

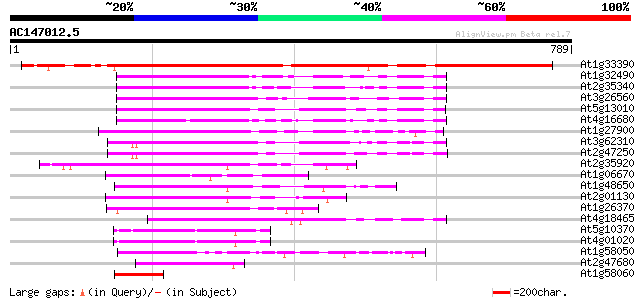
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147012.5 + phase: 0
(789 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g33390 RNA helicase, putative 683 0.0
At1g32490 putative RNA helicase (At1g32490) 189 5e-48
At2g35340 putative pre-mRNA splicing factor RNA helicase 181 2e-45
At3g26560 ATP-dependent RNA helicase, putative 170 3e-42
At5g13010 pre-mRNA splicing factor ATP-dependent RNA helicase -l... 165 1e-40
At4g16680 RNA helicase 164 2e-40
At1g27900 ATP-dependent RNA helicase (At1g27900) 151 1e-36
At3g62310 ATP-dependent RNA helicase-like protein 151 2e-36
At2g47250 putative pre-mRNA splicing factor RNA helicase 149 5e-36
At2g35920 ATP-dependent RNA helicase A like protein 146 5e-35
At1g06670 DEIH-box RNA/DNA helicase 133 3e-31
At1g48650 hypothetical protein 130 3e-30
At2g01130 ATP-dependent RNA like helicase A 125 7e-29
At1g26370 hypothetical protein 123 5e-28
At4g18465 RNA helicase - like protein 118 1e-26
At5g10370 putative protein 117 2e-26
At4g01020 unknown protein 115 7e-26
At1g58050 hypothetical protein 107 2e-23
At2g47680 putative ATP-dependent RNA helicase A 81 3e-15
At1g58060 hypothetical protein 59 1e-08
>At1g33390 RNA helicase, putative
Length = 1237
Score = 683 bits (1762), Expect = 0.0
Identities = 382/765 (49%), Positives = 489/765 (62%), Gaps = 77/765 (10%)
Query: 17 ETSSVEGININEINEAFEMPGSSSMQQTDRFSGYDED-----DNNFDEN--ESDSYDSET 69
+ S G+++ EI EAF+ S Q RFS + ED D N+D++ E D Y+S+
Sbjct: 526 DDGSFGGVDMKEIAEAFD---DDSNNQNSRFSSHGEDPSDIGDGNYDDDFEEEDMYESDE 582
Query: 70 ESELEFNDDDKNNHEGSKNNNNIVDVLGNEGSLASLKAAFENLSGQATLSSSNVNTEDSL 129
+ + E DD EG L +L+AAF L+ + + +V+ E +
Sbjct: 583 DRDWETVDD------------GFASSFVEEGKLDALRAAFNALADK----NGSVSAEPA- 625
Query: 130 DQSKVGREKIARENHDS-------SPGALFVLPLYAMLPAAAQLRVFDGVKEGERLVVVA 182
+ IA EN ++ SPG L VLPLYAML AAQLRVF+ V++ ERLVVVA
Sbjct: 626 -------KSIAAENQEAEQVKNKFSPGKLRVLPLYAMLSPAAQLRVFEEVEKEERLVVVA 678
Query: 183 TNVAETSLTIPGIKYVVDTGREKVKNYDSSNGMETYEVKWISKASAAQRAGRAGRTAAGH 242
TNVAETSLTIPGIKYVVDTGR KVKNYDS GME+YEV WIS+ASA+QRAGRAGRT GH
Sbjct: 679 TNVAETSLTIPGIKYVVDTGRVKVKNYDSKTGMESYEVDWISQASASQRAGRAGRTGPGH 738
Query: 243 CYRLYSSAAFSNEFPEFSPAEVEKVPVHGVVLLLKSMQIKKVANFPFPTSLKAASLLEAE 302
CYRLYSSA FSN F E S E+ KVPV GV+LL+KSM I KV NFPFPT + +++ EAE
Sbjct: 739 CYRLYSSAVFSNIFEESSLPEIMKVPVDGVILLMKSMNIPKVENFPFPTPPEPSAIREAE 798
Query: 303 NCLRALEALDSKDELTLLGKAMALYPLSPRHSRMILTVIKNTR-YKHIRNSSLLLAYAVA 361
CL+ALEALDS LT LGKAM+ YP+SPRHSRM+LTVI+ + ++ ++L+L YAVA
Sbjct: 799 RCLKALEALDSNGGLTPLGKAMSHYPMSPRHSRMLLTVIQMLKETRNYSRANLILGYAVA 858
Query: 362 AAAALSLPNPFVMQYEGNDSNKDSETSEKSRMGDNENNIDKTEKTKRKKLKQTSKVAREK 421
A AALSLPNP +M++EG N+ D + + + +K ++K K+ K AR++
Sbjct: 859 AVAALSLPNPLIMEFEGEKKNESK---------DADKTVKQEDKQRKKDRKEKIKAARDR 909
Query: 422 FRIVSSDALAIAYALQCFEHSQNSVQFCEDNALHFKTMDEMSKLRQQLLRLVFFQNDKGG 481
F SSDAL +AYAL FE S+N + FCE N LH KTMDEMSKL+ QLLRLVF
Sbjct: 910 FSNPSSDALTVAYALHSFEVSENGMGFCEANGLHLKTMDEMSKLKDQLLRLVFNCCKPSE 969
Query: 482 LEQEYSWTHVTLEDVEHVWRV---SSAHYPLPLVEERLICRAICAGWADRVAKRIPISKA 538
E +SWTH T++DVE WR+ +S+ PL EE L+ AICAGWADRVA++
Sbjct: 970 TEDSFSWTHGTIQDVEKSWRITTSTSSKTPLLQNEEELLGEAICAGWADRVARK------ 1023
Query: 539 VDGETISRAGRYQSCMVDESIFIHRWSSVSTVHPEFLVYNELLETKRPNKEGETSAKRAY 598
+RA YQ+C V E +F+HRWSS+ PE LVY+ELL T RP Y
Sbjct: 1024 ------TRATEYQACAVQEPVFLHRWSSLINSAPELLVYSELLLTNRP-----------Y 1066
Query: 599 MHGVTNVDPTWLVENAKSSCIFSPPLTDPRPFYDAQADQVKCWVIPTFGRFCWELPKHSI 658
MHG T V P WLV++AKS C+FS PL DP+P+Y ++ D+V CWV+P+FG WELP HS+
Sbjct: 1067 MHGATRVRPEWLVKHAKSLCVFSAPLKDPKPYYSSEEDRVLCWVVPSFGPHNWELPAHSV 1126
Query: 659 PISNVEHRVQVFAYALLEGQVCTCLKSVRKYMSAPPETILRREALGQKRVGNLISKLNSR 718
I+ R F ALL+G+V TCLKS R ++ PET+L REA G +RVG+L+ L +
Sbjct: 1127 AITEDRDRAAAFGCALLQGEVLTCLKSFRALLAGKPETLLEREAWGLERVGSLVMVLTEK 1186
Query: 719 LIDSSAMLRIVWKQNPRELFSEILDWFQQGFRKHFEELWLQMLGE 763
ID+ LR W+QNP L+SEI WFQ+ FR ++LW ML E
Sbjct: 1187 KIDTLESLRKNWEQNPNVLYSEIEVWFQKKFRHRVKDLWQTMLKE 1231
>At1g32490 putative RNA helicase (At1g32490)
Length = 1044
