

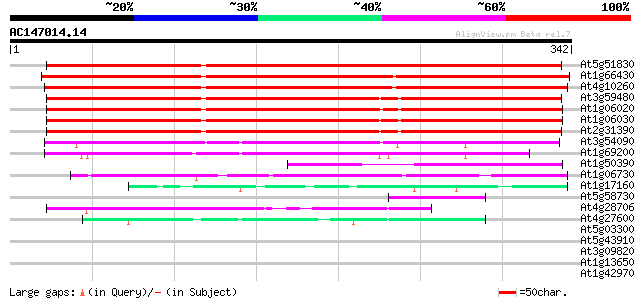
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147014.14 + phase: 0 /pseudo
(342 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g51830 fructokinase 1 404 e-113
At1g66430 putative fructokinase 396 e-111
At4g10260 fructokinase - like protein 369 e-102
At3g59480 fructokinase-like protein 360 e-100
At1g06020 fructokinase, putative 355 2e-98
At1g06030 fructokinase, putative 355 3e-98
At2g31390 putative fructokinase 350 5e-97
At3g54090 fructokinase - like protein 160 9e-40
At1g69200 putative fructokinase 144 7e-35
At1g50390 fructokinase, putative 142 3e-34
At1g06730 sugar kinase, putative 70 1e-12
At1g17160 ribokinase like protein 50 2e-06
At5g58730 unknown protein 48 9e-06
At4g28706 unknown protein 44 1e-04
At4g27600 carbohydrate kinase like protein 44 1e-04
At5g03300 adenosine kinase (MOK16.21) 38 0.009
At5g43910 unknown protein 37 0.012
At3g09820 adenosine kinase like protein 33 0.17
At1g13650 hypothetical protein 32 0.66
At1g42970 putative glyceraldehyde-3-phosphate dehydrogenase (At1... 30 1.9
>At5g51830 fructokinase 1
Length = 343
Score = 404 bits (1039), Expect = e-113
Identities = 201/314 (64%), Positives = 255/314 (81%), Gaps = 2/314 (0%)
Query: 23 LVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVAISRLGGSSAFIGKVGNDEF 82
LVVCFGEM+I+ VPT+ GVSL++A A+KK+P GA A V+V +SRLGGSSAFIGKVG+DEF
Sbjct: 23 LVVCFGEMLIDFVPTVGGVSLAEAPAFKKAPGGAPANVAVGVSRLGGSSAFIGKVGDDEF 82
Query: 83 GHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMFYRNPSADILFRSEEI 142
G ML+DIL+ N VDNSG+ FD +ARTALAF +L+ DG+ EF+F+R+PSAD+L E+
Sbjct: 83 GRMLADILRLNNVDNSGMRFDHNARTALAFVTLRG--DGEREFLFFRHPSADMLLLESEL 140
Query: 143 DKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAPNLTVPLWPSTEAARE 202
DK+LI+KA IFHYGS+SLI+EP RST + A+ AK GS+LSY PNL +PLWPS EAAR+
Sbjct: 141 DKNLIQKAKIFHYGSISLIEEPCRSTQLVAMKIAKAAGSLLSYDPNLRLPLWPSEEAARK 200
Query: 203 GIMSIWNYADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNLKLLLVTEGIKGCRYYTK 262
IMSIWN ADVIK+S +EI LT G+DPYDD ++++KLFH NLKLL+V+EG GCRYYT+
Sbjct: 201 EIMSIWNLADVIKISEDEITFLTGGDDPYDDDVVLQKLFHPNLKLLVVSEGPNGCRYYTQ 260
Query: 263 DFKGWVYGFEVEAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAATVT 322
+FKG V G +V+ +DTTGAGD+FV G L+ L++ + KDEK LREAL FANACGA TVT
Sbjct: 261 EFKGRVGGVKVKPVDTTGAGDAFVSGLLNSLASDLTLLKDEKKLREALLFANACGAITVT 320
Query: 323 GRGAIPSLPTKSSV 336
RGAIP++P+ +V
Sbjct: 321 ERGAIPAMPSMDAV 334
>At1g66430 putative fructokinase
Length = 384
Score = 396 bits (1018), Expect = e-111
Identities = 203/322 (63%), Positives = 246/322 (76%), Gaps = 3/322 (0%)
Query: 20 KGPLVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVAISRLGGSSAFIGKVGN 79
+ P VVCFGEM+I+ VPT G+SL+DA A+KK+P GA A V+V I+RLGGSSAFIGKVG
Sbjct: 62 ESPYVVCFGEMLIDFVPTTSGLSLADAPAFKKAPGGAPANVAVGIARLGGSSAFIGKVGE 121
