

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147178.12 + phase: 0
(163 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
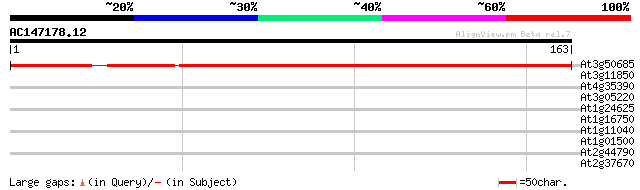
Score E
Sequences producing significant alignments: (bits) Value
At3g50685 unknown protein 212 6e-56
At3g11850 unknown protein (At3g11850) 28 1.7
At4g35390 putative protein 28 2.2
At3g05220 unknown protein 28 2.8
At1g24625 zinc finger protein ZFP7 27 6.3
At1g16750 unknown protein 27 6.3
At1g11040 DnaJ isolog 27 6.3
At1g01500 unknown protein 27 6.3
At2g44790 phytocyanin, blue copper-binding protein II 26 8.3
At2g37670 putative WD-40 repeat protein 26 8.3
>At3g50685 unknown protein
Length = 158
Score = 212 bits (540), Expect = 6e-56
Identities = 107/163 (65%), Positives = 127/163 (77%), Gaps = 5/163 (3%)
Query: 1 MLLFSPISTPKTFTNHLNHPLRKPKSHHVRLKLKITNMAKEGSDTNSSPTEIAAIAGGLI 60
MLL SPIS + H + +R+ ++ ++ AK+ +DT E AAIAG L+
Sbjct: 1 MLLLSPISASLPPSFHRGNLIRRS----IKPLGRVVAKAKDNTDTGGF-LETAAIAGSLV 55
Query: 61 STPVIGWSLYTLKTTGCGLPPGPGGSIGALEGVSYLVVVGIVAWSIYTKTKTGSGLPNGP 120
STPVIGWSLYTLKTTGCGLPPGP G IGALEGVSYLVVVGIV WS+YTKTKTGSGLPNGP
Sbjct: 56 STPVIGWSLYTLKTTGCGLPPGPAGLIGALEGVSYLVVVGIVGWSLYTKTKTGSGLPNGP 115
Query: 121 FGLLGAVEGLSYLALVAIVVVFGLQYFQQGYIPGPLPADQCFG 163
FGLLGAVEGLSYL+++AI+VVFG+Q+ G +PGPLP+DQCFG
Sbjct: 116 FGLLGAVEGLSYLSVLAILVVFGIQFLDNGSVPGPLPSDQCFG 158
>At3g11850 unknown protein (At3g11850)
Length = 504
Score = 28.5 bits (62), Expect = 1.7
Identities = 17/47 (36%), Positives = 23/47 (48%)
Query: 21 LRKPKSHHVRLKLKITNMAKEGSDTNSSPTEIAAIAGGLISTPVIGW 67
+R K+ V LK +++KE P A+IAG L TPV W
Sbjct: 410 MRTEKAQMVLLKEIAQHLSKEVVPQRRLPLRKASIAGPLTFTPVFKW 456
>At4g35390 putative protein
Length = 270
Score = 28.1 bits (61), Expect = 2.2
Identities = 31/89 (34%), Positives = 39/89 (42%), Gaps = 15/89 (16%)
Query: 42 GSDTNSSPTEIAAIAGGLISTPVIGWSLYTLKTTGCGLPP----GPGGSIGALEGVSYLV 97
G+ TN + + AA AGG + T + L TG LPP G GG L G V
Sbjct: 96 GAVTNVTIRQPAAPAGGGVIT--LHGRFDILSLTGTALPPPAPPGAGGLTVYLAGGQGQV 153
Query: 98 VVGIVAWSIYTKTKTGSGLPNGPFGLLGA 126
V G VA GS + +GP L+ A
Sbjct: 154 VGGNVA---------GSLIASGPVVLMAA 173
>At3g05220 unknown protein
Length = 541
Score = 27.7 bits (60), Expect = 2.8
Identities = 27/85 (31%), Positives = 35/85 (40%), Gaps = 16/85 (18%)
Query: 76 GCGLPPGPGGSIGALEGVSYLVVVGIVAWSIYTKTKTGSGLPNGPFGLLGAVEGLSYLAL 135
G G P GPGG +G + + G + G G P GP G +G SY A+
Sbjct: 391 GPGGPMGPGGPMGQGGPMGMMGPGG-------PMSMMGPGGPMGPMG----GQGGSYPAV 439
Query: 136 VAIVVVFGLQYFQQGYIPGPLPADQ 160
+ + G GY PGP A Q
Sbjct: 440 QGLPMSGG-----GGYYPGPPQASQ 459
>At1g24625 zinc finger protein ZFP7
Length = 209
Score = 26.6 bits (57), Expect = 6.3
