

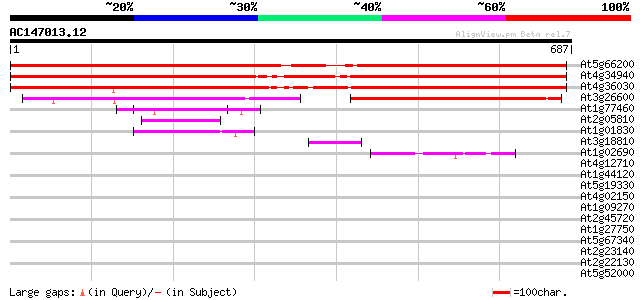
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147013.12 - phase: 0
(687 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g66200 unknown protein 876 0.0
At4g34940 putative protein 838 0.0
At4g36030 unknown protein 787 0.0
At3g26600 unknown protein 202 7e-52
At1g77460 unknown protein 50 4e-06
At2g05810 hypothetical protein 50 5e-06
At1g01830 unknown protein 48 2e-05
At3g18810 protein kinase, putative 44 4e-04
At1g02690 importin alpha, putative 43 7e-04
At4g12710 unknown protein 42 0.001
At1g44120 hypothetical protein 42 0.001
At5g19330 putative protein 41 0.002
At4g02150 AtKAP alpha 39 0.010
At1g09270 putative importin beta binding domain 39 0.013
At2g45720 unknown protein 37 0.028
At1g27750 unknown protein 37 0.028
At5g67340 unknown proteins 37 0.037
At2g23140 hypothetical protein 37 0.037
At2g22130 unknown protein 37 0.037
At5g52000 importin alpha subunit 37 0.048
>At5g66200 unknown protein
Length = 651
Score = 876 bits (2264), Expect = 0.0
Identities = 471/685 (68%), Positives = 538/685 (77%), Gaps = 39/685 (5%)
Query: 1 MGDIVKQILAKPIQLADQVTKAADEASSFKQECSELKSKTEKLATLLRQAARASSDLYER 60
M DIVKQILAKPIQL+DQV KAADEASSFKQEC ELK+KTEKLA LLRQAARAS+DLYER
Sbjct: 1 MADIVKQILAKPIQLSDQVVKAADEASSFKQECGELKAKTEKLAGLLRQAARASNDLYER 60
Query: 61 PTKRIIEETEQVLDKALSLVLKCRANGLMKRVFTIIPAAAFRKTSSHLENSIGDVSWLLR 120
PT+RII++TEQ+L+KALSLVLKCRANGLMKRVFTIIPAAAFRK S LENSIGDVSWLLR
Sbjct: 61 PTRRIIDDTEQMLEKALSLVLKCRANGLMKRVFTIIPAAAFRKMSGQLENSIGDVSWLLR 120
Query: 121 VSAPADDRGGE-YLGLPPIAANEPILCFIWEQIAMLFTGSQEVRSDAAASLVSLARGSDR 179
VSAPA+DRG YLGLPPIAANEPILC IWEQIA+L+TGS E RSDAAASLVSLAR +DR
Sbjct: 121 VSAPAEDRGDAGYLGLPPIAANEPILCLIWEQIAILYTGSLEDRSDAAASLVSLARDNDR 180
Query: 180 YGKLIIEEGGVGPLLKLIKEGKADGQENAARAIGLLGRDAESVEHMIHVGVCSVFAKILK 239
Y KLIIEEGGV PLLKL+KEGK +GQENAARA+GLLGRD ESVEHMIH G CSVF K+LK
Sbjct: 181 YTKLIIEEGGVVPLLKLLKEGKPEGQENAARALGLLGRDPESVEHMIHGGACSVFGKVLK 240
Query: 240 EGPMKVQGVVAWAVSELAANYPKCQELFAQHNIIRLLVGHLAFETVEEHSKYAIVSM-KA 298
EGPMKVQ VVAWA SEL +N+PKCQ++FAQHN IRLLVGHLAFETV+EHSKYAI + KA
Sbjct: 241 EGPMKVQAVVAWATSELVSNHPKCQDVFAQHNAIRLLVGHLAFETVQEHSKYAIATNNKA 300
Query: 299 NSIHAAVVMASNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGER-PRNLHRVI 357
SIH AV +A N NS+S KG + + + HP G++ P +H V+
Sbjct: 301 TSIHHAVALAKENPNSTSATALPKGLDEDQSSIP-----------HPTGKQMPNQMHNVV 349
Query: 358 TSTMAIHAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNY 417
+TMA+ A + + N +Q+ + S+ HQ +
Sbjct: 350 VNTMAVRANPPRKSTSNGVSQSNGVKQPSSV----------------------QQHQNST 387
Query: 418 SHSGINMKGRESEDAETKASMKEMAARALWHLAKGNVAICRSITESRALLCFSVLLEKGP 477
S + K RE ED+ TK +K MAARALW LAKGN IC+SITESRALLCF+VL+EKG
Sbjct: 388 SSAS---KTRELEDSATKCQIKAMAARALWKLAKGNSTICKSITESRALLCFAVLIEKGD 444
Query: 478 EAVQYNSAMALMEITAVAEKDAELRKSAFKPNSPACKAVVDQVLKIIEKADSDLLIPCVK 537
E V+YNSAMALMEITAVAE+DA+LR+SAFKPNSPACKAVVDQVL+IIE ADS+LLIPC++
Sbjct: 445 EEVRYNSAMALMEITAVAEQDADLRRSAFKPNSPACKAVVDQVLRIIEIADSELLIPCIR 504
Query: 538 AIGNLARTFKATETRMIGPLVKLLDEREAEVSREASIALRKFAGSENYLHVDHSNAIISA 597
IGNLARTF+ATETRMIGPLVKLLDERE EV+ EA+ AL KFA + NYLH DHS II A
Sbjct: 505 TIGNLARTFRATETRMIGPLVKLLDEREPEVTGEAAAALTKFACTANYLHKDHSRGIIEA 564
Query: 598 GGAKHLIQLVYFGEQMVQIPALVLLSYIALHVPDSEELALAEVLGVLEWASKQSFMQHDE 657
GG KHL+QL YFGE VQIPAL LL YIAL+VPDSE+LA EVL VLEWASKQS++ E
Sbjct: 565 GGGKHLVQLAYFGEGGVQIPALELLCYIALNVPDSEQLAKDEVLAVLEWASKQSWVTQLE 624
Query: 658 TLEELLQEAKSRLELYQSRGSRGFH 682
+LE LLQEAK L+LYQ RGSRG++
Sbjct: 625 SLEALLQEAKRGLDLYQQRGSRGYN 649
>At4g34940 putative protein
Length = 664
Score = 838 bits (2164), Expect = 0.0
Identities = 454/686 (66%), Positives = 539/686 (78%), Gaps = 26/686 (3%)
Query: 1 MGDIVKQILAKPIQLADQVTKAADEASSFKQECSELKSKTEKLATLLRQAARASSDLYER 60
M DIVKQIL +PIQLADQ+TKA+DEA SF+QEC E+K+KTEKLA LLRQAARAS+DLYER
Sbjct: 1 MADIVKQILVRPIQLADQITKASDEAYSFRQECLEVKAKTEKLAGLLRQAARASNDLYER 60
Query: 61 PTKRIIEETEQVLDKALSLVLKCRANGLMKRVFTIIPAAAFRKTSSHLENSIGDVSWLLR 120
PT+RII++TEQVL KAL+LV KCRA GLMKRVFTIIPAAAFRK + LENSIGDVSWLLR
Sbjct: 61 PTRRIIDDTEQVLFKALALVEKCRATGLMKRVFTIIPAAAFRKITMQLENSIGDVSWLLR 120
Query: 121 VSAPADDRGGEYLGLPPIAANEPILCFIWEQIAMLFTGSQEVRSDAAASLVSLARGSDRY 180
VSA DDR EYLGLPPIAANEPILC IWEQ+A+LFTGS + RSDAAASLVSLAR +DRY
Sbjct: 121 VSASGDDRDDEYLGLPPIAANEPILCLIWEQVAILFTGSLDDRSDAAASLVSLARDNDRY 180
Query: 181 GKLIIEEGGVGPLLKLIKEGKADGQENAARAIGLLGRDAESVEHMIHVGVCSVFAKILKE 240
G+LIIEEGGV LLKL KEGK +GQENAARAIGLLGRD ESVE +++ GVC VFAKILKE
Sbjct: 181 GRLIIEEGGVPSLLKLAKEGKMEGQENAARAIGLLGRDPESVEQIVNAGVCQVFAKILKE 240
Query: 241 GPMKVQGVVAWAVSELAANYPKCQELFAQHNIIRLLVGHLAFETVEEHSKYAIVSMK--A 298
G MKVQ VVAWAVSELA+N+PKCQ+ FAQ+NIIR LV HLAFETV+EHSKYAIVS K
Sbjct: 241 GHMKVQTVVAWAVSELASNHPKCQDHFAQNNIIRFLVSHLAFETVQEHSKYAIVSNKQTL 300
