

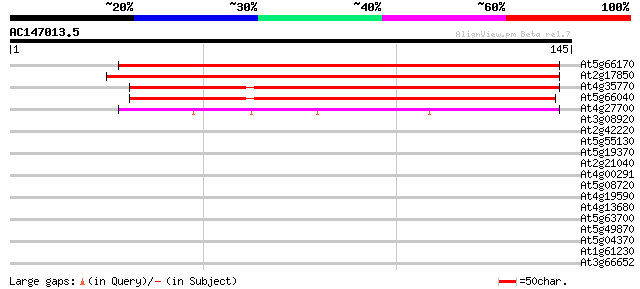
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147013.5 - phase: 0
(145 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g66170 senescence-associated protein sen1-like protein 144 1e-35
At2g17850 putative senescence-associated rhodanese protein 136 3e-33
At4g35770 senescence-associated protein sen1 105 6e-24
At5g66040 senescence-associated protein sen1-like protein; ketoc... 100 4e-22
At4g27700 unknown protein 47 5e-06
At3g08920 unknown protein 39 0.001
At2g42220 unknown protein 39 0.001
At5g55130 molybdopterin synthase sulphurylase (gb|AAD18050.1) 32 0.090
At5g19370 peptidyl-prolyl cis-trans isomerase - like protein 32 0.12
At2g21040 hypothetical protein 30 0.34
At4g00291 unknown protein 28 1.3
At5g08720 unknown protein 28 1.7
At4g19590 putative protein 27 2.9
At4g13680 hypothetical protein 27 3.8
At5g63700 unknown protein 26 6.5
At5g49870 myrosinase binding protein-like 26 6.5
At5g04370 S-adenosyl-L-methionine:salicylic acid carboxyl methyl... 26 6.5
At1g61230 26 6.5
At3g66652 hypothetical protein 26 8.4
>At5g66170 senescence-associated protein sen1-like protein
Length = 136
Score = 144 bits (364), Expect = 1e-35
Identities = 65/114 (57%), Positives = 89/114 (78%)
Query: 29 KVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVK 88
+VV++DV AK L+Q+GH YLDVRT +EF +GH +A KI+NIPY+L+TP+GRVKN F++
Sbjct: 13 EVVSVDVSQAKTLLQSGHQYLDVRTQDEFRRGHCEAAKIVNIPYMLNTPQGRVKNQEFLE 72
Query: 89 QVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPV 142
QVSS + D ++VGCQSG RS AT+EL+A G+K V N+GGGY+ WV + P+
Sbjct: 73 QVSSLLNPADDILVGCQSGARSLKATTELVAAGYKKVRNVGGGYLAWVDHSFPI 126
>At2g17850 putative senescence-associated rhodanese protein
Length = 150
Score = 136 bits (343), Expect = 3e-33
Identities = 63/117 (53%), Positives = 87/117 (73%)
Query: 26 SGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLN 85
S KV+TIDV+ A+ L+ +G+ +LDVRTVEEF+KGHVD+ + N+PY L TP+G+ N N
Sbjct: 2 SEPKVITIDVNQAQKLLDSGYTFLDVRTVEEFKKGHVDSENVFNVPYWLYTPQGQEINPN 61
Query: 86 FVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPV 142
F+K VSS C++ D L++GC+SG RS AT L++ GFK V NM GGY+ WV+ + PV
Sbjct: 62 FLKHVSSLCNQTDHLILGCKSGVRSLHATKFLVSSGFKTVRNMDGGYIAWVNKRFPV 118
>At4g35770 senescence-associated protein sen1
Length = 182
Score = 105 bits (263), Expect = 6e-24
Identities = 50/111 (45%), Positives = 74/111 (66%), Gaps = 2/111 (1%)
Query: 32 TIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVS 91
++ V A+ L Q G+ YLDVRT +EF GH T+ IN+PY+ G VKN +F++QVS
Sbjct: 72 SVPVRVARELAQAGYRYLDVRTPDEFSIGH--PTRAINVPYMYRVGSGMVKNPSFLRQVS 129