Query: 80 DEFGHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMFYRNPSADILFRS 139
DEFG+ML++ILK N V+N G+ FD ARTALAF +L N +G+ EFMFYRNPSAD+L
Sbjct: 122 DEFGYMLANILKDNNVNNDGMRFDPGARTALAFVTLTN--EGEREFMFYRNPSADMLLEE 179
Query: 140 EEIDKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAPNLTVPLWPSTEA 199
E+D LIKKA IFHYGS+SLI EP +S HI A AK G ILSY PNL +PLWPS +
Sbjct: 180 SELDFDLIKKAKIFHYGSISLITEPCKSAHISAAKAAKEAGVILSYDPNLRLPLWPSADN 239
Query: 200 AREGIMSIWNYADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNLKLLLVTEGIKGCRY 259
ARE I+SIW AD+IK+S EEI LT+G DPYDD +++KLFH LKLLLVTEG +GCRY
Sbjct: 240 AREEILSIWETADIIKISEEEIVFLTKGEDPYDDN-VVRKLFHPKLKLLLVTEGPEGCRY 298
Query: 260 YTKDFKGWVYGFEVEAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAA 319
YTKDF G V+G +V+ +DTTGAGD+FV G LS L+ + +DE+ LREAL FANACGA
Sbjct: 299 YTKDFSGRVHGLKVDVVDTTGAGDAFVAGILSQLANDLSLLQDEERLREALMFANACGAL 358
Query: 320 TVTGRGAIPSLPTKSSVLRVML 341
TV RGAIP+LPTK +V +L
Sbjct: 359 TVKVRGAIPALPTKEAVHEALL 380
>At4g10260 fructokinase - like protein
Length = 324
Score = 369 bits (946), Expect = e-102
Identities = 189/319 (59%), Positives = 237/319 (74%), Gaps = 4/319 (1%)
Query: 22 PLVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVAISRLGGSSAFIGKVGNDE 81
PL+V FGEM+I+ VP GVSL+++ + K+P GA A V+ AI++LGG SAFIGK G+DE
Sbjct: 5 PLIVSFGEMLIDFVPDTSGVSLAESTGFLKAPGGAPANVACAITKLGGKSAFIGKFGDDE 64
Query: 82 FGHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMFYRNPSADILFRSEE 141
FGHML +ILK+NGV++ G+ FD +ARTALAF +LK DG+ EFMFYRNPSAD+L + E
Sbjct: 65 FGHMLVNILKKNGVNSEGVCFDTNARTALAFVTLKK--DGEREFMFYRNPSADMLLKESE 122
Query: 142 IDKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAPNLTVPLWPSTEAAR 201
++K LIKKA IFHYGS+SLI EP R+ H+ A+ AK G +LSY PN+ +PLWPSTEAA
Sbjct: 123 LNKDLIKKAKIFHYGSISLISEPCRTAHMAAMKTAKDAGVLLSYDPNVRLPLWPSTEAAI 182
Query: 202 EGIMSIWNYADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNLKLLLVTEGIKGCRYYT 261
EGI SIWN AD+IKVS +E+ LT G+ DD ++ L H LKLL+VT+G KGCRYYT
Sbjct: 183 EGIKSIWNEADIIKVSDDEVTFLTRGDAEKDD--VVLSLMHDKLKLLIVTDGEKGCRYYT 240
Query: 262 KDFKGWVYGFEVEAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAATV 321
K FKG V G+ V+A+DTTGAGDSFVG FL L I DE L+EAL FANACGA
Sbjct: 241 KKFKGRVPGYAVKAVDTTGAGDSFVGAFLVSLGKDGSILDDEGKLKEALAFANACGAVCT 300
Query: 322 TGRGAIPSLPTKSSVLRVM 340
T +GAIP+LPT + ++M
Sbjct: 301 TQKGAIPALPTPADAQKLM 319
>At3g59480 fructokinase-like protein
Length = 326
Score = 360 bits (923), Expect = e-100
Identities = 185/314 (58%), Positives = 238/314 (74%), Gaps = 4/314 (1%)
Query: 23 LVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVAISRLGGSSAFIGKVGNDEF 82
L+V FGEM+I+ VPT+ GVSL+DA + K+P GA A V++AISRLGG +AF+GK+G+DEF
Sbjct: 10 LIVSFGEMLIDFVPTVSGVSLADAPGFIKAPGGAPANVAIAISRLGGRAAFVGKLGDDEF 69
Query: 83 GHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMFYRNPSADILFRSEEI 142
GHML+ ILKQNGV G+ FD ARTALAF +L++ DG+ EFMFYRNPSAD+L R +E+
Sbjct: 70 GHMLAGILKQNGVSAEGINFDTGARTALAFVTLRS--DGEREFMFYRNPSADMLLRPDEL 127
Query: 143 DKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAPNLTVPLWPSTEAARE 202
+ +I+ A +FHYGS+SLI EP RS H++A+ AK G++LSY PNL +PLWPS E A++
Sbjct: 128 NLDVIRSAKVFHYGSISLIVEPCRSAHLKAMEVAKEAGALLSYDPNLRLPLWPSKEEAQK 187
Query: 203 GIMSIWNYADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNLKLLLVTEGIKGCRYYTK 262
I+SIW+ A+VIKVS EE+ LT G+D DD+ + L+H NLKLLLVT G KGCRYYTK
Sbjct: 188 QILSIWDKAEVIKVSDEELMFLT-GSDKVDDETAL-SLWHSNLKLLLVTLGEKGCRYYTK 245
Query: 263 DFKGWVYGFEVEAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAATVT 322
F+G V F V+A+DTTGAGDSFVG L + + + +DE LRE L ANACGA T T
Sbjct: 246 SFRGSVDPFHVDAVDTTGAGDSFVGALLCKIVDDRAVLEDEARLREVLRLANACGAITTT 305
Query: 323 GRGAIPSLPTKSSV 336
+GAIP+LPT+S V
Sbjct: 306 KKGAIPALPTESEV 319
>At1g06020 fructokinase, putative
Length = 345
Score = 355 bits (911), Expect = 2e-98
Identities = 183/315 (58%), Positives = 234/315 (74%), Gaps = 4/315 (1%)
Query: 23 LVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVAISRLGGSSAFIGKVGNDEF 82
L+V FGEM+I+ VPT+ GVSLS++ + K+P GA A V++A+SRLGG +AF+GK+G+D+F
Sbjct: 10 LIVSFGEMLIDFVPTVSGVSLSESPGFLKAPGGAPANVAIAVSRLGGRAAFVGKLGDDDF 69
Query: 83 GHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMFYRNPSADILFRSEEI 142
GHML+ IL++NGVD+ G+ FDE ARTALAF +L++ DG+ EFMFYRNPSAD+L R +E+
Sbjct: 70 GHMLAGILRKNGVDDQGINFDEGARTALAFVTLRS--DGEREFMFYRNPSADMLLRPDEL 127
Query: 143 DKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAPNLTVPLWPSTEAARE 202
+ LI+ A +FHYGS+SLI EP RS H++A+ AK G++LSY PNL PLWPS E AR
Sbjct: 128 NLELIRSAKVFHYGSISLITEPCRSAHMKAMEVAKEAGALLSYDPNLREPLWPSPEEART 187
Query: 203 GIMSIWNYADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNLKLLLVTEGIKGCRYYTK 262
IMSIW+ AD+IKVS E+ LTE N DDK M L+H NLKLLLVT G KGC Y+TK
Sbjct: 188 QIMSIWDKADIIKVSDVELEFLTE-NKTMDDKTAM-SLWHPNLKLLLVTLGEKGCTYFTK 245
Query: 263 DFKGWVYGFEVEAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAATVT 322
F G V F V+A+DTTGAGDSFVG L + + + +DE LR+ L FANACGA T T
Sbjct: 246 KFHGSVETFHVDAVDTTGAGDSFVGALLQQIVDDQSVLEDEARLRKVLRFANACGAITTT 305
Query: 323 GRGAIPSLPTKSSVL 337
+GAIP+LPT L
Sbjct: 306 KKGAIPALPTDIEAL 320
>At1g06030 fructokinase, putative
Length = 329
Score = 355 bits (910), Expect = 3e-98
Identities = 183/315 (58%), Positives = 236/315 (74%), Gaps = 4/315 (1%)
Query: 23 LVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVAISRLGGSSAFIGKVGNDEF 82
LVV FGEM+I+ VPT GVSLS++ + K+P GA A V++A+SRLGG +AF+GK+G+DEF
Sbjct: 11 LVVSFGEMLIDFVPTESGVSLSESSGFLKAPGGAPANVAIAVSRLGGRAAFVGKLGDDEF 70
Query: 83 GHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMFYRNPSADILFRSEEI 142
GHML+ IL++N VD+ G+ FD+ ARTALAF +L++ DG+ EFMFYRNPSAD+L R +E+
