
Number of extensions: 358202
Number of successful extensions: 2135
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 2092
Number of HSP's gapped (non-prelim): 41
length of query: 465
length of database: 11,318,596
effective HSP length: 103
effective length of query: 362
effective length of database: 8,566,539
effective search space: 3101087118
effective search space used: 3101087118
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147178.14