Score = 189 bits (480), Expect = 5e-48
Identities = 143/464 (30%), Positives = 223/464 (47%), Gaps = 91/464 (19%)
Query: 151 LFVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVKNYD 210
L + P+YA LP+ Q ++F+ EG R VV+ATN+AETSLTI GIKYVVD G K+K+Y+
Sbjct: 646 LIICPIYANLPSELQAKIFEPTPEGARKVVLATNIAETSLTIDGIKYVVDPGFSKMKSYN 705
Query: 211 SSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYSSAAFSNEFPEFSPAEVEKVPVH 270
GME+ + ISKASA QRAGRAGRT+ G CYRLY++ ++N+ E + EV++ +
Sbjct: 706 PRTGMESLLITPISKASATQRAGRAGRTSPGKCYRLYTAFNYNNDLEENTVPEVQRTNLA 765
Query: 271 GVVLLLKSMQIKKVANFPFPTSLKAASLLEAENCLRALEALDSKDELTLLGKAMALYPLS 330
VVL LKS+ I + NF F A +L+++ L AL AL+ ELT G+ MA +PL
Sbjct: 766 SVVLALKSLGIHDLINFDFMDPPPAEALVKSLELLFALGALNKLGELTKAGRRMAEFPLD 825
Query: 331 PRHSRMILTVIKNTRYKHIRNSSLLLAYAVAAAAALSLPNPFVMQYEGNDSNKDSETSEK 390
P S+MI + + +YK ++ AA LS+ + + + D+
Sbjct: 826 PMLSKMI---VVSDKYK-------CSDEIISIAAMLSIGGSIFYRPKDKQVHADN----- 870
Query: 391 SRMGDNENNIDKTEKTKRKKLKQTSKVAREKFRIVSSDALAIAYALQCFEHSQNSVQFCE 450
+RM + N+ D +A+ ++ + S Q+C
Sbjct: 871 ARMNFHTGNV--------------------------GDHIALLKVYSSWKETNFSTQWCY 904
Query: 451 DNALHFKTMDEMSKLRQQLLRLVFFQNDKGGLEQEYSWTHVTLEDVEHVWRVSSAHYPLP 510
+N + ++M +R QL +G LE+ L +++ V
Sbjct: 905 ENYIQVRSMKRARDIRDQL---------EGLLERVEIDISSNLNELDSV----------- 944
Query: 511 LVEERLICRAICAGWADRVAKRIPISKAVDGETISRAGRYQSCMVDESIFIHRWSSVSTV 570
++I AG+ AK + + G Y++ +++ IH S +S V
Sbjct: 945 -------RKSIVAGFFPHTAK------------LQKNGSYRTVKHPQTVHIHPNSGLSQV 985
Query: 571 HPEFLVYNELLETKRPNKEGETSAKRAYMHGVTNVDPTWLVENA 614
P ++VY+EL+ T + YM VT + P WL+E A
Sbjct: 986 LPRWVVYHELVLT-----------SKEYMRQVTELKPEWLIELA 1018
>At2g35340 putative pre-mRNA splicing factor RNA helicase
Length = 1087
Score = 181 bits (458), Expect = 2e-45
Identities = 145/465 (31%), Positives = 220/465 (47%), Gaps = 93/465 (20%)
Query: 151 LFVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVKNYD 210
L + P+YA LP+ Q ++F+ EG R VV+ATN+AETSLTI GIKYVVD G K+K+Y+
Sbjct: 689 LIICPIYANLPSELQAKIFEPTPEGARKVVLATNIAETSLTIDGIKYVVDPGFSKMKSYN 748
Query: 211 SSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYSSAAFSNEFPEFSPAEVEKVPVH 270
GME+ V ISKASA QR GRAGRT+ G CYRLY++ + N+ + + E+++ +
Sbjct: 749 PRTGMESLLVTPISKASATQRTGRAGRTSPGKCYRLYTAFNYYNDLEDNTVPEIQRTNLA 808
Query: 271 GVVLLLKSMQIKKVANFPFPTSLKAASLLEAENCLRALEALDSKDELTLLGKAMALYPLS 330
VVL LKS+ I + NF F + +L+++ L AL AL+ ELT G+ MA +PL
Sbjct: 809 SVVLSLKSLGIHNLLNFDFMDPPPSEALIKSLELLFALGALNQLGELTKAGRRMAEFPLD 868
Query: 331 PRHSRMILTVIKNTRYKHIRNSSLLLAYAVAAAAALSL-PNPFVMQYEGNDSNKDSETSE 389
P S+MI + + +YK ++ AA LS+ P+ F Y D ++ +
Sbjct: 869 PMLSKMI---VVSDKYK-------CSDEIISIAAMLSIGPSIF---YRPKDKQVHADNAM 915
Query: 390 KSRMGDNENNIDKTEKTKRKKLKQTSKVAREKFRIVSSDALAIAYALQCFEHSQNSVQFC 449
K+ N D +A ++ + S Q+C
Sbjct: 916 KNFHVGN-----------------------------VGDHIAFLKIYNSWKETNYSTQWC 946
Query: 450 EDNALHFKTMDEMSKLRQQLLRLVFFQNDKGGLEQEYSWTHVTLEDVEHVWRVSSAHYPL 509
+N + ++M +R QL L LE VE VSS L
Sbjct: 947 YENYIQVRSMKRARDIRDQLEGL--------------------LERVE--IDVSSNANEL 984
Query: 510 PLVEERLICRAICAGWADRVAKRIPISKAVDGETISRAGRYQSCMVDESIFIHRWSSVST 569
+ ++I AG+ AK + + G Y++ +++ IH S +S
Sbjct: 985 DSIR-----KSIVAGFFPHTAK------------LQKNGSYRTVKHPQTVHIHPASGLSQ 1027
Query: 570 VHPEFLVYNELLETKRPNKEGETSAKRAYMHGVTNVDPTWLVENA 614
V P ++VY++L+ T + YM VT + P WL+E A
Sbjct: 1028 VLPRWVVYHQLVLT-----------SKEYMRQVTELKPEWLIEIA 1061
>At3g26560 ATP-dependent RNA helicase, putative
Length = 1168
Score = 170 bits (430), Expect = 3e-42
Identities = 135/464 (29%), Positives = 213/464 (45%), Gaps = 93/464 (20%)
Query: 151 LFVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVKNYD 210
L +LP+Y+ LP+ Q R+FD G+R VVVATN+AE SLTI GI YVVD G K Y+
Sbjct: 757 LIILPVYSALPSEMQSRIFDPPPPGKRKVVVATNIAEASLTIDGIYYVVDPGFAKQNVYN 816
Query: 211 SSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYSSAAFSNEFPEFSPAEVEKVPVH 270
G+E+ + IS+ASA QRAGRAGRT G CYRLY+ +A+ NE P S E++++ +
Sbjct: 817 PKQGLESLVITPISQASAKQRAGRAGRTGPGKCYRLYTESAYRNEMPPTSIPEIQRINLG 876
Query: 271 GVVLLLKSMQIKKVANFPFPTSLKAASLLEAENCLRALEALDSKDELTLLGKAMALYPLS 330