Sbjct: 71 GHMLAGILRKNDVDDQGINFDKGARTALAFVTLRS--DGEREFMFYRNPSADMLLRPDEL 128
Query: 143 DKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAPNLTVPLWPSTEAARE 202
+ LI+ A +FHYGS+SLI EP RS H++A+ AK G++LSY PNL PLWPS E AR+
Sbjct: 129 NLELIRSAKVFHYGSISLITEPCRSAHMKAMEVAKEAGALLSYDPNLREPLWPSPEEARK 188
Query: 203 GIMSIWNYADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNLKLLLVTEGIKGCRYYTK 262
IMSIW+ AD+IKVS E+ LT GN DD+ M L+H NLKLLLVT G GCRYYTK
Sbjct: 189 QIMSIWDKADIIKVSDVELEFLT-GNKTIDDETAM-SLWHPNLKLLLVTLGENGCRYYTK 246
Query: 263 DFKGWVYGFEVEAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAATVT 322
DF G V F V+A+DTTGAGDSFVG L+ + + + ++E+ LR+ L FANACGA T T
Sbjct: 247 DFHGSVETFHVDAVDTTGAGDSFVGALLNQIVDDQSVLEEEERLRKVLRFANACGAITTT 306
Query: 323 GRGAIPSLPTKSSVL 337
+GAIP+LPT L
Sbjct: 307 KKGAIPALPTDCEAL 321
>At2g31390 putative fructokinase
Length = 325
Score = 350 bits (899), Expect = 5e-97
Identities = 181/314 (57%), Positives = 235/314 (74%), Gaps = 4/314 (1%)
Query: 23 LVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVAISRLGGSSAFIGKVGNDEF 82
L+V FGEM+I+ VPT GVSL++A + K+P GA A V++A+SRLGG SAF+GK+G+DEF
Sbjct: 9 LIVSFGEMLIDFVPTESGVSLAEAPGFLKAPGGAPANVAIAVSRLGGRSAFVGKLGDDEF 68
Query: 83 GHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMFYRNPSADILFRSEEI 142
GHML+ IL++NGVD+ G+ FD ARTALAF +L+ DG EFMFYRNPSAD+L R +E+
Sbjct: 69 GHMLAGILRKNGVDDQGINFDTGARTALAFVTLRA--DGDREFMFYRNPSADMLLRPDEL 126
Query: 143 DKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAPNLTVPLWPSTEAARE 202
+ LI+ A +FHYGS+SLI EP RS H++A+ AK G++LSY PNL PLWPS E A+
Sbjct: 127 NLDLIRSAKVFHYGSISLIVEPCRSAHLKAMEVAKEAGALLSYDPNLREPLWPSKEEAKT 186
Query: 203 GIMSIWNYADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNLKLLLVTEGIKGCRYYTK 262
IMSIW+ A++IKVS E+ LT G++ DD+ + L+H NLKLLLVT G KGCRYYTK
Sbjct: 187 QIMSIWDKAEIIKVSDVELEFLT-GSNKIDDETAL-TLWHPNLKLLLVTLGEKGCRYYTK 244
Query: 263 DFKGWVYGFEVEAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAATVT 322
FKG V F V A+DTTGAGDSFVG L+ + + + +DE+ LR+ L FANACGA T T
Sbjct: 245 TFKGAVDPFHVNAVDTTGAGDSFVGALLNQIVDDRSVLEDEERLRKVLRFANACGAITTT 304
Query: 323 GRGAIPSLPTKSSV 336
+GAIP+LP+ + V
Sbjct: 305 KKGAIPALPSDAEV 318
>At3g54090 fructokinase - like protein
Length = 471
Score = 160 bits (405), Expect = 9e-40
Identities = 112/359 (31%), Positives = 173/359 (47%), Gaps = 48/359 (13%)
Query: 22 PLVVCFGEMMINLVPTID--------------GVSLSDAEAYKKSPAGATAIVSVAISRL 67
PLV CFG + VP + + D + ++P G + V+++ RL
Sbjct: 102 PLVCCFGAVQKEFVPVVRVHDNPMHPDMYSQWKMLQWDPPEFGRAPGGPPSNVAISHVRL 161
Query: 68 GGSSAFIGKVGNDEFGHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMF 127
GG +AF+GKVG D+FG L ++ Q V + FDE+++TA +K D GK
Sbjct: 162 GGRAAFMGKVGEDDFGDELVLMMNQERVQTRAVKFDENSKTACTRVKIKFKD-GKMMAET 220
Query: 128 YRNPSADILFRSEEIDKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAP 187