Query: 92 SSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPV 142
S K D +++GC+SG+ SF A+++LL GF + ++ GGY+ W N+LPV
Sbjct: 130 SHFRKHDEIIIGCESGQMSFMASTDLLTAGFTAITDIAGGYVAWTENELPV 180
>At5g66040 senescence-associated protein sen1-like protein;
ketoconazole resistance protein-like
Length = 120
Score = 100 bits (248), Expect = 4e-22
Identities = 52/110 (47%), Positives = 69/110 (62%), Gaps = 2/110 (1%)
Query: 32 TIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVS 91
++ V A +L+ GH YLDVRT EEF +GH A IN+PY+ G KN +F++QVS
Sbjct: 10 SVSVTVAHDLLLAGHRYLDVRTPEEFSQGH--ACGAINVPYMNRGASGMSKNPDFLEQVS 67
Query: 92 SSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLP 141
S + D ++VGCQSG RS AT++LL GF V ++ GGY W N LP
Sbjct: 68 SHFGQSDNIIVGCQSGGRSIKATTDLLHAGFTGVKDIVGGYSAWAKNGLP 117
>At4g27700 unknown protein
Length = 224
Score = 46.6 bits (109), Expect = 5e-06
Identities = 39/146 (26%), Positives = 63/146 (42%), Gaps = 32/146 (21%)
Query: 29 KVVTIDVHAAKNLIQTGH-IYLDVRTVEEFEKGH------VDATKIINIPYLLDTPK--- 78
+V ++DV A+ L + + + LDVR E++ GH V+ ++I D +
Sbjct: 73 RVRSVDVKEAQRLQKENNFVILDVRPEAEYKAGHPPGAINVEMYRLIREWTAWDIARRLG 132
Query: 79 --------GRVKNLNFVKQVSSSCDKEDCLVVGCQSG--------------KRSFSATSE 116
G +N F++ V + DKE ++V C S RS A
Sbjct: 133 FAFFGIFSGTEENPEFIQSVEAKLDKEAKIIVACSSAGTMKPTQNLPEGQQSRSLIAAYL 192
Query: 117 LLADGFKNVHNMGGGYMEWVSNKLPV 142
L+ +G+KNV ++ GG W LPV
Sbjct: 193 LVLNGYKNVFHLEGGIYTWGKEGLPV 218
>At3g08920 unknown protein
Length = 214
Score = 38.9 bits (89), Expect = 0.001
Identities = 35/141 (24%), Positives = 62/141 (43%), Gaps = 27/141 (19%)
Query: 29 KVVTIDVHAAKNLIQT-GHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKG-------- 79
KV ++ A ++ + G+I LDVR E EK V + +++P ++ P
Sbjct: 44 KVRAVEPKEANAVVASEGYILLDVRPAWEREKARVKGS--LHVPLFVEDPDNGPITLLKK 101
Query: 80 -------------RVKNLN---FVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFK 123
R +N ++ V + DKE ++V C G RS +A S+L +G+K
Sbjct: 102 WIHLGYIGLWTGQRFTMINDEFALRVVEAVPDKESKVLVVCGEGLRSLAAVSKLHGEGYK 161
Query: 124 NVHNMGGGYMEWVSNKLPVIQ 144
++ + GG+ P I+
Sbjct: 162 SLGWLTGGFNRVSEGDFPEIE 182
>At2g42220 unknown protein
Length = 234
Score = 38.5 bits (88), Expect = 0.001
Identities = 31/120 (25%), Positives = 54/120 (44%), Gaps = 22/120 (18%)
Query: 28 AKVVTIDVHAAKNLI-QTGHIYLDVRTVEEFEKGHVDATKIINI-PYLLDTPKGRV---- 81
A++ ++ AK LI + G+ +DVR +FE+ H+ + I + Y D G +
Sbjct: 47 AELKFVNAEEAKQLIAEEGYSVVDVRDKTQFERAHIKSCSHIPLFIYNEDNDIGTIIKRT 106
Query: 82 ----------------KNLNFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNV 125
N F+K V + ++ L++ CQ G RS +A S L G++N+