L +K+M I + +F F + +L+ A L +L ALD + LT LG+ MA +PL
Sbjct: 877 MTTLTMKAMGINDLLSFDFMDPPQPQALISAMEQLYSLGALDEEGLLTKLGRKMAEFPLE 936
Query: 331 PRHSRMILTVIKNTRYKHIRNSSLLLAYAVAAAAALSLPNPFVMQYEGNDSNKDSETSEK 390
P S+M+L + I + A + N F E K ++ +K
Sbjct: 937 PPLSKMLLASVDLGCSDEI----------LTMIAMIQTGNIFYRPRE-----KQAQADQK 981
Query: 391 SRMGDNENNIDKTEKTKRKKLKQTSKVAREKFRIVSSDALAIAYALQCFEHSQNSVQFCE 450
R KF D L + + ++ S +C
Sbjct: 982 ----------------------------RAKFFQPEGDHLTLLAVYEAWKAKNFSGPWCF 1013
Query: 451 DNALHFKTMDEMSKLRQQLLRLVFFQNDKGGLEQEYSWTHVTLEDVEHVWRVSSAHYPLP 510
+N + +++ +R+QLL ++ DK L+ + + T
Sbjct: 1014 ENFIQSRSLRRAQDVRKQLLSIM----DKYKLDVVTAGKNFT------------------ 1051
Query: 511 LVEERLICRAICAGWADRVAKRIPISKAVDGETISRAGRYQSCMVDESIFIHRWSSVSTV 570
I +AI AG+ A++ P Y++ + ++ ++IH S++
Sbjct: 1052 -----KIRKAITAGFFFHGARKDP------------QEGYRTLVENQPVYIHPSSALFQR 1094
Query: 571 HPEFLVYNELLETKRPNKEGETSAKRAYMHGVTNVDPTWLVENA 614
P++++Y++L+ T + YM VT +DP WLVE A
Sbjct: 1095 QPDWVIYHDLVMT-----------TKEYMREVTVIDPKWLVELA 1127
>At5g13010 pre-mRNA splicing factor ATP-dependent RNA helicase -like
protein
Length = 1226
Score = 165 bits (417), Expect = 1e-40
Identities = 128/464 (27%), Positives = 209/464 (44%), Gaps = 95/464 (20%)
Query: 151 LFVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVKNYD 210
L +LP+Y+ LPA Q ++F ++G R +VATN+AETSLT+ GI YV+DTG K+K ++
Sbjct: 775 LLILPIYSQLPADLQAKIFQKPEDGARKCIVATNIAETSLTVDGIYYVIDTGYGKMKVFN 834
Query: 211 SSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYSSAAFSNEFPEFSPAEVEKVPVH 270
GM+ +V IS+A++ QRAGRAGRT G CYRLY+ +A+ NE E+++ +
Sbjct: 835 PRMGMDALQVFPISRAASDQRAGRAGRTGPGTCYRLYTESAYLNEMLPSPVPEIQRTNLG 894
Query: 271 GVVLLLKSMQIKKVANFPFPTSLKAASLLEAENCLRALEALDSKDELTLLGKAMALYPLS 330
VVLLLKS++I + +F F ++L + L L AL++ LT LG M +PL
Sbjct: 895 NVVLLLKSLKIDNLLDFDFMDPPPQENILNSMYQLWVLGALNNVGGLTDLGWKMVEFPLD 954
Query: 331 PRHSRMILTVIKNTRYKHIRNSSLLLAYAVAAAAALSLPNPFVMQYEGNDSNKDSETSEK 390
P ++M+L + + + + LS+P+ F
Sbjct: 955 PPLAKMLL----------MGERLDCIDEVLTIVSMLSVPSVFF----------------- 987
Query: 391 SRMGDNENNIDKTEKTKRKKLKQTSKVAREKFRIVSSDALAIAYALQCFEHSQNSVQFCE 450
+ K+ + S AREKF + SD L + Q ++ +C
Sbjct: 988 ----------------RPKERAEESDAAREKFFVPESDHLTLLNVYQQWKEHDYRGDWCN 1031
Query: 451 DNALHFKTMDEMSKLRQQLLRLVFFQNDKGGLEQEYSWTHVTLEDVEHVWRVSSAHYPLP 510
D+ L K + + ++R QLL ++ + L W +
Sbjct: 1032 DHYLQVKGLRKAREVRSQLLDIL-------------KQLKIELRSCGPDWDI-------- 1070
Query: 511 LVEERLICRAICAGWADRVAKRIPISKAVDGETISRAGRYQSCMVDESIFIHRWSSVSTV 570
+ +AIC+ + A+ + G Y +C +H S++ +
Sbjct: 1071 ------VRKAICSAYFHNSAR------------LKGVGEYVNCRTGMPCHLHPSSALYGL 1112
Query: 571 --HPEFLVYNELLETKRPNKEGETSAKRAYMHGVTNVDPTWLVE 612
P+++VY+EL+ T + YM T+V+P WL E
Sbjct: 1113 GYTPDYVVYHELILT-----------TKEYMQCATSVEPHWLAE 1145
>At4g16680 RNA helicase
Length = 883
Score = 164 bits (415), Expect = 2e-40
Identities = 138/465 (29%), Positives = 217/465 (45%), Gaps = 94/465 (20%)
Query: 151 LFVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVKNYD 210
+ + P+Y+ LP Q +VF+ +G R VV+ATN+AETSLTI G+KYV+D G K+ +Y+
Sbjct: 464 IIICPIYSNLPTPLQAKVFEPAPKGTRKVVLATNIAETSLTIDGVKYVIDPGYCKINSYN 523
Query: 211 SSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYSSAAFSNEFPEFSPAEVEKVPVH 270
GME+ V ISKASAAQRAGR+GRT G C+RLY+ + + E+++ +
Sbjct: 524 PRTGMESLLVTPISKASAAQRAGRSGRTGPGKCFRLYN----IKDLEPTTIPEIQRANLA 579
Query: 271 GVVLLLKSMQIKKVANFPFPTSLKAASLLEAENCLRALEALDSKDELTLLGKAMALYPLS 330
VVL LKS+ I+ V NF F +LL+A L AL ALD E+T +G+ M +P+
Sbjct: 580 SVVLTLKSLGIQDVFNFDFMDPPPENALLKALELLYALGALDEIGEITKVGERMVEFPVD 639
Query: 331 PRHSRMILTVIKNTRYKHIRNSSLLLAYAVAAAAALSLPNPFVMQYEGNDSNKDSETSEK 390
P S+MI + + +YK + + AA LS+ N + ++K
Sbjct: 640 PMLSKMI---VGSEKYKCSKE-------IITIAAMLSVGNSVFYR-----PKNQQVFADK 684
Query: 391 SRMGDNENNIDKTEKTKRKKLKQTSKVAREKFRIVSSDALAIAYALQCFEHSQNSVQFCE 450
+RM E+ TE D +A+ ++ S Q+C
Sbjct: 685 ARMDFYED----TENV--------------------GDHIALLRVYNSWKEENYSTQWCC 720
Query: 451 DNALHFKTMDEMSKLRQQLLRLVFFQNDKGGLEQEYSWTHVTLEDVEHVWRVSSAHYPLP 510
+ + K+M +R QLL L+ +K G+E ++S L