+ P D LF SE ++ +++K+A IFH+ S L +ST AI ++K G ++ +
Sbjct: 221 VKEPPEDSLFASE-LNLAVLKEARIFHFNSEVLTSPTMQSTLFTAIQWSKKFGGLIFFDL 279
Query: 188 NLTVPLWPSTEAAREGIMSIWNYADVIKVSVEEIRILTEGNDPYDDKM------------ 235
NL +PLW S R+ I WN A++I+VS +E+ L + D Y+ +
Sbjct: 280 NLPLPLWRSRNETRKLIKKAWNEANIIEVSQQELEFLLD-EDYYERRRNYTPQYFAEDFD 338
Query: 236 --------------IMKKLFHHNLKLLLVTEGIKGCRYYTKDFKGWVYGFEVEAI----- 276
+K L+H LKLL+VT+G YYT F G V G E I
Sbjct: 339 QTKNRRDYYHYTPEEIKSLWHDKLKLLVVTDGTLRLHYYTPTFDGVVIGTEDVLITPFTC 398
Query: 277 DTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAATVTGRGAIPSLPTKSS 335
D TG+GD+ V G + L+ +++D+ ++ L FA A G GA+ PT+S+
Sbjct: 399 DRTGSGDAVVAGIMRKLTTCPEMFEDQDVMERQLRFAVAAGIIAQWTIGAVRGFPTESA 457
>At1g69200 putative fructokinase
Length = 614
Score = 144 bits (363), Expect = 7e-35
Identities = 100/329 (30%), Positives = 163/329 (49%), Gaps = 36/329 (10%)
Query: 22 PLVVCFGEMMINLVPTIDGVS----------LSDA----EAYKKSPAGATAIVSVAISRL 67
PLV CFG VP+ + + DA E Y ++P G V++A++ L
Sbjct: 205 PLVCCFGSAQHAFVPSGRPANRLLDYELHERMRDAKWAPEKYIRAPGGCAGGVAIALASL 264
Query: 68 GGSSAFIGKVGNDEFGHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMF 127
GG AF+GK+G D++G + L V + D TA + ++K S G+ +
Sbjct: 265 GGKVAFMGKLGADDYGQAMLYYLNVCKVQTRSVKIDGKRVTACS--TMKISKRGRLKSTC 322
Query: 128 YRNPSADILFRSEEIDKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAP 187
+ + D L +SE I+ ++K+A +F++ + SL+ + ST I+AI +K G+++ Y
Sbjct: 323 IKPCAEDSLSKSE-INVDVLKEAKMFYFSTHSLLDKKMMSTTIQAIKISKQLGNVIFYDL 381
Query: 188 NLTVPLWPSTEAAREGIMSIWNYADVIKVSVEEIRIL----------TEGND----PYDD 233
NL +PLW S+E + I +WN ADVI+++ +E+ L TE ND +
Sbjct: 382 NLPLPLWHSSEETKSFIQEVWNLADVIEITKQELEFLCGIEPTEEFDTENNDISKFVHYP 441
Query: 234 KMIMKKLFHHNLKLLLVTEGIKGCRYYTKDFKGWVYGFEVEAI-----DTTGAGDSFVGG 288
+++L+H NLK+L VT G YYTK+ G V G E I D + +GD V G
Sbjct: 442 PETVEQLWHENLKVLFVTNGTSKIHYYTKEHNGAVSGMEDVPITPFTRDMSASGDGIVAG 501
Query: 289 FLSILSAHKHIYKDEKILREALDFANACG 317
+ +L+ + ++ L +A CG
Sbjct: 502 LIRMLTVQPDLMNNKGYLERTARYAIECG 530
>At1g50390 fructokinase, putative
Length = 146
Score = 142 bits (357), Expect = 3e-34
Identities = 80/168 (47%), Positives = 98/168 (57%), Gaps = 31/168 (18%)
Query: 170 IEAINYAKMCGSILSYAPNLTVPLWPSTEAAREGIMSIWNYADVIKVSVEEIRILTEGND 229
++A AK G++LSY PNL PLWPS E AR IMSIW+ AD+IK
Sbjct: 1 MKATEEAKEAGALLSYDPNLREPLWPSPEEARTQIMSIWDKADIIK-------------- 46
Query: 230 PYDDKMIMKKLFHHNLKLLLVTEGIKGCRYYTKDFKGWVYGFEVEAIDTTGAGDSFVGGF 289
LLLVT G KGCRYYTKDF G V F V+A+DTTGAGDSFVG
Sbjct: 47 -----------------LLLVTLGEKGCRYYTKDFHGSVETFHVDAVDTTGAGDSFVGAL 89
Query: 290 LSILSAHKHIYKDEKILREALDFANACGAATVTGRGAIPSLPTKSSVL 337
L+ + + + ++E+ LR+ L ANACGA T T +GAIP+LPT L
Sbjct: 90 LNQIVDDQSVLEEEERLRKVLRIANACGAITTTKKGAIPALPTDCEAL 137
>At1g06730 sugar kinase, putative
Length = 488
Score = 70.5 bits (171), Expect = 1e-12