Sbjct: 107 VHNNFSGLFFGLPFTKVNPEFLKSVRNEFSQDSKLLLVCQEGLRSAAAASRLEEAGYENI 166
>At5g55130 molybdopterin synthase sulphurylase (gb|AAD18050.1)
Length = 464
Score = 32.3 bits (72), Expect = 0.090
Identities = 28/107 (26%), Positives = 43/107 (40%), Gaps = 20/107 (18%)
Query: 46 HIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKN-LNFVKQV----------SSSC 94
H+ LDVR + KI+++P L+ P ++ LN + + SC
Sbjct: 362 HVLLDVRPSHHY--------KIVSLPDSLNIPLANLETRLNELTSALKEKGNGHANTESC 413
Query: 95 DKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLP 141
VV C+ G S A L GF + ++ GG W +N P
Sbjct: 414 TNPSVFVV-CRRGNDSQRAVQYLRESGFDSAKDIIGGLEAWAANVNP 459
>At5g19370 peptidyl-prolyl cis-trans isomerase - like protein
Length = 299
Score = 32.0 bits (71), Expect = 0.12
Identities = 29/125 (23%), Positives = 53/125 (42%), Gaps = 5/125 (4%)
Query: 7 NLLPRCLAFFLLLVFVLCSSGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATK 66
N + RC F L + + S V I V + +Q + + ++ E ++
Sbjct: 165 NQVVRCRTQFGLHLLQVLSEREPVKDIQVEELHSKMQDPVFMDEAQLIDVREPNEIE--- 221
Query: 67 IINIPYLLDTPKGRVKNLNFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVH 126
I ++P P + + ++S + E V C+ G RS + L + GFK+V+
Sbjct: 222 IASLPGFKVFPLRQFGT--WAPDITSKLNPEKDTFVLCKVGGRSMQVANWLQSQGFKSVY 279
Query: 127 NMGGG 131
N+ GG
Sbjct: 280 NITGG 284
>At2g21040 hypothetical protein
Length = 277
Score = 30.4 bits (67), Expect = 0.34
Identities = 14/33 (42%), Positives = 20/33 (60%)
Query: 18 LLVFVLCSSGAKVVTIDVHAAKNLIQTGHIYLD 50
+L+ + S V T+DV+ AK + TGH YLD
Sbjct: 176 VLLSICFYSVEDVETVDVYTAKGFLSTGHRYLD 208
>At4g00291 unknown protein
Length = 497
Score = 28.5 bits (62), Expect = 1.3
Identities = 20/76 (26%), Positives = 33/76 (43%), Gaps = 4/76 (5%)
Query: 73 LLDTPKGRVKNLNFVKQVSSSCDKEDCLVVGCQSGKRSFSATSE----LLADGFKNVHNM 128
LLD PK ++F + D + +G S R+FS+ S+ ++A G +
Sbjct: 41 LLDGPKASPSMISFSSNIRLHNDAKPFNYLGHSSYARAFSSKSDDFGSIVASGVTGSGDG 100
Query: 129 GGGYMEWVSNKLPVIQ 144
G +WV V+Q
Sbjct: 101 NGNGNDWVEKAKDVLQ 116
>At5g08720 unknown protein
Length = 719
Score = 28.1 bits (61), Expect = 1.7
Identities = 12/43 (27%), Positives = 25/43 (57%)
Query: 45 GHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFV 87
G I+L+ R ++ H++A ++++ LD+P GR + + V
Sbjct: 141 GRIWLEQRGLQRALYWHIEARVVLDLHECLDSPNGRELHFSMV 183
>At4g19590 putative protein
Length = 345
Score = 27.3 bits (59), Expect = 2.9
Identities = 19/53 (35%), Positives = 25/53 (46%), Gaps = 3/53 (5%)
Query: 75 DTPKGRVKNLNFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHN 127
+TP+ KN N SSSCD++ L V S R S+ S + NV N