Sbjct: 721 EKFIQSKSMKRARDIRDQLLGLL----NKIGVE------------------LTSNPNDLD 758
Query: 511 LVEERLICRAICAGWADRVAKRIPISKAVDGETISRAGRYQSCMVDESIFIHRWSSVSTV 570
++ +AI AG+ AK + + G Y+ +++++H S +
Sbjct: 759 AIK-----KAILAGFFPHSAK------------LQKNGSYRRVKEPQTVYVHPNSGLFGA 801
Query: 571 HP-EFLVYNELLETKRPNKEGETSAKRAYMHGVTNVDPTWLVENA 614
P ++LVY+EL+ T + YM T + P WL+E A
Sbjct: 802 SPSKWLVYHELVLT-----------TKEYMRHTTEMKPEWLIEIA 835
>At1g27900 ATP-dependent RNA helicase (At1g27900)
Length = 700
Score = 151 bits (382), Expect = 1e-36
Identities = 130/491 (26%), Positives = 224/491 (45%), Gaps = 83/491 (16%)
Query: 126 EDSLDQSKVGREKIARENHDSSPGALFVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNV 185
+D +++ E+ R + S + PL+ LP Q+RVF R +V+TN+
Sbjct: 223 QDDIEKLVSRLEEKVRSLAEGSCMDAIIYPLHGSLPPEMQVRVFSPPPPNCRRFIVSTNI 282
Query: 186 AETSLTIPGIKYVVDTGREKVKNYDSSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYR 245
AETSLT+ G+ YV+D+G K + Y+ S+GM + +V ISK A QRAGRAGRT G CYR
Sbjct: 283 AETSLTVDGVVYVIDSGYVKQRQYNPSSGMFSLDVIQISKVQANQRAGRAGRTRPGKCYR 342
Query: 246 LYSSAAFSNEFPEFSPAEVEKVPVHGVVLLLKSMQIKKV--ANFPFPTSLKAASLLEAEN 303
LY A + ++F + + E+++ + G VL LKS+ + + F F + + SL +A
Sbjct: 343 LYPLAVYRDDFLDATIPEIQRTSLAGSVLYLKSLDLPDIDILKFDFLDAPSSESLEDALK 402
Query: 304 CLRALEALDSKDELTLLGKAMALYPLSPRHSRMILTVIKNTRYKHIRNSSLLLAYAVAAA 363
L ++A+D +T +G+ M+ PL P SR ++ N + L+ A+
Sbjct: 403 QLYFIDAIDENGAITRIGRTMSDLPLEPSLSRTLIEA----------NETGCLSQALTVV 452
Query: 364 AALSLPNPFVMQYEGNDSNKDSETSEKSRMGDNENNIDKTEKTKRKKLKQTSKVAREKFR 423
A LS + S+ SEK R D ++N+
Sbjct: 453 AMLSAETTLLPAR--------SKPSEKKRKHDEDSNLPNGSGY----------------- 487
Query: 424 IVSSDALAIAYALQCFEHSQNSVQFCEDNALHFKTMDEMSKLRQQLLRLVFFQNDKGGLE 483
D + + + ++ + +C++N + + M + +R+QL +++
Sbjct: 488 ---GDHIQLLQIFESWDRTNYDPVWCKENGMQVRGMVFVKDVRRQLCQIM---------- 534
Query: 484 QEYSWTHVTLEDVEHVWRVSSAHYPLPLVEERLICRAICAGWADRVAKRIPISKAVDGET 543
Q+ S + +V R SS+ + R + +A+C G A+++A+R+
Sbjct: 535 QKISKDRL---EVGADGRKSSSRD-----DYRKLRKALCVGNANQIAERMLRHNGY---- 582
Query: 544 ISRAGRYQSCMVDESIFIHRWSSVST----VHPEFLVYNELLETKRPNKEGETSAKRAYM 599
R +QS +V +H S +S + P ++VY+EL+ T RP +M
Sbjct: 583 --RTLSFQSQLVQ----VHPSSVLSADNDGMMPNYVVYHELISTTRP-----------FM 625
Query: 600 HGVTNVDPTWL 610
V VD W+
Sbjct: 626 RNVCAVDMAWV 636
>At3g62310 ATP-dependent RNA helicase-like protein
Length = 726
Score = 151 bits (381), Expect = 2e-36
Identities = 134/486 (27%), Positives = 220/486 (44%), Gaps = 99/486 (20%)
Query: 138 KIARE--NHDSSPGALFVLPLYAMLPAAAQLRVFDG----VKEGE---RLVVVATNVAET 188
KI +E N G + V+PLY+ LP A Q ++FD V EG R +VV+TN+AET
Sbjct: 294 KINKEVGNLGDQVGPIKVVPLYSTLPPAMQQKIFDPAPEPVTEGGPPGRKIVVSTNIAET 353
Query: 189 SLTIPGIKYVVDTGREKVKNYDSSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYS 248
SLTI GI YV+D G K K Y+ +E+ V ISKASA QR+GRAGRT G C+RLY+
Sbjct: 354 SLTIDGIVYVIDPGFAKQKVYNPRIRVESLLVSPISKASAHQRSGRAGRTRPGKCFRLYT 413
Query: 249 SAAFSNEFPEFSPAEVEKVPVHGVVLLLKSMQIKKVANFPFPTSLKAASLLEAENCLRAL 308
+F+N+ + E+ + + VL LK + I + +F F +L+ A L L
Sbjct: 414 EKSFNNDLQPQTYPEILRSNLANTVLTLKKLGIDDLVHFDFMDPPAPETLMRALEVLNYL 473
Query: 309 EALDSKDELTLLGKAMALYPLSPRHSRMILTVIKNTRYKHIRNSSLLLAYAVAAAAALSL 368
ALD LT G+ M+ +PL P+ ++M++ + I ++ +A LS+
Sbjct: 474 GALDDDGNLTKTGEIMSEFPLDPQMAKMLIVSPEFNCSNEI----------LSVSAMLSV 523
Query: 369 PNPFVMQYEGNDSNKDSETSEKSRMGDNENNIDKTEKTKRKKLKQTSKVAREKFRIVSSD 428
PN F+ + ++ ++ + A+ +F + D
Sbjct: 524 PNCFI---------------------------------RPREAQKAADEAKARFGHIEGD 550
Query: 429 ALAIAYALQCFEHSQNSVQFCEDNALHFKTMDEMSKLRQQLLRLVFFQNDKGGLEQEYSW 488
L + F+ + +C +N ++ + M +RQQL+R++ N K
Sbjct: 551 HLTLLNVYHAFKQNNEDPNWCYENFINNRAMKSADNVRQQLVRIMSRFNLK--------- 601
Query: 489 THVTLEDVEHVWRVSSAHYPLPLVEERLICRAICAGWADRVAKRIPISKAVDGETISRAG 548
+ D +S Y + I +A+ AG+ +VA + R G
Sbjct: 602 --MCSTD------FNSRDYYIN------IRKAMLAGYFMQVAH------------LERTG 635
Query: 549 RYQSCMVDESIFIHRWSSVSTVHPEFLVYNELLETKRPNKEGETSAKRAYMHGVTNVDPT 608
Y + ++ + +H S+ PE+++YNE + T R ++ VT++
Sbjct: 636 HYLTVKDNQVVHLHP-SNCLDHKPEWVIYNEYVLT-----------SRNFIRTVTDIRGE 683
Query: 609 WLVENA 614
WLV+ A
Sbjct: 684 WLVDVA 689