Identities = 83/320 (25%), Positives = 134/320 (40%), Gaps = 34/320 (10%)
Query: 38 IDGVSLSDAEAYKKSPAGATAIVSVAISRLGGSSAFIGKVGNDEFGHMLSDILKQNGVDN 97
+D +S+S + K AG +++A +RLG IG VG++ +G L D+L + G+
Sbjct: 116 MDELSMSPPDK-KYWEAGGNCNMAIAAARLGLHCVAIGHVGDEIYGEFLLDVLHEEGIGT 174
Query: 98 SGLLFDEHARTALAF---------------HSLKNSDDGKPEFMFYRNPSADILFRSEEI 142
L + + +F H + D K E F + I S+E+
Sbjct: 175 VALNGGTNEKDTSSFCETLICWVLVDPLQRHGFCSRADFKEEPAF-----SWITDLSDEV 229
Query: 143 DKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAPN-LTVPLWPSTEAAR 201
++ + +F G + S S + I+YA G+ + + P L T R
Sbjct: 230 KMAIRQSKVLFCNGYD--FDDFSPSFIMSTIDYAAKVGTAIFFDPGPRGKSLSKGTPDER 287
Query: 202 EGIMSIWNYADVIKVSVEEIRILTEGNDPYD-DKMIMKKLFHHNLKLLLVTEGIKGCRYY 260
+ +DV+ ++ EE+ LT +P + I++ K ++V G KG
Sbjct: 288 RALAHFLRMSDVLLLTSEEVEALTGIRNPVKAGQEILRN--GKGTKWVIVKMGPKGSILV 345
Query: 261 TKDFKGWVYGFEVEAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAAT 320
TK F+VE +DT G GDSFV +A Y L L ANA GAAT
Sbjct: 346 TKSSVSVAPAFKVEVVDTVGCGDSFV-------AAIALGYIRNMPLVNTLTIANAVGAAT 398
Query: 321 VTGRGAIPSLPTKSSVLRVM 340
G GA ++ + V+ +M
Sbjct: 399 AMGCGAGRNVAKRHQVVDLM 418
>At1g17160 ribokinase like protein
Length = 379
Score = 49.7 bits (117), Expect = 2e-06
Identities = 68/284 (23%), Positives = 112/284 (38%), Gaps = 48/284 (16%)
Query: 73 FIGKVGNDEFGHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMFYRNPS 132
F+G++G D G ++++ L G D G+ D S+ N G M +
Sbjct: 124 FVGRLGEDAHGKLIAEAL---GDDGCGVHLDY-------VRSVNNEPTGHAVVMLQSDGQ 173
Query: 133 ADILFRS-----------EEIDKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGS 181
I+ + D +++ A G V L +E S +I+ K G
Sbjct: 174 NSIIIVGGANMKAWPEIMSDDDLEIVRNA-----GIVLLQREIPDSINIQVAKAVKKAG- 227
Query: 182 ILSYAPNLTVPLWPSTEAAREGIMSIWNYADVIKVSVEEIRILTEGNDPYDDKMIMKKLF 241
P + T E + SI D++ + E+ LT +++
Sbjct: 228 ----VPVILDVGGMDTPIPNELLDSI----DILSPNETELSRLTGMPTETFEQISQAVAK 279
Query: 242 HHNL--KLLLVTEGIKGCRYYTKDFKGWVYGF--EVEAIDTTGAGDSFVGGF-LSILSAH 296
H L K +LV G KG + + K + +DTTGAGD+F F ++++
Sbjct: 280 CHKLGVKQVLVKLGSKGSALFIQGEKPIQQSIIPAAQVVDTTGAGDTFTAAFAVAMVEGK 339
Query: 297 KHIYKDEKILREALDFANACGAATVTGRGAIPSLPTKSSVLRVM 340
H E L FA A + V +GAIPS+P + SVL+++
Sbjct: 340 SH--------EECLRFAAAAASLCVQVKGAIPSMPDRKSVLKLL 375
>At5g58730 unknown protein
Length = 353
Score = 47.8 bits (112), Expect = 9e-06
Identities = 25/59 (42%), Positives = 35/59 (58%)
Query: 232 DDKMIMKKLFHHNLKLLLVTEGIKGCRYYTKDFKGWVYGFEVEAIDTTGAGDSFVGGFL 290
D+ + M +L ++VT G KGCR Y KD + V F + +D TGAGDSF+GG +
Sbjct: 199 DEALFMDVEQMKHLCCVVVTNGEKGCRIYHKDDEMTVPPFLAKQVDPTGAGDSFLGGLI 257
>At4g28706 unknown protein
Length = 401
Score = 44.3 bits (103), Expect = 1e-04
Identities = 53/239 (22%), Positives = 97/239 (40%), Gaps = 21/239 (8%)
Query: 23 LVVCFGEMMINLVPTIDGVSLSD----AEAYKKSPAGATAIVSVAISRLGGSSAFIGKVG 78