Sbjct: 246 ETPQATNKNTN---GASSSCDRDSSLNVNFDSSFRRDSSLSRMSYFNSGNVAN 295
>At4g13680 hypothetical protein
Length = 354
Score = 26.9 bits (58), Expect = 3.8
Identities = 15/51 (29%), Positives = 26/51 (50%)
Query: 51 VRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVSSSCDKEDCLV 101
V + E H K I++P L+ P + + + V +SS ++EDC+V
Sbjct: 108 VLRLREVNLNHETDRKRISLPPLVTLPHCQTQYVTNVAMSTSSPEEEDCVV 158
>At5g63700 unknown protein
Length = 571
Score = 26.2 bits (56), Expect = 6.5
Identities = 18/54 (33%), Positives = 23/54 (42%)
Query: 84 LNFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVS 137
L +KQ S DK V ++G R F A L G KN + G + VS
Sbjct: 418 LELLKQNDSFVDKVVGSFVKVKNGPRDFMAYQILQVTGIKNADDQSEGVLLHVS 471
>At5g49870 myrosinase binding protein-like
Length = 601
Score = 26.2 bits (56), Expect = 6.5
Identities = 19/50 (38%), Positives = 26/50 (52%), Gaps = 2/50 (4%)
Query: 21 FVLCSSGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFE-KGHVDATKIIN 69
F L +SG K+V +A KNL G YL T ++ E KG D + + N
Sbjct: 265 FKLTASGMKIVGFHGYAEKNLSSLG-AYLTPLTPKKSECKGITDGSNVWN 313
>At5g04370 S-adenosyl-L-methionine:salicylic acid carboxyl
methyltransferase-like protein
Length = 396
Score = 26.2 bits (56), Expect = 6.5
Identities = 15/48 (31%), Positives = 24/48 (49%), Gaps = 7/48 (14%)
Query: 71 PYLLDTPKGRVKNLNFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELL 118
P L+ K + NLNF + + +GC SG+ +F A SE++
Sbjct: 66 PVLVKNTKDLMINLNFPTYIKVAD-------LGCSSGQNTFLAMSEII 106
>At1g61230
Length = 546
Score = 26.2 bits (56), Expect = 6.5
Identities = 15/65 (23%), Positives = 28/65 (43%), Gaps = 10/65 (15%)
Query: 21 FVLCSSGAKVVTIDVHAAKNLIQTGHIYLDV----------RTVEEFEKGHVDATKIINI 70
F L G K++ +A KNLI G + V + +E ++ G D + + +
Sbjct: 218 FSLADKGKKIIGFHGYAEKNLISLGAYFTTVSVTKSVCHGSKIIESWDDGVFDGIRKVYV 277
Query: 71 PYLLD 75
Y ++
Sbjct: 278 SYSIN 282
>At3g66652 hypothetical protein
Length = 980
Score = 25.8 bits (55), Expect = 8.4
Identities = 10/22 (45%), Positives = 14/22 (63%)
Query: 78 KGRVKNLNFVKQVSSSCDKEDC 99
+G VK + V + SS CD +DC
Sbjct: 47 EGEVKKFDVVAKDSSPCDDDDC 68
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,219,324
Number of Sequences: 26719
Number of extensions: 124708
Number of successful extensions: 320
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 301
Number of HSP's gapped (non-prelim): 19
length of query: 145
length of database: 11,318,596
effective HSP length: 90
effective length of query: 55
effective length of database: 8,913,886
effective search space: 490263730
effective search space used: 490263730
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC147013.5