>At2g47250 putative pre-mRNA splicing factor RNA helicase
Length = 729
Score = 149 bits (377), Expect = 5e-36
Identities = 130/487 (26%), Positives = 221/487 (44%), Gaps = 99/487 (20%)
Query: 138 KIARE--NHDSSPGALFVLPLYAMLPAAAQLRVFDG----VKEGE---RLVVVATNVAET 188
KI +E N G + V+PLY+ LP A Q ++FD + EG R +VV+TN+AET
Sbjct: 298 KINKEVSNLGDQVGPVKVVPLYSTLPPAMQQKIFDPAPVPLTEGGPAGRKIVVSTNIAET 357
Query: 189 SLTIPGIKYVVDTGREKVKNYDSSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYS 248
SLTI GI YV+D G K K Y+ +E+ V ISKASA QR+GRAGRT G C+RLY+
Sbjct: 358 SLTIDGIVYVIDPGFAKQKVYNPRIRVESLLVSPISKASAHQRSGRAGRTRPGKCFRLYT 417
Query: 249 SAAFSNEFPEFSPAEVEKVPVHGVVLLLKSMQIKKVANFPFPTSLKAASLLEAENCLRAL 308
+F+N+ + E+ + + VL LK + I + +F F +L+ A L L
Sbjct: 418 EKSFNNDLQPQTYPEILRSNLANTVLTLKKLGIDDLVHFDFMDPPAPETLMRALEVLNYL 477
Query: 309 EALDSKDELTLLGKAMALYPLSPRHSRMILTVIKNTRYKHIRNSSLLLAYAVAAAAALSL 368
ALD + LT G+ M+ +PL P+ S+M++ + I ++ +A LS+
Sbjct: 478 GALDDEGNLTKTGEIMSEFPLDPQMSKMLIVSPEFNCSNEI----------LSVSAMLSV 527
Query: 369 PNPFVMQYEGNDSNKDSETSEKSRMGDNENNIDKTEKTKRKKLKQTSKVAREKFRIVSSD 428
PN FV + ++ ++ + A+ +F + D
Sbjct: 528 PNCFV---------------------------------RPREAQKAADEAKARFGHIDGD 554
Query: 429 ALAIAYALQCFEHSQNSVQFCEDNALHFKTMDEMSKLRQQLLRLVFFQNDKGGLEQEYSW 488
L + ++ + +C +N ++ + M +RQQL+R++ ++
Sbjct: 555 HLTLLNVYHAYKQNNEDPNWCFENFVNNRAMKSADNVRQQLVRIM----------SRFNL 604
Query: 489 THVTLEDVEHVWRVSSAHYPLPLVEERLICRAICAGWADRVAKRIPISKAVDGETISRAG 548
+ + + V+ I +A+ AG+ +VA + R G
Sbjct: 605 KMCSTDFNSRDYYVN-------------IRKAMLAGYFMQVAH------------LERTG 639
Query: 549 RYQSCMVDESIFIHRWSSVSTVHPEFLVYNELLETKRPNKEGETSAKRAYMHGVTNVDPT 608
Y + ++ + +H S+ PE+++YNE + T R ++ VT++
Sbjct: 640 HYLTVKDNQVVHLHP-SNCLDHKPEWVIYNEYVLT-----------TRNFIRTVTDIRGE 687
Query: 609 WLVENAK 615
WLV+ A+
Sbjct: 688 WLVDVAQ 694
>At2g35920 ATP-dependent RNA helicase A like protein
Length = 995
Score = 146 bits (368), Expect = 5e-35
Identities = 131/466 (28%), Positives = 207/466 (44%), Gaps = 68/466 (14%)
Query: 43 QTDRFSGYDEDDNNFDENESDSYDSETESELE-----FNDDDKNNHE---GSKNNNNIVD 94
+ R++ D N+ + S E+ES+ + F D D N+H S N++
Sbjct: 421 EKSRYNIKSSDSGNY-QGSSRGRRRESESKKDDLTTLFEDIDINSHYKSYSSATRNSLEA 479
Query: 95 VLGNEGSLASLKAAFENLSGQATLSSSNVNTEDSLDQSKVGREKIARENHDSSPGALFVL 154
G + + ++A E++ + V + SK+ EKI N VL
Sbjct: 480 WSGAQIDVDLVEATIEHICRLEGGGAILVFLTGWDEISKL-LEKINMNNFLGDSSKFLVL 538
Query: 155 PLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVKNYDSSNG 214
PL+ +P Q +FD +R +V+ATN+AE+S+TI + YVVD G+ K +YD+ N
Sbjct: 539 PLHGSMPTVNQREIFDRPPPNKRKIVLATNIAESSITIDDVVYVVDCGKAKETSYDALNK 598
Query: 215 METYEVKWISKASAAQRAGRAGRTAAGHCYRLYSSAAFSNEFPEFSPAEVEKVPVHGVVL 274
+ WISKASA QR GRAGR AG CYRLY + + FP++ E+ + P+ + L
Sbjct: 599 VACLLPSWISKASAHQRRGRAGRVQAGVCYRLYPKVIY-DAFPQYQLPEIIRTPLQELCL 657
Query: 275 LLKSMQIKKVANFPFPTSLKAASLLEAENC---LRALEALDSKDELTLLGKAMALYPLSP 331
+KS+Q+ + +F +L+ L EN L+ + AL+ +ELT LG+ + P+ P
Sbjct: 658 HIKSLQVGSIGSF-LAKALQPPDALAVENAIELLKTIGALNDVEELTPLGRHLCTLPVDP 716
Query: 332 RHSRMILTVIKNTRYKHIRNSSLLLAYAVAAAAALSLPNPFVMQYEGNDSNKDSETSEKS 391
+M+L I + A+ AAAL+ +PFV+ N+ E E
Sbjct: 717 NIGKMLL----------IGAIFQCVNPALTIAAALAYRSPFVLPL-----NRKEEADEAK 761
Query: 392 R--MGDNENNIDKTEKTKRKKLKQTSKVAREKFRIVSSDALAIAYALQCFEHSQ---NSV 446
R GD+ SD +A+ A + + ++ N
Sbjct: 762 RYFAGDS-----------------------------CSDHIALLKAYEGYRDAKRGGNEK 792
Query: 447 QFCEDNALHFKTMDEMSKLRQQLLRLV----FFQNDKGGLEQEYSW 488
FC N L T+ M +R Q L L+ F K +YS+
Sbjct: 793 DFCWQNFLSPVTLRMMEDMRNQFLDLLSDIGFVDKSKPNAYNQYSY 838
>At1g06670 DEIH-box RNA/DNA helicase
Length = 1576
Score = 133 bits (335), Expect = 3e-31
Identities = 93/296 (31%), Positives = 153/296 (51%), Gaps = 28/296 (9%)
Query: 136 REKIARENHDSSPGALFVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGI 195
+EK+ + + +L L++ +PA Q +VF+ G R +V+ATN+AE+++TI +
Sbjct: 592 KEKLLDDRFFAHSAKFIILCLHSRVPAEEQKKVFNRPPRGCRKIVLATNIAESAVTIDDV 651
Query: 196 KYVVDTGREKVKNYDSSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYSSAAFSNE 255
YV+D+GR K K+YD N + T + W+SKA+A QRAGRAGR AG CY LYS ++