+V+ G + ++ + T+D +D + + K G A +RLG +S I KV
Sbjct: 48 IVLGCGGIAVDFLATVDSYPQADDKIRSTSLKVQGGGNAANALTCAARLGLNSRLISKVA 107
Query: 79 NDEFGHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMFYRNPSADILFR 138
ND G + + L+ +GVD S ++ + + + + N + +P
Sbjct: 108 NDSQGKGMLEELEADGVDTSFIVVSKEGNSPFTYILVDNQTKTRTCIHTPGDPPMLPTDL 167
Query: 139 SEEIDKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAPNLTVPLWPSTE 198
S+ S + +A+I ++ V L H A+ AK A +P+ TE
Sbjct: 168 SQSSMLSALDRASIVYF-DVRL--------HETALVIAK-------EASRKKIPILVDTE 211
Query: 199 AAREGIMSIWNYADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNLKLLLVTEGIKGC 257
R+G+ + +AD + + TE + ++ L LK ++VT G GC
Sbjct: 212 KKRDGLDDLLPFADYVVCPENFPQTWTEVSST-PGALVSMLLRLPKLKFVIVTSGEHGC 269
Score = 31.6 bits (70), Expect = 0.66
Identities = 20/55 (36%), Positives = 27/55 (48%), Gaps = 4/55 (7%)
Query: 274 EAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAATVTGRGAIP 328
E +DTTGAGD+F+G L + A EK+L A A C + R +P
Sbjct: 339 ELVDTTGAGDAFIGAVLYAICAG---MPPEKMLPFAAQVA-GCSCRALGARTGLP 389
>At4g27600 carbohydrate kinase like protein
Length = 471
Score = 43.9 bits (102), Expect = 1e-04
Identities = 55/259 (21%), Positives = 98/259 (37%), Gaps = 27/259 (10%)
Query: 45 DAEAYKKSPAGATAIVSVAISRLGGSS--------AFIGKVGNDEFGHMLSDILKQNGVD 96
D +YK + G+ + VA++RLG S A G +G D G L++ V+
Sbjct: 168 DGCSYKAAAGGSLSNTLVALARLGSQSICDRPLNVAMAGSIGGDPLGSFYGTKLRRANVN 227
Query: 97 N-SGLLFDEHARTALAFHSLKNSDDGKPEFMFYRNPSADILFRSEEIDKSLIKKATIFHY 155
S + D T + + D + + Y+ S+ + + S SLI K +F
Sbjct: 228 FLSAPIKDGTTGTVIVL----TTPDAQRTMLAYQGTSSVVNYDS--CLASLIAKTNVFVV 281
Query: 156 GSVSLIKEPSRSTHIEAINYAKMCGSILSYAPNLTVPLWPSTEAAREGIMSIW----NYA 211
+ T +A A G++++ + W NYA
Sbjct: 282 EGYLFELPDTIRTITKACEEAHRNGALVAVTAS-------DVSCIERHYDDFWDIVGNYA 334
Query: 212 DVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNLKLLLVTEGIKGCRYYTKDFKGWVYGF 271
D++ + +E R + + + + H + + VT+GI G K ++
Sbjct: 335 DIVFANSDEARAFCHFSAE-ESPISATRYMSHFVPFVSVTDGINGSYIGVKGEAIYIPPS 393
Query: 272 EVEAIDTTGAGDSFVGGFL 290
+DT GAGD++ G L
Sbjct: 394 PCVPVDTCGAGDAYASGIL 412
>At5g03300 adenosine kinase (MOK16.21)
Length = 345
Score = 37.7 bits (86), Expect = 0.009
Identities = 58/256 (22%), Positives = 100/256 (38%), Gaps = 41/256 (16%)
Query: 66 RLGGSSAFIGKVGNDEFGHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEF 125
++ G+++++G +G D++G + GV N DE A T + E
Sbjct: 80 QIPGATSYMGSIGKDKYGEAMKKDATAAGV-NVHYYEDESAPTGTCGVCVVGG-----ER 133
Query: 126 MFYRNPSADILFRSEEIDK----SLIKKATIFHYGSVSLIKEPSR----STHIEAINYAK 177
N SA ++ + + K +L++KA ++ L P S H A N K
Sbjct: 134 SLIANLSAANCYKVDHLKKPENWALVEKAKFYYIAGFFLTVSPESIQLVSEHAAANN--K 191
Query: 178 MCGSILS--------------YAPNLTVPLWPSTEAAREGIMSIWNYADVIKVSVEEIRI 223
+ LS + P + TEA + W DV +++++ I
Sbjct: 192 VFTMNLSAPFICEFFKDVQEKFLPYMDFVFGNETEARTFSRVHGWETEDVEQIAIK-ISQ 250