Identities = 14/34 (41%), Positives = 20/34 (58%), Gaps = 2/34 (5%)
Query: 18 NHPLRKPKSHHVRLKLKITNMAKEGSDTNSSPTE 51
N L KP+S+HV L LK+ + + DT S+ E
Sbjct: 22 NQDLSKPESNHVSLDLKLNDTFND--DTKSTKCE 53
>At1g16750 unknown protein
Length = 529
Score = 26.6 bits (57), Expect = 6.3
Identities = 22/89 (24%), Positives = 43/89 (47%), Gaps = 4/89 (4%)
Query: 8 STPKTFTNHLNHPLRKPKSHHVR--LKLKITNMAKEGSDTNSSPTEIAAIAGGLISTPVI 65
+T + T ++ PL +P+ + LKLK+++ ++ + P A++ G+ S+P +
Sbjct: 368 ATIEYMTLKMSPPLHRPQIALLLSILKLKVSDEQRQAGISTPEPLVSFALSCGMHSSPAV 427
Query: 66 GWSLYTLKTTGCGLPPGPGGSIGALEGVS 94
+Y+ + G L I A GVS
Sbjct: 428 --RIYSAENVGEELEEAQKDYIQASVGVS 454
>At1g11040 DnaJ isolog
Length = 438
Score = 26.6 bits (57), Expect = 6.3
Identities = 16/44 (36%), Positives = 21/44 (47%), Gaps = 6/44 (13%)
Query: 75 TGCGLPPGPGGSIGALEGVSYLVVVGIVAWSIYTKTKTGSGLPN 118
TGC L S+ L G S + VG V + + K G G+PN
Sbjct: 361 TGCKL------SVPLLSGESMSITVGDVIFHGFEKAIKGQGMPN 398
>At1g01500 unknown protein
Length = 327
Score = 26.6 bits (57), Expect = 6.3
Identities = 21/94 (22%), Positives = 30/94 (31%), Gaps = 27/94 (28%)
Query: 21 LRKPKSHHVRLKLKITNMAKEGSDTNSSPTEIAAIAG----------------------- 57
+R P S LKL+ + +E S+ TE + G
Sbjct: 82 VRVPSSQTASLKLRRDRVDRESSEVTYVSTETVRVTGCVDFEVYDNEDMVLCGNLDRIEG 141
Query: 58 ----GLISTPVIGWSLYTLKTTGCGLPPGPGGSI 87
G +S P GW + G G GP S+
Sbjct: 142 AWNNGTVSDPKTGWGMDCYIAMGNGHVSGPSASV 175
>At2g44790 phytocyanin, blue copper-binding protein II
Length = 202
Score = 26.2 bits (56), Expect = 8.3
Identities = 21/61 (34%), Positives = 27/61 (43%), Gaps = 9/61 (14%)
Query: 43 SDTNSSPTEIAAIAGGLISTPVIGWSLYTLKTTGCGL--PPGPGGSIGALEGVSYLVVVG 100
S T +PT + G TP T T G G PP P + GA +GV V+VG
Sbjct: 139 SSTPGTPTTPESPPSGGSPTP-------TTPTPGAGSTSPPPPPKASGASKGVMSYVLVG 191
Query: 101 I 101
+
Sbjct: 192 V 192
>At2g37670 putative WD-40 repeat protein
Length = 903
Score = 26.2 bits (56), Expect = 8.3
Identities = 12/31 (38%), Positives = 18/31 (57%), Gaps = 1/31 (3%)
Query: 64 VIGWS-LYTLKTTGCGLPPGPGGSIGALEGV 93
V+ WS L+ + T C P G G IG+ +G+
Sbjct: 581 VVEWSDLHEMVTAACYTPDGQGALIGSHKGI 611
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.140 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,194,648
Number of Sequences: 26719
Number of extensions: 191471
Number of successful extensions: 397
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 392
Number of HSP's gapped (non-prelim): 10
length of query: 163
length of database: 11,318,596
effective HSP length: 92
effective length of query: 71
effective length of database: 8,860,448
effective search space: 629091808
effective search space used: 629091808
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC147178.12