Sbjct: 652 VYVIDSGRMKEKSYDPYNDVSTLQSSWVSKANAKQRAGRAGRCQAGICYHLYSKLRAAS- 710
Query: 256 FPEFSPAEVEKVPVHGVVLLLKSMQ--------IKKVANFPFPTSLKAASLLEAENCLRA 307
PE+ EV ++PV + L +K + ++K+ + P S++ A ++ L+
Sbjct: 711 LPEYRVPEVMRMPVDELCLQVKMLDPNCNVNDFLQKLMDPPVAQSIENALII-----LKD 765
Query: 308 LEALDSKDELTLLGKAMALYPLSPRHSRMI-LTVIKNTRYKHIRNSSLLLAYAVAAAAAL 366
+ AL ++ELT LG+ P+ PR S+MI ++ N L A+ A A
Sbjct: 766 IGALTPEEELTELGQKFGQLPVHPRISKMIYFAILVN-----------CLDPALILACAA 814
Query: 367 SLPNPFVMQYEGNDSNKDSETSEK--SRMGDNENNIDKTEKTKRKKLKQTSKVARE 420
+PF M D K + + S GD+ +++ + K + S A+E
Sbjct: 815 DEKDPFTMPLSPGDRKKAAAAKHELASLYGDHSDHLATVAAFQCWKNAKASGQAKE 870
>At1g48650 hypothetical protein
Length = 1167
Score = 130 bits (327), Expect = 3e-30
Identities = 108/405 (26%), Positives = 181/405 (44%), Gaps = 76/405 (18%)
Query: 148 PGALFVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVK 207
P + +L + + ++ Q +FD EG R +V+ATN+AETS+TI + YV+D G+ K
Sbjct: 577 PNKVLLLACHGSMASSEQRLIFDRPPEGIRKIVLATNMAETSITINDVVYVIDCGKAKET 636
Query: 208 NYDSSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYSSAAFSNEFPEFSPAEVEKV 267
+YD+ N WISKA+A QR GRAGR G CY LY + F ++ E+ +
Sbjct: 637 SYDALNNTPCLLPSWISKAAARQRRGRAGRVMPGECYHLYPRCVY-EAFADYQQPELLRT 695
Query: 268 PVHGVVLLLKSMQIKKVANFPFPTSLKAASLLEAENC---LRALEALDSKDELTLLGKAM 324
P+ + L +KS+ + ++ F +L+ L +N L+ + ALD + LT LGK +
Sbjct: 696 PLQSLCLQIKSLGLGSISEF-LSRALQPPEALSVQNAVEYLKIIGALDDDENLTPLGKNL 754
Query: 325 ALYPLSPRHSRM-ILTVIKNTRYKHIRNSSLLLAYAVAAAAALSLPNPFVMQYEGNDSNK 383
++ P+ P+ +M IL I N L + A LS+ +PF+M ++ D
Sbjct: 755 SMLPVEPKLGKMLILGAIFN-----------CLDPVMTVVAGLSVRDPFLMPFDKKD--- 800
Query: 384 DSETSEKSRMGDNENNIDKTEKTKRKKLKQTSKVAREKFRIVS-SDALAIAYALQCF--- 439
++ AR KF SD L + A +
Sbjct: 801 ------------------------------LAETARSKFSGRDYSDHLTLVRAYNGWKDA 830
Query: 440 EHSQNSVQFCEDNALHFKTMDEMSKLRQQLLRLVFFQNDKGGLEQEYSWTHVTLEDVEHV 499
E + + +C N L +T+ M +R+Q L+ +E S ++++E
Sbjct: 831 ERTHSGYDYCWKNFLSSQTLKAMDSMRKQFFNLL----------KEAS----LIDNIEGC 876
Query: 500 WRVSSAHYPLPLVEERLICRAICAGWADRVAKRIPISKAVDGETI 544
++S +E L+ ICAG V + K++ +T+
Sbjct: 877 SKLSH--------DEHLVRAIICAGMFPGVCSVVNKEKSITLKTM 913
>At2g01130 ATP-dependent RNA like helicase A
Length = 899
Score = 125 bits (315), Expect = 7e-29
Identities = 90/346 (26%), Positives = 162/346 (46%), Gaps = 55/346 (15%)
Query: 136 REKIARENHDSSPGALFVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGI 195
+EK+ +P + +L + + Q +F+ G R +V+ATN+AETS+TI +
Sbjct: 301 KEKLQIHPIFGNPDLVMLLACHGSMETFEQRLIFEEPASGVRKIVLATNIAETSITINDV 360
Query: 196 KYVVDTGREKVKNYDSSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYSSAAFSNE 255
+V+D G+ K +YD+ N WISK SA QR GRAGR G CY LY + +
Sbjct: 361 AFVIDCGKAKETSYDALNNTPCLLPSWISKVSAQQRRGRAGRVRPGQCYHLYPKCVY-DA 419
Query: 256 FPEFSPAEVEKVPVHGVVLLLKSMQIKKVANFPFPTSLKAASLLEAENC---LRALEALD 312
F E+ E+ + P+H + L +KS+ + ++ F +L++ LL + L+ + ALD
Sbjct: 420 FAEYQLPEILRTPLHSLCLQIKSLNLGSISEF-LSRALQSPELLAVQKAIAFLKIIGALD 478
Query: 313 SKDELTLLGKAMALYPLSPRHSRMILTVIKNTRYKHIRNSSLL--LAYAVAAAAALSLPN 370
++LT LG+ ++ P+ P+ +M++ ++L L + AA LS+ +
Sbjct: 479 ENEDLTTLGRYLSKLPMEPKLGKMLIL------------GAILGCLDPILTVAAGLSVRD 526
Query: 371 PFVMQYEGNDSNKDSETSEKSRMGDNENNIDKTEKTKRKKLKQTSKVAREKFRIVSSDAL 430
PF+ T + K K ++ A+ +F SD L
Sbjct: 527 PFL-----------------------------TPQDK----KDLAEAAKSQFSRDHSDHL 553
Query: 431 AIAYALQCFEHSQNS---VQFCEDNALHFKTMDEMSKLRQQLLRLV 473
A+ A + ++ ++ +C N L +++ + LR++ L+
Sbjct: 554 ALVRAYEGWKKAEEESAVYDYCWKNFLSIQSLRAIDSLRKEFFSLL 599
>At1g26370 hypothetical protein
Length = 710
Score = 123 bits (308), Expect = 5e-28
Identities = 94/330 (28%), Positives = 158/330 (47%), Gaps = 46/330 (13%)
Query: 137 EKIARENHDSSPG---ALFVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIP 193
E++ +E + P L L +++ LP+ Q++VF G R V++ATN+AETS+TIP
Sbjct: 301 ERLVQERLQNIPEDKRKLLPLAIFSALPSEQQMKVFAPAPTGFRKVILATNIAETSITIP 360
Query: 194 GIKYVVDTGREKVKNYDSSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYSSAAFS 253
GI+YV+D G K ++YD S GME+ +V SKA QR+GRAGR G +RLY F
Sbjct: 361 GIRYVIDPGFVKARSYDPSKGMESLDVVPASKAQTLQRSGRAGREGPGKSFRLYPEREF- 419