Query: 224 LTEGNDPYDDKMIMKKLFHHNLKLLLVTEGIKGCRYYTKDFKGWVYGFEVEAIDTTGAGD 283
L + Y ++ + ++V E K +Y + + +DT GAGD
Sbjct: 251 LPKATGTYKRTTVITQ----GADPVVVAEDGKVKKYPVIPLP------KEKLVDTNGAGD 300
Query: 284 SFVGGFLSILSAHKHI 299
+FVGGF+S L K I
Sbjct: 301 AFVGGFMSQLVKEKSI 316
>At5g43910 unknown protein
Length = 365
Score = 37.4 bits (85), Expect = 0.012
Identities = 25/61 (40%), Positives = 29/61 (46%), Gaps = 7/61 (11%)
Query: 274 EAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAATVTGRGAIPSLPTK 333
E IDTTGAGD+F G L L L E L FA+ A G GA SLP +
Sbjct: 302 ELIDTTGAGDAFTGALLYGLCT-------GMALEEMLTFASRVAACCCRGLGARTSLPYR 354
Query: 334 S 334
+
Sbjct: 355 T 355
Score = 36.6 bits (83), Expect = 0.020
Identities = 20/81 (24%), Positives = 40/81 (48%), Gaps = 4/81 (4%)
Query: 22 PLVVCFGEMMINLVPTIDGVSLSDAE----AYKKSPAGATAIVSVAISRLGGSSAFIGKV 77
P+V+ G++ ++ + T+ + D + ++K G T ++RLG + KV
Sbjct: 13 PIVLGCGQLCLDYLVTVASFPIPDQKIRGTSFKVQGGGNTGNALTCVARLGLPCRILAKV 72
Query: 78 GNDEFGHMLSDILKQNGVDNS 98
+D G + + L+ +GVD S
Sbjct: 73 ADDSHGRYMVEELESSGVDTS 93
>At3g09820 adenosine kinase like protein
Length = 344
Score = 33.5 bits (75), Expect = 0.17
Identities = 16/24 (66%), Positives = 18/24 (74%)
Query: 276 IDTTGAGDSFVGGFLSILSAHKHI 299
+DT GAGD+FVGGFLS L K I
Sbjct: 292 VDTNGAGDAFVGGFLSQLVHGKGI 315
>At1g13650 hypothetical protein
Length = 281
Score = 31.6 bits (70), Expect = 0.66
Identities = 30/114 (26%), Positives = 52/114 (45%), Gaps = 13/114 (11%)
Query: 116 KNSDDGKPEFMFYRNPSADILFRSEE----IDKSLIKKATIFHYGSVSLIKEPSRSTHIE 171
KN+ +G E M + NP +I SE D +L+K+ +G +S + S
Sbjct: 77 KNNTNGPEENMVHHNPHLEITEESEADNSCDDNNLLKRNLPNGFGEISFCEAKS------ 130
Query: 172 AINYAKMCGSILSYAPNLTVPL--WPSTEAAREGIMSIWNYADVIKVSVEEIRI 223
+++Y CG LS + NL++ ++ A + S WN + V EE ++
Sbjct: 131 SLDYITYCGP-LSGSENLSIRSDGTSASSFALPILQSEWNSSPVRMGKAEETQL 183
>At1g42970 putative glyceraldehyde-3-phosphate dehydrogenase
(At1g42970)
Length = 447
Score = 30.0 bits (66), Expect = 1.9
Identities = 17/71 (23%), Positives = 37/71 (51%)
Query: 20 KGPLVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVAISRLGGSSAFIGKVGN 79
+ PL + + E+ I++V GV + A K AGA+ ++ A ++ ++ V
Sbjct: 160 RDPLKLPWAELGIDIVIEGTGVFVDGPGAGKHIQAGASKVIITAPAKGADIPTYVMGVNE 219
Query: 80 DEFGHMLSDIL 90
++GH +++I+
Sbjct: 220 QDYGHDVANII 230
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,673,719
Number of Sequences: 26719
Number of extensions: 325252
Number of successful extensions: 797
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 742
Number of HSP's gapped (non-prelim): 35
length of query: 342
length of database: 11,318,596
effective HSP length: 100
effective length of query: 242
effective length of database: 8,646,696
effective search space: 2092500432
effective search space used: 2092500432
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147014.14