Query: 299 NSIHAAVVMASNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVIT 358
+SIH VVMASN N + KK +D +SH + P +H +I
Sbjct: 301 SSIH-TVVMASNTNPAD-----KKENNEQD-------ETKSNISHPLSNQTPSQMHSLIA 347
Query: 359 STMAIHAA--SKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRN 416
+T+A+ + S G+ + N+N + SN + N G+ +GN+ H
Sbjct: 348 NTLAMKGSGPSSGSGSGSGSGTNKNQIKQSNQQHQNHTKGGS------NPRGNNPTH--- 398
Query: 417 YSHSGINMKGRESEDAETKASMKEMAARALWHLAKGNVAICRSITESRALLCFSVLLEKG 476
S G ++KGRE ED TKA MK MAARALW L++GN+ ICRSITESRALLCF+VLLEKG
Sbjct: 399 VSLMGTSIKGREYEDPATKAQMKAMAARALWQLSRGNLQICRSITESRALLCFAVLLEKG 458
Query: 477 PEAVQYNSAMALMEITAVAEKDAELRKSAFKPNSPACKAVVDQVLKIIEKADSDLLIPCV 536
+ V+ SA+A+MEIT VAE+ ELR+SAFKP SPA KAVV+Q+LK+IE DLLIPC+
Sbjct: 459 DDEVKSYSALAMMEITDVAEQYPELRRSAFKPTSPAAKAVVEQLLKVIENEILDLLIPCI 518
Query: 537 KAIGNLARTFKATETRMIGPLVKLLDEREAEVSREASIALRKFAGSENYLHVDHSNAIIS 596
K+IG+L+RTF+ATETR+IGPLVKLLDEREAE++ EA++AL KF+ +EN+L +HS AII+
Sbjct: 519 KSIGSLSRTFRATETRIIGPLVKLLDEREAEIAMEAAVALIKFSCTENFLRDNHSKAIIA 578
Query: 597 AGGAKHLIQLVYFGEQMVQIPALVLLSYIALHVPDSEELALAEVLGVLEWASKQSFMQHD 656
AGGAKHLIQLVYFGEQMVQ+PAL+LL YIAL+VPDSE LA EVL VLEW++KQ+ +
Sbjct: 579 AGGAKHLIQLVYFGEQMVQVPALMLLCYIALNVPDSETLAQEEVLVVLEWSTKQAHLVEA 638
Query: 657 ETLEELLQEAKSRLELYQSRGSRGFH 682
T++E+L EAKSRLELYQSRGSRGFH
Sbjct: 639 PTIDEILPEAKSRLELYQSRGSRGFH 664
>At4g36030 unknown protein
Length = 670
Score = 787 bits (2033), Expect = 0.0
Identities = 428/688 (62%), Positives = 520/688 (75%), Gaps = 25/688 (3%)
Query: 1 MGDIVKQILAKPIQLADQVTKAADEASSFKQECSELKSKTEKLATLLRQAARASSDLYER 60
MGD+ KQIL++PIQLADQV KA DEA+ KQEC+++KSKTEKLA LLRQAARASSDLYER
Sbjct: 1 MGDLAKQILSRPIQLADQVVKAGDEATINKQECADIKSKTEKLAALLRQAARASSDLYER 60
Query: 61 PTKRIIEETEQVLDKALSLVLKCRANGLMKRVFTIIPAAAFRKTSSHLENSIGDVSWLLR 120
PT+RI+++TE VL+KAL++V +CR +G + R+F IIPAAAFRK S LENS+GDVSWLLR
Sbjct: 61 PTRRILDDTENVLEKALTMVQRCRDDGYIMRLFNIIPAAAFRKMISQLENSVGDVSWLLR 120
Query: 121 VSAPA---DDRGGEYLGLPPIAANEPILCFIWEQIAMLFTGSQEVRSDAAASLVSLARGS 177
VS PA DD G YLGLPPIAANEPILC IWEQIA+L TGS E +SDAAASL SLAR +
Sbjct: 121 VSTPAGNDDDEGFGYLGLPPIAANEPILCLIWEQIAVLMTGSPEDKSDAAASLASLARDN 180
Query: 178 DRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAIGLLGRDAESVEHMIHVGVCSVFAKI 237
DRY KLI+EEGGV PLLKL+KEGK DGQENAAR IGLLGRD ESVEHMI +GVCSV + I
Sbjct: 181 DRYVKLIVEEGGVNPLLKLVKEGKIDGQENAARTIGLLGRDPESVEHMIQLGVCSVLSSI 240
Query: 238 LKEGPMKVQGVVAWAVSEL-AANYPKCQELFAQHNIIRLLVGHLAFETVEEHSKYAIVSM 296
LKEG MKVQ VVAWAVSEL + N+ KCQELFAQ+N+IRLLV HLAFETV+EHSKYA+V+
Sbjct: 241 LKEGSMKVQAVVAWAVSELVSGNHAKCQELFAQNNVIRLLVSHLAFETVQEHSKYAVVAG 300
Query: 297 KANSIHAAVVMASNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRV 356
+A S+H AVVMAS ++S NL P E +D + H VS +H +
Sbjct: 301 RATSMHHAVVMASKISSSKENL-PALNEEEDD-------DNHIGVS----SPMTNQMHSI 348
Query: 357 ITSTMAIHAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRN 416
+ +TMA+ A G+++N + + + + + + + K HQ
Sbjct: 349 VATTMAMKAV----GSGSKSNLSSRFVTGDDDKPPEKIPEKSYSMSSQIKAYGSIAHQ-- 402
Query: 417 YSHSGINMKGRESEDAETKASMKEMAARALWHLAKGNVAICRSITESRALLCFSVLLEKG 476
S + +GRE ED TK MK MAARALW LA GN +ICR ITESRALLCF+VLL+KG
Sbjct: 403 -SRNASVTRGRELEDPVTKTYMKAMAARALWKLAVGNSSICRVITESRALLCFAVLLDKG 461
Query: 477 PEAVQYNSAMALMEITAVAEKDAELRKSAFKPNSPACKAVVDQVLKIIEKAD--SDLLIP 534
E +YN+AMA+MEITAVAE++A+LR+SAF+ SPACKAVVDQ+ +I+E AD SDLLIP
Sbjct: 462 DEETKYNTAMAIMEITAVAEENADLRRSAFRRTSPACKAVVDQLFRIVENADAGSDLLIP 521
Query: 535 CVKAIGNLARTFKATETRMIGPLVKLLDEREAEVSREASIALRKFAGSENYLHVDHSNAI 594
CV++IGNLARTFK+ ET MI PLVKLLD+ E +++ E +IAL KFA +N+L +HS I
Sbjct: 522 CVRSIGNLARTFKSAETHMIVPLVKLLDDGEPDLAAEVAIALAKFATEDNFLGKEHSRTI 581
Query: 595 ISAGGAKHLIQLVYFGEQMVQIPALVLLSYIALHVPDSEELALAEVLGVLEWASKQSFMQ 654
I AGG+K L+QL YFGE QIPA+VLLSY+A++VPDSE+LA EVL VLEW+SKQ+ +
Sbjct: 582 IEAGGSKLLVQLAYFGENGAQIPAMVLLSYVAMNVPDSEQLAKDEVLTVLEWSSKQANVL 641
Query: 655 HDETLEELLQEAKSRLELYQSRGSRGFH 682
DE +E LL EAKSRLELYQSRGSRGFH
Sbjct: 642 EDEDMEALLYEAKSRLELYQSRGSRGFH 669
>At3g26600 unknown protein
Length = 615
Score = 202 bits (513), Expect = 7e-52
Identities = 119/259 (45%), Positives = 169/259 (64%), Gaps = 2/259 (0%)
Query: 418 SHSGINMKGRESEDAETKASMKEMAARALWHLAKGNVAICRSITESRALLCFSVLLEKGP 477
S +G K R++E+ E K +K A ALW LA+GNVA R ITE++ LL + ++EK
Sbjct: 346 SRTGNFKKERDNENPEVKHELKVNCAEALWMLARGNVANSRRITETKGLLSLAKIVEKEV 405
Query: 478 EAVQYNSAMALMEITAVAEKDAELRKSAFKPNSPACKAVVDQVLKIIEKADSDLL-IPCV 536
+QYN M LMEITA AE A+LR++AFK NSPA KAV+DQ+L II+ DS +L IP +
Sbjct: 406 GELQYNCLMTLMEITAAAESSADLRRAAFKTNSPAAKAVIDQMLWIIKDVDSPILKIPAI 465
Query: 537 KAIGNLARTFKATETRMIGPLVKLLDEREAEVSREASIALRKFAGSENYLHVDHSNAIIS 596
++IG+LARTF A ETRMI PLV+ L EV+ A I+L+KF EN+L +HS II
Sbjct: 466 QSIGSLARTFPARETRMIKPLVEKLGSSNQEVAITAVISLQKFVCPENFLCAEHSKNIIE 525
Query: 597 AGGAKHLIQLVYFGEQMVQIPALVLLSYIALHVPDSEELALAEVLGVLEWASKQSFMQHD 656