Query: 254 NEFPEFSPAEVEKVPVHGVVLLLKSMQIKKVANFPFPTSLKAASLLEAENCLRALEALDS 313
+ + + E+++ + ++L LK++ I + F F ++++A L +L AL
Sbjct: 420 EKLEDSTKPEIKRCNLSNIILQLKALGIDDIVGFDFIDKPSRGAIIKALAELHSLGALAD 479
Query: 314 KDEL-TLLGKAMALYPLSPRHSRMILTVIKNTRYKHIRNSSLLLAYAVAAAAALSLPNPF 372
+L +G M+ PL P +S+ ++ + N L + A LS+ + F
Sbjct: 480 DGKLENPVGYQMSRLPLEPVYSKALI----------LANQFNCLEEMLITVAVLSVESIF 529
Query: 373 VMQYEGNDSNKDSETS-------------------------EKSRMGDNENNIDKTEKTK 407
Y+ + +++ TS EK + + NNIDK K
Sbjct: 530 ---YDPREKREEARTSKNHFASVEGDHLTYLSVYRESDEFLEKRKAAGSGNNIDKIMKKW 586
Query: 408 RKK---LKQTSKVAREKFRIVSSDALAIAY 434
K+ ++ K AR+ +R + I +
Sbjct: 587 CKENYVNSRSLKHARDIYRQIREHVEQIGF 616
Score = 33.9 bits (76), Expect = 0.36
Identities = 19/67 (28%), Positives = 32/67 (47%), Gaps = 11/67 (16%)
Query: 548 GRYQSCMVDESIFIHRWSSVSTVHPEFLVYNELLETKRPNKEGETSAKRAYMHGVTNVDP 607
G Y++ E + IH S + PE +++NEL++T + Y+ +T +D
Sbjct: 647 GTYRALESGEVVHIHPTSVLFRAKPECVIFNELMQT-----------SKKYIKNLTIIDS 695
Query: 608 TWLVENA 614
WL E A
Sbjct: 696 LWLSELA 702
>At4g18465 RNA helicase - like protein
Length = 720
Score = 118 bits (295), Expect = 1e-26
Identities = 108/432 (25%), Positives = 190/432 (43%), Gaps = 59/432 (13%)
Query: 194 GIKYVVDTGREKVKNYDSSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYSSAAFS 253
G+ YV+D+G K K Y+ + +E+ V ISKASA QR+GRAGR G CYRLY+ F
Sbjct: 324 GVVYVIDSGFSKQKFYNPISDIESLVVAPISKASARQRSGRAGRVRPGKCYRLYTEDYFL 383
Query: 254 NEFPEFSPAEVEKVPVHGVVLLLKSMQIKKVANFPFPTSLKAASLLEAENCLRALEALDS 313
N+ P E+++ + V+ LK++ I + F +P + +++ A L +L+ LD
Sbjct: 384 NQMPGEGIPEMQRSNLVSTVIQLKALGIDNILGFDWPAPPSSEAMIRALEVLYSLQILDD 443
Query: 314 KDELTL-LGKAMALYPLSPRHSRMILTVIKNTRYKHIRNSSLLLAYAVAAAAALSLPNPF 372
+LT G +A PL P S+MIL + I + +L+ V A L F
Sbjct: 444 DAKLTSPTGFQVAELPLDPMISKMILASSELGCSHEIITIAAVLSVQVKALYPFFLCLKF 503
Query: 373 VM-QYEGNDSN--KDSETSEKSRMGD---NENNIDKTEKTK----RKKLKQTSKVAREKF 422
++ Y N+ K + + D N N++ + +++ A+ +F
Sbjct: 504 LLFIYIRNNRTYCKFVIICFRLELWDLKLNRCNLEYVSMQSVWIIARGVQKEQDEAKLRF 563
Query: 423 RIVSSDALAIAYALQCFEHSQNSVQFCEDNALHFKTMDEMSKLRQQLLRLVFFQNDKGGL 482
D + + F S+ Q+C N L++++M ++ ++R QL R+
Sbjct: 564 AAAEGDHVTFLNVYKGFLESKKPTQWCYKNFLNYQSMKKVVEIRDQLKRIA--------- 614
Query: 483 EQEYSWTHVTLEDVEHVWRVSSAHYPLPLVEERLICRAICAGWADRVAKRIPISKAVDGE 542
+TL+ + + + +A+ AG+ + P S V
Sbjct: 615 ----RRLGITLKSCDG--------------DMEAVRKAVTAGFFANACRLEPHSNGV--- 653
Query: 543 TISRAGRYQSCMVDESIFIHRWSSVSTVHPEFLVYNELLETKRPNKEGETSAKRAYMHGV 602
Y++ E ++IH S + V+P+++VY ++ T +R YM V
Sbjct: 654 -------YKTIRGSEEVYIHPSSVLFRVNPKWVVYQSIVST-----------ERQYMRNV 695
Query: 603 TNVDPTWLVENA 614
++P+WL E A
Sbjct: 696 VTINPSWLTEVA 707
>At5g10370 putative protein
Length = 1751
Score = 117 bits (294), Expect = 2e-26
Identities = 80/223 (35%), Positives = 120/223 (52%), Gaps = 10/223 (4%)
Query: 147 SPGALFVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKV 206
+P A+ LPL+ L Q RVF G R V+ ATN+AETSLTIPG+KYV+D+G K
Sbjct: 546 TPSAI-ALPLHGKLSFEEQFRVFQN-HPGRRKVIFATNIAETSLTIPGVKYVIDSGMVKE 603
Query: 207 KNYDSSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYSSAAFSNEFPEFSPAEVEK 266
Y+ GM +V +S++SA QRAGRAGRT G CYRLYS F + P E+ +
Sbjct: 604 SKYEPRTGMSILKVCRVSQSSARQRAGRAGRTEPGRCYRLYSKNDFDSMNLNQEP-EIRR 662
Query: 267 VPVHGVVLLLKSMQIKKVANFPFPTSLKAASLLEAENCLRALEALDSKD---ELTLLGKA 323
V + +L + ++ + +A F F + ++ A L L A+ K+ ELT G
Sbjct: 663 VHLGVALLRMLALGVNNIAEFNFVDAPVPEAIAMAVQNLVQLGAVVEKNGVHELTQEGHC 722
Query: 324 MALYPLSPRHSRMILTVIKNTRYKHIRNSSLLLAYAVAAAAAL 366
+ L P+ ++IL ++ K ++LA +A A+++
Sbjct: 723 LVKLGLEPKLGKLILGCFRHRMGK----EGIVLAAVMANASSI 761
>At4g01020 unknown protein
Length = 1787
Score = 115 bits (289), Expect = 7e-26
Identities = 80/223 (35%), Positives = 119/223 (52%), Gaps = 10/223 (4%)
Query: 147 SPGALFVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKV 206
+P A+ LPL+ L Q VF G R V+ ATN+AETSLTIPG+KYV+D+G K
Sbjct: 543 APSAI-ALPLHGKLSFEEQFMVFQNYP-GRRKVIFATNIAETSLTIPGVKYVIDSGMVKE 600
Query: 207 KNYDSSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYSSAAFSNEFPEFSPAEVEK 266
Y+ GM +V +S++SA QRAGRAGRT G CYRLYS F + P E+ +
Sbjct: 601 SKYEPRTGMSILKVCQVSQSSARQRAGRAGRTEPGRCYRLYSKTDFDSMNLNQEP-EIRR 659
Query: 267 VPVHGVVLLLKSMQIKKVANFPFPTSLKAASLLEAENCLRALEALDSKD---ELTLLGKA 323
V + +L + ++ I +A F F + ++ A L L A+ K+ ELT G
Sbjct: 660 VHLGVALLRMLALGIDNIAAFEFVDAPVPEAIAMAIQNLVQLGAVVEKNGVLELTQEGHC 719
Query: 324 MALYPLSPRHSRMILTVIKNTRYKHIRNSSLLLAYAVAAAAAL 366
+ L P+ ++IL ++ K ++LA +A A+++
Sbjct: 720 LVKLGLEPKLGKLILGCFRHRMGK----EGIVLAAVMANASSI 758
>At1g58050 hypothetical protein
Length = 1393
Score = 107 bits (268), Expect = 2e-23
Identities = 121/449 (26%), Positives = 198/449 (43%), Gaps = 79/449 (17%)
Query: 152 FVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVKNYDS 211
++LPL++ + + Q +VF +G R V++ATN+AETS+TI + YV+D+G+ K Y+
Sbjct: 915 WLLPLHSSIASTEQKKVFLRPPKGIRKVIIATNIAETSITIEDVVYVIDSGKHKENRYNP 974
Query: 212 SNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYSSAAFSNEFPEFSPAEVEKVPVHG 271
+ + W+SKA+A QR GRAGR GHC+ LY+ F + E
Sbjct: 975 HKKLSSMVEDWVSKANARQRMGRAGRVKPGHCFSLYTRHRFEKLMRPYQVPE-------- 1026
Query: 272 VVLLLKSMQIKKVANFPFPTSLKAASLLEAENCLRALEALDSKDELTLLGKAMALYPLSP 331
+L++++ P +++ +A LL L + AL+ +ELT LG +A P+
Sbjct: 1027 ---MLRALEP------PSESAINSAILL-----LHKVGALEGDEELTPLGHHLAKLPVDL 1072
Query: 332 RHSRMILTVIKNTRYKHIRNSSLLLAYAVAAAAALS-LPNPFVMQYEGNDSNKD----SE 386
+M+L Y I L+ ++ AA LS +PFV Y ++ N D +
Sbjct: 1073 LIGKMLL-------YGGIFG---CLSPILSIAAFLSCCKSPFV--YAKDEQNVDRVKLAL 1120
Query: 387 TSEKSRMGDNENNIDKTEKTKRKKLKQTSKVAREKF-RIVSSDALAIAYALQCFEHSQNS 445
S+K N NN D R+ VA EK+ RI+ A +
Sbjct: 1121 LSDKLESSSNLNNND------RQSDHLLMVVAYEKWVRILHEQGFKAAES---------- 1164
Query: 446 VQFCEDNALHFKTMDEMSKLRQQL------LRLVFFQNDKGGLEQEYSWTHVTLEDVEHV 499
FCE L+ M M + R + + L+ KG ++ + V D
Sbjct: 1165 --FCESKFLNSSVMRMMRERRVEFGMLLADIGLINLPKGKGRRKENFD---VWFSDKTQP 1219
Query: 500 WRVSSAHYPLPLVEERLICRAICAGWADRVAKRIPISKAVDGETISRAGRYQSCMVDESI 559
+ + S P V + ++C +C A+ + R ++K + ET A + +
Sbjct: 1220 FNMYSQE---PEVVKAILCAGLCPNIAEGLVNR--LTKPAE-ETQRYAVWHDG---KREV 1270
Query: 560 FIHRWS---SVSTVHPEFLVYNELLETKR 585
IHR S + F+V+ E LETK+
Sbjct: 1271 HIHRNSINKNCKAFQYPFIVFLEKLETKK 1299
>At2g47680 putative ATP-dependent RNA helicase A
Length = 1015
Score = 80.9 bits (198), Expect = 3e-15
Identities = 52/163 (31%), Positives = 86/163 (51%), Gaps = 10/163 (6%)
Query: 177 RLVVVATNVAETSLTIPGIKYVVDTGREKVKNYDSSNGMETYEVKWISKASAAQRAGRAG 236
R V++ATN+AE+S+TIP + YV+D+ R +D S + ++ W+S++ A QR GR G
Sbjct: 322 RKVILATNIAESSVTIPKVAYVIDSCRSLQVFWDPSRKRDAVQLVWVSRSQAEQRRGRTG 381
Query: 237 RTAAGHCYRLYSSAAFSNEFPEFSPAEVEKVPVHGVVLLLKSMQIKKV--ANFPFPTSLK 294
RT G YRL S AF N+ E P + K+ + VL + + + + AN ++
Sbjct: 382 RTCDGEVYRLVPS-AFFNKLEEHEPPSILKLSLRQQVLHICCTESRAINDANALLAKAMD 440
Query: 295 AASLLEAENCLRALEALDS-------KDELTLLGKAMALYPLS 330
++ LR L ++ + + E T G+ +A +PLS
Sbjct: 441 PPDPDVVDDALRMLLSIQALRKSPRGRYEPTFYGRLLASFPLS 483
>At1g58060 hypothetical protein
Length = 1389
Score = 58.5 bits (140), Expect = 1e-08
Identities = 27/69 (39%), Positives = 45/69 (65%)
Query: 148 PGALFVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVK 207
P A ++LPL++ + ++ Q +VF +G R V+ ATN+AETS+TI + YV+D+G+ K
Sbjct: 958 PAADWLLPLHSSIASSEQRKVFLRPPKGLRKVIAATNIAETSITIDDVVYVIDSGKHKEN 1017
Query: 208 NYDSSNGME 216
Y+ +E
Sbjct: 1018 RYNPQKALE 1026
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.130 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,532,068
Number of Sequences: 26719
Number of extensions: 762261
Number of successful extensions: 4984
Number of sequences better than 10.0: 179
Number of HSP's better than 10.0 without gapping: 61
Number of HSP's successfully gapped in prelim test: 121
Number of HSP's that attempted gapping in prelim test: 4272
Number of HSP's gapped (non-prelim): 506
length of query: 789
length of database: 11,318,596
effective HSP length: 107
effective length of query: 682
effective length of database: 8,459,663
effective search space: 5769490166
effective search space used: 5769490166
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC147012.5