G L++L+ EQ +Q+ L LL Y++++ + ++L A+VL VLE A + + +Q+
Sbjct: 526 YGAIPLLMKLIRNVEQQMQLQCLALLCYLSVNASNHQQLEQAKVLTVLEGAERLAGLQNM 585
Query: 657 ETLEELLQEAKSRLELYQS 675
E L EL+ +A +L LY +
Sbjct: 586 E-LRELVSKAIYQLSLYNA 603
Score = 197 bits (502), Expect = 1e-50
Identities = 128/352 (36%), Positives = 199/352 (56%), Gaps = 15/352 (4%)
Query: 16 ADQVTKAADEASSFKQECSELKSKTEKLATLLRQAAR----ASSDLYERPTKRIIEETEQ 71
A+++ A DEA SFK EC E+ + ++LA +LR R +S +Y+RP +R+I + ++
Sbjct: 19 AERLRVAVDEAESFKTECGEVGKQVDRLAQMLRTLVRFVSSSSQQVYDRPIRRVIVDVKK 78
Query: 72 VLDKALSLVLKCRANGLMKRVFTIIPAAAFRKTSSHLENSIGDVSWLLRVSAPADD---R 128
L++ +LV KCR + +++RV TII AA FRK + LE+S GDV W+L V D
Sbjct: 79 NLERGFALVRKCRRHNIIRRVCTIINAADFRKVINLLESSNGDVKWILSVFDSDGDGSFG 138
Query: 129 GGEYLGLPPIAANEPILCFIWEQIAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEG 188
GG + LPPIA N+PIL ++W +A + G + DAA L SLA +DR K+I++EG
Sbjct: 139 GGIVISLPPIATNDPILPWVWSLVASIQMGKLVDKIDAANQLGSLAGDNDRNKKIIVDEG 198
Query: 189 GVGPLLKLIKE-GKADGQENAARAIGLLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQG 247
GV PLL+L+KE A+GQ AA A+GLL D + V +++ + ++L + ++VQ
Sbjct: 199 GVSPLLRLLKESSSAEGQIAAATALGLLACDEDKVRSIVNELGVPIIVQVLGDSSVRVQI 258
Query: 248 VVAWAVSELAANYPKCQELFAQHNIIRLLVGHLAFETVEEHSKYAIVSMKANSIHAAVVM 307
VA V+ +A + P Q+ FA+ ++I+ LV L+ + + I K NSIH+ V M
Sbjct: 259 KVATLVARMAEHDPVAQDEFARQSVIKPLVTLLSLDVFVDD----IHLSKHNSIHSLVQM 314
Query: 308 AS--NNNNSSSNLNPKKGTENEDGVVVGG-GNKHGRVSHHPLGERPRNLHRV 356
+ SS P K +++ +GG G++ G E P H +
Sbjct: 315 NKEVEKDPSSKLYRPLKSSKSNVYRDIGGSGSRTGNFKKERDNENPEVKHEL 366
>At1g77460 unknown protein
Length = 2110
Score = 50.1 bits (118), Expect = 4e-06
Identities = 34/118 (28%), Positives = 55/118 (45%), Gaps = 3/118 (2%)
Query: 152 IAMLFTGSQEVRSDAAASLVSLAR---GSDRYGKLIIEEGGVGPLLKLIKEGKADGQENA 208
I +L +Q V A +L +L+R ++ K I EG + L+KL K + ENA
Sbjct: 656 IKLLTNNTQNVAKQVARALDALSRPVKNNNNKKKSYIAEGDIKSLIKLAKNSSIESAENA 715
Query: 209 ARAIGLLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWAVSELAANYPKCQEL 266
A+ L D + + V S F +IL +G + + + A+ +L N+P C L
Sbjct: 716 VSALANLLSDPDIAAEALAEDVVSAFTRILADGSPEGKRNASRALHQLLKNFPVCDVL 773
Score = 42.4 bits (98), Expect = 9e-04
Identities = 35/190 (18%), Positives = 87/190 (45%), Gaps = 14/190 (7%)
Query: 131 EYLGLPPIAANEPILCFIWEQIAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGV 190
+++G+ I E ++ +W+Q+++ + V +L +L D Y +L +E GV
Sbjct: 132 DHIGMK-IFITEGVVPTLWDQLSLKGNQDKVVEGYVTGALRNLCGVDDGYWRLTLEGSGV 190
Query: 191 GPLLKLIKEGKADGQENAARAIG-LLGRDAESVEHMIHVGVCSVFAKILKE-GPMKVQGV 248
++ L+ + Q NAA + L+ +S++ +++ GV ++L++ + V+
Sbjct: 191 DIVVSLLSSDNPNSQANAASLLARLVLSFCDSIQKILNSGVVKSLIQLLEQKNDINVRAS 250
Query: 249 VAWAVSELAANYPKCQELFAQHNIIRLLVGHLAF-----------ETVEEHSKYAIVSMK 297
A A+ L+AN + ++ + L+ + ++++EH+ A+ ++
Sbjct: 251 AADALEALSANSDEAKKCVKDAGGVHALIEAIVAPSKECMQGKHGQSLQEHATGALANVF 310
Query: 298 ANSIHAAVVM 307
H + +
Sbjct: 311 GGMRHLIIYL 320
Score = 38.5 bits (88), Expect = 0.013
Identities = 22/69 (31%), Positives = 36/69 (51%)
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
+ +L TGSQ+ + DAA L +L S+ + GG+ L L+K G + QE +A+
Sbjct: 493 VQLLETGSQKAKEDAACILWNLCCHSEEIRDCVERAGGIPAFLWLLKTGGPNSQETSAKT 552
Query: 212 IGLLGRDAE 220
+ L A+
Sbjct: 553 LVKLVHTAD 561
Score = 30.0 bits (66), Expect = 4.5
Identities = 21/74 (28%), Positives = 39/74 (52%), Gaps = 1/74 (1%)
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
I+ML G+ + + A+ L L + D K+++ G + PLL ++K G + ++ AA A
Sbjct: 62 ISMLRNGTTLAKVNVASILCVLCKDKDLRLKVLLG-GCIPPLLSVLKSGTMETRKAAAEA 120
Query: 212 IGLLGRDAESVEHM 225
I + S +H+
Sbjct: 121 IYEVSSAGISNDHI 134
>At2g05810 hypothetical protein
Length = 580
Score = 49.7 bits (117), Expect = 5e-06
Identities = 30/97 (30%), Positives = 51/97 (51%)
Query: 162 VRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAIGLLGRDAES 221
+R A A++ L S K + E+GG+GPLL+L++ G + + AA AI + D +
Sbjct: 218 IREHALAAVSLLTSSSADSRKTVFEQGGLGPLLRLLETGSSPFKTRAAIAIEAITADPAT 277
Query: 222 VEHMIHVGVCSVFAKILKEGPMKVQGVVAWAVSELAA 258
+ G +V + + G +VQ +A A+S +AA
Sbjct: 278 AWAISAYGGVTVLIEACRSGSKQVQEHIAGAISNIAA 314
Score = 33.9 bits (76), Expect = 0.31
Identities = 46/205 (22%), Positives = 83/205 (40%), Gaps = 18/205 (8%)
Query: 109 ENSIGDVSWLLRVSAPADDRGGEYLGLPPIAANEPILCFIWEQIAMLFTGSQEVRSDAAA 168
E+++ VS L SA + E GL P+ + +L TGS ++ AA
Sbjct: 220 EHALAAVSLLTSSSADSRKTVFEQGGLGPL-------------LRLLETGSSPFKTRAAI 266
Query: 169 SLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAIGLLGRDAESVEHMIHV 228
++ ++ I GGV L++ + G QE+ A AI + E +
Sbjct: 267 AIEAIT-ADPATAWAISAYGGVTVLIEACRSGSKQVQEHIAGAISNIAAVEEIRTTLAEE 325
Query: 229 GVCSVFAKILKEGPMKVQGVVAWAVSELAANYPKCQELFAQHNIIRLLVGHLAFETVE-- 286
G V ++L G VQ A +S ++++ ++L + ++ HL E+
Sbjct: 326 GAIPVLIQLLISGSSSVQEKTANFISLISSSGEYYRDLIVRERGGLQILIHLVQESSNPD 385
Query: 287 --EHSKYAIVSMKANSIHAAVVMAS 309
EH A+ + A + V+ +S
Sbjct: 386 TIEHCLLALSQISAMETVSRVLSSS 410
>At1g01830 unknown protein
Length = 574
Score = 47.8 bits (112), Expect = 2e-05
Identities = 39/152 (25%), Positives = 69/152 (44%), Gaps = 4/152 (2%)
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
+ L T + + A +L+S+ S + +I EG + PL++LI+ G + +E AA A
Sbjct: 215 LVQLLTATSTRIREKAVNLISVLAESGHCDEWLISEGVLPPLVRLIESGSLETKEKAAIA 274
Query: 212 IGLLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWAVSELAANYPKCQELFAQHN 271
I L E+ + G + + K G Q A A+ ++A + ++L A+
Sbjct: 275 IQRLSMTEENAREIAGHGGITPLIDLCKTGDSVSQAASAAALKNMSA-VSELRQLLAEEG 333
Query: 272 IIRL---LVGHLAFETVEEHSKYAIVSMKANS 300
IIR+ L+ H EH + ++ A S
Sbjct: 334 IIRVSIDLLNHGILLGSREHMAECLQNLTAAS 365
>At3g18810 protein kinase, putative
Length = 700
Score = 43.5 bits (101), Expect = 4e-04
Identities = 24/68 (35%), Positives = 32/68 (46%), Gaps = 3/68 (4%)
Query: 367 SKQPNEGNEANQNQNILANSNTPNGNGLG-NGNGNGNDGGKQGNHNNHQRN--YSHSGIN 423
S Q N N+ N N N+N NGN N NGN D GN+NN N ++G N
Sbjct: 66 SPQGNNNNDGNNGNNNNDNNNNNNGNNNNDNNNGNNKDNNNNGNNNNGNNNNGNDNNGNN 125
Query: 424 MKGRESED 431
G +++
Sbjct: 126 NNGNNNDN 133
Score = 40.8 bits (94), Expect = 0.003
Identities = 20/53 (37%), Positives = 25/53 (46%), Gaps = 2/53 (3%)
Query: 371 NEGNEANQNQN--ILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSG 421
N GN N N N N+N N N N NGN N+G +NN N ++ G
Sbjct: 88 NNGNNNNDNNNGNNKDNNNNGNNNNGNNNNGNDNNGNNNNGNNNDNNNQNNGG 140
Score = 37.7 bits (86), Expect = 0.022
Identities = 28/103 (27%), Positives = 35/103 (33%), Gaps = 32/103 (31%)
Query: 309 SNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAASK 368
+N NN++ N N G N D G NK
Sbjct: 76 NNGNNNNDNNNNNNGNNNNDNN--NGNNKDNN---------------------------- 105
Query: 369 QPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHN 411
N GN N N N ++N N NG N N N N+GG N +
Sbjct: 106 --NNGNNNNGNNNNGNDNNGNNNNGNNNDNNNQNNGGGSNNRS 146
Score = 36.2 bits (82), Expect = 0.063
Identities = 18/61 (29%), Positives = 24/61 (38%)
Query: 371 NEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESE 430
N N N N N N + N GN N N+ G N NN+ N + + G S
Sbjct: 84 NNNNNNGNNNNDNNNGNNKDNNNNGNNNNGNNNNGNDNNGNNNNGNNNDNNNQNNGGGSN 143
Query: 431 D 431
+
Sbjct: 144 N 144
Score = 35.4 bits (80), Expect = 0.11
Identities = 21/62 (33%), Positives = 29/62 (45%), Gaps = 6/62 (9%)
Query: 371 NEGNEANQNQNILANSNTPNGNGL---GNG---NGNGNDGGKQGNHNNHQRNYSHSGINM 424
N N+ N N N N++ NGN NG NGN N+G +NN+ N ++ N
Sbjct: 79 NNNNDNNNNNNGNNNNDNNNGNNKDNNNNGNNNNGNNNNGNDNNGNNNNGNNNDNNNQNN 138
Query: 425 KG 426
G
Sbjct: 139 GG 140
Score = 30.8 bits (68), Expect = 2.6
Identities = 25/110 (22%), Positives = 36/110 (32%), Gaps = 33/110 (30%)
Query: 309 SNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAASK 368
+NNNN+ +N N N+D G N
Sbjct: 84 NNNNNNGNNNNDNNNGNNKDNNNNGNNNNGNN---------------------------- 115
Query: 369 QPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYS 418
N GN+ N N N N N + N NG G+ N + N+ + + S
Sbjct: 116 --NNGNDNNGNNN---NGNNNDNNNQNNGGGSNNRSPPPPSRNSDRNSPS 160
>At1g02690 importin alpha, putative
Length = 538
Score = 42.7 bits (99), Expect = 7e-04
Identities = 47/183 (25%), Positives = 77/183 (41%), Gaps = 18/183 (9%)
Query: 442 AARALWHLAKGNVAICRSITESRALLCFSVLLEKGPEAVQYNSAMALMEITAVAEKDAEL 501
AA AL ++A G R I +S A+ F LL E V+ + AL + + K
Sbjct: 140 AAWALTNIASGTSENTRVIIDSGAVPLFVKLLSSASEEVREQAVWALGNVAGDSPK---- 195
Query: 502 RKSAFKPNSPACKAVVDQVLKIIEKADSDLLIPCVKAIGNLAR-----TFKATETRMIGP 556
+ + +C+A++ + + E + +L + N R F+ T+ +
Sbjct: 196 ----CRDHVLSCEAMMSLLAQFHEHSKLSMLRNATWTLSNFCRGKPQPAFEQTKAALPA- 250
Query: 557 LVKLLDEREAEVSREASIALRKFAGSENYLHVDHSNAIISAGGAKHLIQLVYFGEQMVQI 616
L +LL + EV +AS AL + N + +I AG L+QL+ V I
Sbjct: 251 LERLLHSTDEEVLTDASWALSYLSDGTN----EKIQTVIDAGVIPRLVQLLAHPSPSVLI 306
Query: 617 PAL 619
PAL
Sbjct: 307 PAL 309
Score = 37.0 bits (84), Expect = 0.037
Identities = 29/129 (22%), Positives = 58/129 (44%), Gaps = 4/129 (3%)
Query: 161 EVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAIGLLGRDAE 220
+++ +AA +L ++A G+ ++II+ G V +KL+ + +E A A+G + D+
Sbjct: 135 QLQFEAAWALTNIASGTSENTRVIIDSGAVPLFVKLLSSASEEVREQAVWALGNVAGDSP 194
Query: 221 SV-EHMIHV-GVCSVFAKILKEGPMKVQGVVAWAVSELAANYPKCQELFAQHNIIRLLVG 278
+H++ + S+ A+ + + + W +S P Q F Q +
Sbjct: 195 KCRDHVLSCEAMMSLLAQFHEHSKLSMLRNATWTLSNFCRGKP--QPAFEQTKAALPALE 252
Query: 279 HLAFETVEE 287
L T EE
Sbjct: 253 RLLHSTDEE 261
Score = 33.9 bits (76), Expect = 0.31
Identities = 39/155 (25%), Positives = 74/155 (47%), Gaps = 8/155 (5%)
Query: 154 MLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAIG 213
+L + +EV +DA+ +L L+ G++ + +I+ G + L++L+ A R IG
Sbjct: 254 LLHSTDEEVLTDASWALSYLSDGTNEKIQTVIDAGVIPRLVQLLAHPSPSVLIPALRTIG 313
Query: 214 -LLGRDAESVEHMIHVGVCSVFAKILKEGPMK-VQGVVAWAVSEL-AANYPKCQELFAQH 270
++ D + +I +LK K ++ W +S + A N + QE+F Q
Sbjct: 314 NIVTGDDIQTQAVISSQALPGLLNLLKNTYKKSIKKEACWTISNITAGNTSQIQEVF-QA 372
Query: 271 NIIRLLVGHL---AFETVEEHSKYAIVSMKANSIH 302
IIR L+ L FE +++ + +AI + + H
Sbjct: 373 GIIRPLINLLEIGEFE-IKKEAVWAISNATSGGNH 406
>At4g12710 unknown protein
Length = 402
Score = 42.0 bits (97), Expect = 0.001
Identities = 46/180 (25%), Positives = 86/180 (47%), Gaps = 13/180 (7%)
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
+ MLF+ + + R + +L++LA ++R I++ G V PL++++K A +E A A
Sbjct: 96 VPMLFSSNVDARHASLLALLNLAVRNERNKIEIVKAGAVPPLIQILKLHNASLRELATAA 155
Query: 212 IGLLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWAVSELAANYPKCQELFAQHN 271
I L + +I GV + ++L G VQG V AV+ L N C+E A
Sbjct: 156 ILTLSAAPANKAMIISSGVPPLLIQMLSSG--TVQGKVD-AVTAL-HNLSACKEYSAP-- 209
Query: 272 IIRLLVGHLAFETVEEHSKYAIVSMKANSIHAAVVMASNNNNSSSNLNPKKGTENEDGVV 331
I+ + ++E K++ + KA ++ ++ S + ++ T EDG++
Sbjct: 210 ILDAKAVYPLIHLLKECKKHSKFAEKATALVEMILSHSEDGRNAI-------TSCEDGIL 262
Score = 32.0 bits (71), Expect = 1.2
Identities = 18/51 (35%), Positives = 29/51 (56%), Gaps = 1/51 (1%)
Query: 166 AAASLVSLARGS-DRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAIGLL 215
A +L+SL R D+Y KLI++EG + LL +G + ++ A + LL
Sbjct: 279 AVGALLSLCRSDRDKYRKLILKEGAIPGLLSSTVDGTSKSRDRARVLLDLL 329
>At1g44120 hypothetical protein
Length = 2114
Score = 41.6 bits (96), Expect = 0.001
Identities = 38/163 (23%), Positives = 73/163 (44%), Gaps = 5/163 (3%)
Query: 138 IAANEPILCFIWEQIAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLI 197
I E ++ +W+Q+ + V +L +L D + L +E+GGV +LKL+
Sbjct: 140 IFVTEGVVPSLWDQLKTGKKQDKTVEGHLVGALRNLCGDKDGFWALTLEDGGVDIILKLL 199
Query: 198 KEGKADGQENAARAIGLLGR-DAESVEHMIHVGVCSVFAKIL-KEGPMKVQGVVAWAVSE 255
+ Q NAA + L R S+ + G V ++L +E + V+ V A+
Sbjct: 200 QSSNPVSQSNAASLLARLIRIFTSSISKVEESGAVQVLVQLLGEENSVFVRASVVNALEA 259
Query: 256 LAANYPKCQELFAQHNIIRLLVGHL---AFETVEEHSKYAIVS 295
+ + + + + I LL+ + + E+VEE ++ + S
Sbjct: 260 ITSKSEEAITVARDLDGIHLLISAVVASSKESVEEETERVLQS 302
Score = 33.1 bits (74), Expect = 0.53
Identities = 33/116 (28%), Positives = 55/116 (46%), Gaps = 9/116 (7%)
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
I++L +G+ + ++A+ L L + + K++I G + PLL L+K D + A A
Sbjct: 64 ISLLRSGTLLAKLNSASVLTVLCKDKNVRSKILIG-GCIPPLLSLLKSDSVDAKRVVAEA 122
Query: 212 I---GLLGRDAESVEHMIHV--GVCSVFAKILKEGPMK---VQGVVAWAVSELAAN 259
I L G D ++V I V GV LK G + V+G + A+ L +
Sbjct: 123 IYEVSLCGMDGDNVGTKIFVTEGVVPSLWDQLKTGKKQDKTVEGHLVGALRNLCGD 178
Score = 30.8 bits (68), Expect = 2.6
Identities = 20/70 (28%), Positives = 37/70 (52%), Gaps = 1/70 (1%)
Query: 152 IAMLFTG-SQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAAR 210
+ +L TG SQ+ + DA +++L S+ + + G + LL L+K G QE++A
Sbjct: 495 LQILETGVSQKAKDDAVRVILNLCCHSEEIRLCVEKAGAIPALLGLLKNGGPKSQESSAN 554
Query: 211 AIGLLGRDAE 220
+ L + A+
Sbjct: 555 TLLKLIKTAD 564
>At5g19330 putative protein
Length = 636
Score = 41.2 bits (95), Expect = 0.002
Identities = 39/157 (24%), Positives = 67/157 (41%), Gaps = 2/157 (1%)
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
I +L + E + +AA L A I++ G V PL+++++ +E +A A
Sbjct: 289 IGLLSSCCPESQREAALLLGQFASTDSDCKVHIVQRGAVRPLIEMLQSPDVQLKEMSAFA 348
Query: 212 IGLLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWAVSELAANYPKCQELFAQHN 271
+G L +DA + + H G K+L +Q A+A+ LA N +
Sbjct: 349 LGRLAQDAHNQAGIAHSGGLGPLLKLLDSRNGSLQHNAAFALYGLADNEDNVSDFIRVGG 408
Query: 272 IIRLLVGHLAFETVEEHSKYAIVSMKANSIHAAVVMA 308
I +L G + V H Y ++ + SI V +A
Sbjct: 409 IQKLQDGEFIVQ-VLRHLLY-LMRISEKSIQRRVALA 443
Score = 38.5 bits (88), Expect = 0.013
Identities = 36/158 (22%), Positives = 67/158 (41%), Gaps = 28/158 (17%)
Query: 495 AEKDAELRKSAFKPNSPACKAVVDQ------VLKIIEKADSDLLIPCVKAIGNLARTFK- 547
++++A L F CK + Q ++++++ D L A+G LA+
Sbjct: 299 SQREAALLLGQFASTDSDCKVHIVQRGAVRPLIEMLQSPDVQLKEMSAFALGRLAQDAHN 358
Query: 548 ---ATETRMIGPLVKLLDEREAEVSREASIALRKFAGSENYLHVDHSNAIISAGG----- 599
+ +GPL+KLLD R + A+ AL A +E D+ + I GG
Sbjct: 359 QAGIAHSGGLGPLLKLLDSRNGSLQHNAAFALYGLADNE-----DNVSDFIRVGGIQKLQ 413
Query: 600 --------AKHLIQLVYFGEQMVQIPALVLLSYIALHV 629
+HL+ L+ E+ +Q + L+++ L +
Sbjct: 414 DGEFIVQVLRHLLYLMRISEKSIQRRVALALAHLWLEL 451
Score = 38.1 bits (87), Expect = 0.016
Identities = 42/154 (27%), Positives = 68/154 (43%), Gaps = 14/154 (9%)
Query: 161 EVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQEN---------AARA 211
EV +A +L LA + Y KLI+++G + L+ L+K K DG + AA A
Sbjct: 123 EVEKGSAFALGLLAIKPE-YQKLIVDKGALPHLVNLLKRNK-DGSSSRAVNSVIRRAADA 180
Query: 212 IGLLGRDAESVEHMIHV-GVCSVFAKILKEGPMKVQGVVAWAVSELAANYPKCQELFAQH 270
I L + S++ + V G ++L+ KVQ A A+ LA + +
Sbjct: 181 ITNLAHENSSIKTRVRVEGGIPPLVELLEFSDSKVQRAAAGALRTLAFKNDDNKNQIVEC 240
Query: 271 NIIRLLVGHLAFETVEEHSKYAIVSMKANSIHAA 304
N + L+ L E H Y V + N +H++
Sbjct: 241 NALPTLILMLGSEDAAIH--YEAVGVIGNLVHSS 272
Score = 35.8 bits (81), Expect = 0.082
Identities = 50/230 (21%), Positives = 96/230 (41%), Gaps = 20/230 (8%)
Query: 422 INMKGRESEDAETKA--SMKEMAARALWHLAKGNVAICRSITESRALLCFSVLLEKGPEA 479
+N+ R + + ++A S+ AA A+ +LA N +I + + LLE
Sbjct: 155 VNLLKRNKDGSSSRAVNSVIRRAADAITNLAHENSSIKTRVRVEGGIPPLVELLEFSDSK 214
Query: 480 VQYNSAMALMEITAVAEKDAELRKSAFKPNSPACKAVVDQVLKIIEKADSDLLIPCVKAI 539
VQ +A AL + + K C A+ +L ++ D+ + V I
Sbjct: 215 VQRAAAGALRTLA--------FKNDDNKNQIVECNALPTLIL-MLGSEDAAIHYEAVGVI 265
Query: 540 GNLARTFKATETRMIG-----PLVKLLDEREAEVSREASIALRKFAGSENYLHVDHSNAI 594
GNL + + ++ P++ LL E REA++ L +FA +++ D I
Sbjct: 266 GNLVHSSPHIKKEVLTAGALQPVIGLLSSCCPESQREAALLLGQFASTDS----DCKVHI 321
Query: 595 ISAGGAKHLIQLVYFGEQMVQIPALVLLSYIALHVPDSEELALAEVLGVL 644
+ G + LI+++ + ++ + L +A + +A + LG L
Sbjct: 322 VQRGAVRPLIEMLQSPDVQLKEMSAFALGRLAQDAHNQAGIAHSGGLGPL 371
Score = 34.7 bits (78), Expect = 0.18
Identities = 34/136 (25%), Positives = 64/136 (47%), Gaps = 14/136 (10%)
Query: 526 KADSDLLIPCVKAIGNLARTFKATETRM-----IGPLVKLLDEREAEVSREASIALRKFA 580
+A + ++ AI NLA + +TR+ I PLV+LL+ +++V R A+ ALR A
Sbjct: 168 RAVNSVIRRAADAITNLAHENSSIKTRVRVEGGIPPLVELLEFSDSKVQRAAAGALRTLA 227
Query: 581 GSENYLHVDHSNAIISAGGAKHLIQLVYFGEQMVQIPALVLLSYIALHVPDSEEL----- 635
+ + D+ N I+ LI ++ + + A+ ++ + P ++
Sbjct: 228 ----FKNDDNKNQIVECNALPTLILMLGSEDAAIHYEAVGVIGNLVHSSPHIKKEVLTAG 283
Query: 636 ALAEVLGVLEWASKQS 651
AL V+G+L +S
Sbjct: 284 ALQPVIGLLSSCCPES 299
>At4g02150 AtKAP alpha
Length = 531
Score = 38.9 bits (89), Expect = 0.010
Identities = 49/183 (26%), Positives = 73/183 (39%), Gaps = 18/183 (9%)
Query: 442 AARALWHLAKGNVAICRSITESRALLCFSVLLEKGPEAVQYNSAMALMEITAVAEKDAEL 501
AA AL ++A G I ES A+ F LL E V+ + AL + + K +L
Sbjct: 139 AAWALTNIASGTSENTNVIIESGAVPIFIQLLSSASEDVREQAVWALGNVAGDSPKCRDL 198
Query: 502 RKSAFKPNSPACKAVVDQVLKIIEKADSDLLIPCVKAIGNLAR-----TFKATETRMIGP 556
S A+ + + E +L + N R F+ T+ +
Sbjct: 199 VLSY--------GAMTPLLSQFNENTKLSMLRNATWTLSNFCRGKPPPAFEQTQPAL-PV 249
Query: 557 LVKLLDEREAEVSREASIALRKFAGSENYLHVDHSNAIISAGGAKHLIQLVYFGEQMVQI 616
L +L+ + EV +A AL + + N D A+I AG LIQL+ V I
Sbjct: 250 LERLVQSMDEEVLTDACWALSYLSDNSN----DKIQAVIEAGVVPRLIQLLGHSSPSVLI 305
Query: 617 PAL 619
PAL
Sbjct: 306 PAL 308
Score = 33.9 bits (76), Expect = 0.31
Identities = 17/66 (25%), Positives = 35/66 (52%)
Query: 161 EVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAIGLLGRDAE 220
+++ +AA +L ++A G+ +IIE G V ++L+ D +E A A+G + D+
Sbjct: 134 KLQFEAAWALTNIASGTSENTNVIIESGAVPIFIQLLSSASEDVREQAVWALGNVAGDSP 193
Query: 221 SVEHMI 226
++
Sbjct: 194 KCRDLV 199
Score = 33.1 bits (74), Expect = 0.53
Identities = 21/105 (20%), Positives = 48/105 (45%), Gaps = 2/105 (1%)
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEG--KADGQENAA 209
I +L S V A ++ ++ G D ++++++ + LL L+K K+ +E
Sbjct: 293 IQLLGHSSPSVLIPALRTIGNIVTGDDLQTQMVLDQQALPCLLNLLKNNYKKSIKKEACW 352
Query: 210 RAIGLLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWAVS 254
+ +A+ ++ +I G+ +L+ +V+ AW +S
Sbjct: 353 TISNITAGNADQIQAVIDAGIIQSLVWVLQSAEFEVKKEAAWGIS 397
Score = 32.7 bits (73), Expect = 0.69
Identities = 21/96 (21%), Positives = 46/96 (47%), Gaps = 2/96 (2%)
Query: 184 IIEEGGVGPLLKLI-KEGKADGQENAARAI-GLLGRDAESVEHMIHVGVCSVFAKILKEG 241
+++ G V ++K + ++ Q AA A+ + +E+ +I G +F ++L
Sbjct: 114 VVQSGVVPRVVKFLSRDDFPKLQFEAAWALTNIASGTSENTNVIIESGAVPIFIQLLSSA 173
Query: 242 PMKVQGVVAWAVSELAANYPKCQELFAQHNIIRLLV 277
V+ WA+ +A + PKC++L + + L+
Sbjct: 174 SEDVREQAVWALGNVAGDSPKCRDLVLSYGAMTPLL 209
Score = 29.6 bits (65), Expect = 5.9
Identities = 33/132 (25%), Positives = 55/132 (41%), Gaps = 4/132 (3%)
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEG-KADGQENAAR 210
I +L + S++VR A +L ++A S + L++ G + PLL E K NA
Sbjct: 167 IQLLSSASEDVREQAVWALGNVAGDSPKCRDLVLSYGAMTPLLSQFNENTKLSMLRNATW 226
Query: 211 AIGLLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWAVSELAAN-YPKCQELFAQ 269
+ R V ++++ +V WA+S L+ N K Q +
Sbjct: 227 TLSNFCRGKPPPAFEQTQPALPVLERLVQSMDEEVLTDACWALSYLSDNSNDKIQAVIEA 286
Query: 270 HNIIRL--LVGH 279
+ RL L+GH
Sbjct: 287 GVVPRLIQLLGH 298
>At1g09270 putative importin beta binding domain
Length = 538
Score = 38.5 bits (88), Expect = 0.013
Identities = 21/103 (20%), Positives = 53/103 (51%), Gaps = 2/103 (1%)
Query: 161 EVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAIGLLGRDAE 220
+++ +AA +L ++A G+ + +++IE+G V +KL+ D +E A A+G + D+
Sbjct: 138 QLQFEAAWALTNVASGTSDHTRVVIEQGAVPIFVKLLTSASDDVREQAVWALGNVAGDSP 197
Query: 221 SVEHMI--HVGVCSVFAKILKEGPMKVQGVVAWAVSELAANYP 261
+ +++ + + + A++ + + + W +S P
Sbjct: 198 NCRNLVLNYGALEPLLAQLNENSKLSMLRNATWTLSNFCRGKP 240
Score = 34.7 bits (78), Expect = 0.18
Identities = 44/189 (23%), Positives = 72/189 (37%), Gaps = 30/189 (15%)
Query: 442 AARALWHLAKGNVAICRSITESRALLCFSVLLEKGPEAVQYNSAMALMEITAVAEKDAEL 501
AA AL ++A G R + E A+ F LL + V+ + AL +
Sbjct: 143 AAWALTNVASGTSDHTRVVIEQGAVPIFVKLLTSASDDVREQAVWALGNVAG-------- 194
Query: 502 RKSAFKPNSPACKAVV-------DQVLKIIEKADSDLLIPCVKAIGNLARTFKATETRMI 554
+SP C+ +V + ++ E + +L + N R T +
Sbjct: 195 -------DSPNCRNLVLNYGALEPLLAQLNENSKLSMLRNATWTLSNFCRGKPPTPFEQV 247
Query: 555 GPLVKLLDE----REAEVSREASIALRKFAGSENYLHVDHSNAIISAGGAKHLIQLVYFG 610
P + +L + + EV +A AL + N D A+I AG L++L+
Sbjct: 248 KPALPILRQLIYLNDEEVLTDACWALSYLSDGPN----DKIQAVIEAGVCPRLVELLGHQ 303
Query: 611 EQMVQIPAL 619
V IPAL
Sbjct: 304 SPTVLIPAL 312
Score = 32.0 bits (71), Expect = 1.2
Identities = 17/70 (24%), Positives = 32/70 (45%), Gaps = 5/70 (7%)
Query: 225 MIHVGVCSVFAKILKEGPMKVQGVVAWAVSELAANYPKCQELFAQHNIIRLLVGHLAFET 284
+I G +F K+L V+ WA+ +A + P C+ L + + L+ L
Sbjct: 161 VIEQGAVPIFVKLLTSASDDVREQAVWALGNVAGDSPNCRNLVLNYGALEPLLAQL---- 216
Query: 285 VEEHSKYAIV 294
E+SK +++
Sbjct: 217 -NENSKLSML 225
Score = 30.8 bits (68), Expect = 2.6
Identities = 35/157 (22%), Positives = 67/157 (42%), Gaps = 6/157 (3%)
Query: 154 MLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAIG 213
+++ +EV +DA +L L+ G + + +IE G L++L+ A R +G
Sbjct: 257 LIYLNDEEVLTDACWALSYLSDGPNDKIQAVIEAGVCPRLVELLGHQSPTVLIPALRTVG 316
Query: 214 -LLGRDAESVEHMIHVGVCSVFAKILKEGPMK-VQGVVAWAVSELAANYPKCQELFAQHN 271
++ D + +I GV +L + K ++ W +S + A E
Sbjct: 317 NIVTGDDSQTQFIIESGVLPHLYNLLTQNHKKSIKKEACWTISNITAGNKLQIEAVVGAG 376
Query: 272 IIRLLVGHL---AFETVEEHSKYAIVSMKANSIHAAV 305
II LV HL A +++ + +AI + + H +
Sbjct: 377 IILPLV-HLLQNAEFDIKKEAAWAISNATSGGSHEQI 412
>At2g45720 unknown protein
Length = 553
Score = 37.4 bits (85), Expect = 0.028
Identities = 25/90 (27%), Positives = 44/90 (48%), Gaps = 1/90 (1%)
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
I +L +GS + A SL ++ S+ + I+ GGVGPL+++ K G + Q +A
Sbjct: 238 IRLLESGSIVAKEKAVISLQRMSISSET-SRSIVGHGGVGPLIEICKTGDSVSQSASACT 296
Query: 212 IGLLGRDAESVEHMIHVGVCSVFAKILKEG 241
+ + E +++ G+ V IL G
Sbjct: 297 LKNISAVPEVRQNLAEEGIVKVMINILNCG 326
Score = 34.7 bits (78), Expect = 0.18
Identities = 39/131 (29%), Positives = 63/131 (47%), Gaps = 12/131 (9%)
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADG-QENAAR 210
+ +L +GS + AA+++ +A ++ K +I E G PLL + E KA G +E AA+
Sbjct: 399 VHVLKSGSIGAQQAAASTICRIATSNET--KRMIGESGCIPLLIRMLEAKASGAREVAAQ 456
Query: 211 AIGLL---GRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWAVSELAA--NYPKCQE 265
AI L R+ V+ SV + ++ P +AVS LAA + KC++
Sbjct: 457 AIASLVTVPRNCREVKR----DEKSVTSLVMLLEPSPGNSAKKYAVSGLAALCSSRKCKK 512
Query: 266 LFAQHNIIRLL 276
L H + L
Sbjct: 513 LMVSHGAVGYL 523
>At1g27750 unknown protein
Length = 1975
Score = 37.4 bits (85), Expect = 0.028
Identities = 18/48 (37%), Positives = 24/48 (49%), Gaps = 1/48 (2%)
Query: 368 KQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQR 415
+QP E + + N N N+ NG G G G G GG + NHN+ R
Sbjct: 1917 EQPQEKSNESSNNNSEVNTEAENGGGRGRGRGRRGGGGGR-NHNHKDR 1963
>At5g67340 unknown proteins
Length = 707
Score = 37.0 bits (84), Expect = 0.037
Identities = 19/51 (37%), Positives = 29/51 (56%)
Query: 158 GSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENA 208
GS + +A A+L+ L S ++ +I EG + PL+ L K G A G+E A
Sbjct: 639 GSARGKENATAALLQLCTHSPKFCNNVIREGVIPPLVALTKSGTARGKEKA 689
Score = 33.9 bits (76), Expect = 0.31
Identities = 22/65 (33%), Positives = 35/65 (53%), Gaps = 2/65 (3%)
Query: 179 RYGKLII-EEGGVGPLLKLIKEGKADGQENAARA-IGLLGRDAESVEHMIHVGVCSVFAK 236
R GK+ I EEGG+ L+++++ G A G+ENA A + L + ++I GV
Sbjct: 617 REGKIAIGEEGGIPVLVEVVELGSARGKENATAALLQLCTHSPKFCNNVIREGVIPPLVA 676
Query: 237 ILKEG 241
+ K G
Sbjct: 677 LTKSG 681
Score = 33.9 bits (76), Expect = 0.31
Identities = 24/110 (21%), Positives = 53/110 (47%), Gaps = 2/110 (1%)
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEG-KADGQENAAR 210
+++L++ + +++DA L++L+ +D LI E G + PL+ ++K G + + N+A
Sbjct: 469 VSLLYSTDERIQADAVTCLLNLSI-NDNNKSLIAESGAIVPLIHVLKTGYLEEAKANSAA 527
Query: 211 AIGLLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWAVSELAANY 260
+ L E + G +L G + + A A+ L+ ++
Sbjct: 528 TLFSLSVIEEYKTEIGEAGAIEPLVDLLGSGSLSGKKDAATALFNLSIHH 577
>At2g23140 hypothetical protein
Length = 924
Score = 37.0 bits (84), Expect = 0.037
Identities = 18/54 (33%), Positives = 31/54 (57%)
Query: 158 GSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
GS + +AAA+L+ L+ S R+ ++++EG V PL+ L + G +E A
Sbjct: 837 GSARGKENAAAALLQLSTNSGRFCNMVLQEGAVPPLVALSQSGTPRAREKKPTA 890
Score = 35.0 bits (79), Expect = 0.14
Identities = 25/126 (19%), Positives = 60/126 (46%), Gaps = 2/126 (1%)
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
+ +L++ + +A +L++L+ +D K I + G + PL+ +++ G ++ +EN+A
Sbjct: 668 VELLYSTDSATQENAVTALLNLSI-NDNNKKAIADAGAIEPLIHVLENGSSEAKENSAAT 726
Query: 212 IGLLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWAVSELAANYPKCQELFAQHN 271
+ L E+ + G +L G + + A A+ L+ + + + + Q
Sbjct: 727 LFSLSVIEENKIKIGQSGAIGPLVDLLGNGTPRGKKDAATALFNLSI-HQENKAMIVQSG 785
Query: 272 IIRLLV 277
+R L+
Sbjct: 786 AVRYLI 791
Score = 31.6 bits (70), Expect = 1.5
Identities = 16/45 (35%), Positives = 29/45 (63%), Gaps = 4/45 (8%)
Query: 168 ASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAI 212
A+L ++ G + G +EGG+ L+++++ G A G+ENAA A+
Sbjct: 809 ANLATIPEGRNAIG----QEGGIPLLVEVVELGSARGKENAAAAL 849
>At2g22130 unknown protein
Length = 2048
Score = 37.0 bits (84), Expect = 0.037
Identities = 46/203 (22%), Positives = 87/203 (42%), Gaps = 17/203 (8%)
Query: 83 CRANGLMKRVFT--IIPAAAFRKTSSHLENSIGDVSWLLRVSAPADDRGG--EYLGLPPI 138
C+ N L +V IP SS +E I + VS GG +++G I
Sbjct: 82 CKENELRVKVLLGGCIPPLLGLLKSSSVEGQIAAAKTIYAVS-----EGGVKDHVG-SKI 135
Query: 139 AANEPILCFIWEQIAMLFTGSQEVRSDA--AASLVSLARGSDRYGKLIIEEGGVGPLLKL 196
+ E ++ +W+Q+ +G+++ D +L +L+ ++ + I GGV L+KL
Sbjct: 136 FSTEGVVPVLWDQLR---SGNKKGEVDGLLTGALKNLSSTTEGFWSETIRAGGVDVLVKL 192
Query: 197 IKEGKADGQENAARAIG-LLGRDAESVEHMIHVGVCSVFAKILKEG-PMKVQGVVAWAVS 254
+ G++ N + ++ DA ++ + K+L G V+ A A+
Sbjct: 193 LTSGQSSTLSNVCFLLACMMMEDASVCSSVLTADITKQLLKLLGSGNEAPVRAEAAAALK 252
Query: 255 ELAANYPKCQELFAQHNIIRLLV 277
L+A + + A N I +L+
Sbjct: 253 SLSAQSKEAKREIANSNGIPVLI 275
Score = 35.0 bits (79), Expect = 0.14
Identities = 21/64 (32%), Positives = 33/64 (50%)
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
+ +L TGS + R D+A L +L S+ + V LL L+K G +G+E AA+
Sbjct: 489 VQILETGSAKAREDSATILRNLCNHSEDIRACVESADAVPALLWLLKNGSPNGKEIAAKT 548
Query: 212 IGLL 215
+ L
Sbjct: 549 LNHL 552
Score = 34.7 bits (78), Expect = 0.18
Identities = 32/113 (28%), Positives = 56/113 (49%), Gaps = 7/113 (6%)
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
+++L +GS V+ AA L SL + ++ K+++ G + PLL L+K +GQ AA+
Sbjct: 60 VSLLRSGSVGVKIQAATVLGSLCKENELRVKVLLG-GCIPPLLGLLKSSSVEGQIAAAKT 118
Query: 212 IGLLG----RDAESVEHMIHVGVCSVFAKILKEGPMK--VQGVVAWAVSELAA 258
I + +D + GV V L+ G K V G++ A+ L++
Sbjct: 119 IYAVSEGGVKDHVGSKIFSTEGVVPVLWDQLRSGNKKGEVDGLLTGALKNLSS 171
Score = 34.3 bits (77), Expect = 0.24
Identities = 24/106 (22%), Positives = 49/106 (45%), Gaps = 1/106 (0%)
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
I++L S++ + A A L L+ +D I GG+ PL+++++ G A +E++A
Sbjct: 447 ISLLGLSSEQQQECAVALLCLLSNENDESKWAITAAGGIPPLVQILETGSAKAREDSATI 506
Query: 212 IGLLGRDAESVEHMIH-VGVCSVFAKILKEGPMKVQGVVAWAVSEL 256
+ L +E + + +LK G + + A ++ L
Sbjct: 507 LRNLCNHSEDIRACVESADAVPALLWLLKNGSPNGKEIAAKTLNHL 552
Score = 30.0 bits (66), Expect = 4.5
Identities = 32/143 (22%), Positives = 61/143 (42%), Gaps = 6/143 (4%)
Query: 509 NSPACKAVVDQVLKIIEKADSDLLIPCVKAIGNLARTFKATETRMIGPLVKLLDEREAEV 568
NS A + +V + + + +L+ + + ++A + R L+ L +E
Sbjct: 397 NSDAKRLLVGLITMAVNEVQDELVKALLMLCNHEGSLWQALQGREGIQLLISLLGLSSEQ 456
Query: 569 SREASIALRKFAGSENYLHVDHSN-AIISAGGAKHLIQLVYFGEQMVQIPALVLLSYIAL 627
+E ++AL +EN D S AI +AGG L+Q++ G + + +L +
Sbjct: 457 QQECAVALLCLLSNEN----DESKWAITAAGGIPPLVQILETGSAKAREDSATILRNLCN 512
Query: 628 HVPDSEE-LALAEVLGVLEWASK 649
H D + A+ + L W K
Sbjct: 513 HSEDIRACVESADAVPALLWLLK 535
>At5g52000 importin alpha subunit
Length = 441
Score = 36.6 bits (83), Expect = 0.048
Identities = 34/154 (22%), Positives = 75/154 (48%), Gaps = 6/154 (3%)
Query: 154 MLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAIG 213
+L + ++V +A +L L+ GS+ + +IE G V L+++++ A IG
Sbjct: 178 LLHSHDEDVLKNACMALCHLSEGSEDGIQSVIEAGFVPKLVQILQLPSPVVLVPALLTIG 237
Query: 214 -LLGRDAESVEHMIHVGVCSVFAKIL-KEGPMKVQGVVAWAVSELAANYPKCQELFAQHN 271
+ + + + +I+ G + + +L + K++ W +S + A + + N
Sbjct: 238 AMTAGNHQQTQCVINSGALPIISNMLTRNHENKIKKCACWVISNITAGTKEQIQSVIDAN 297
Query: 272 IIRLLVGHLAFET---VEEHSKYAIVSMKANSIH 302
+I +LV +LA +T +++ + +AI +M N H
Sbjct: 298 LIPILV-NLAQDTDFYMKKEAVWAISNMALNGSH 330
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.131 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,608,338
Number of Sequences: 26719
Number of extensions: 626978
Number of successful extensions: 3633
Number of sequences better than 10.0: 115
Number of HSP's better than 10.0 without gapping: 49
Number of HSP's successfully gapped in prelim test: 70
Number of HSP's that attempted gapping in prelim test: 3103
Number of HSP's gapped (non-prelim): 391
length of query: 687
length of database: 11,318,596
effective HSP length: 106
effective length of query: 581
effective length of database: 8,486,382
effective search space: 4930587942
effective search space used: 4930587942
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 64 (29.3 bits)
Medicago: description of AC147013.12


