

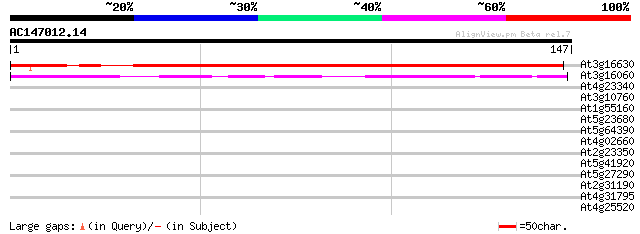
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147012.14 + phase: 0
(147 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g16630 kinesin-like protein 199 6e-52
At3g16060 kinesin like protein 45 1e-05
At4g23340 unknown protein 32 0.12
At3g10760 unknown protein 29 1.0
At1g55160 unknown protein 28 2.3
At5g23680 unknown protein 27 3.0
At5g64390 HEN4 27 3.9
At4g02660 putative protein 27 3.9
At2g23350 putative poly(A) binding protein 27 3.9
At5g41920 SCARECROW gene regulator-like protein 26 6.7
At5g27290 unknown protein 26 6.7
At2g31190 unknown protein 26 6.7
At4g31795 WRKY like transcription factor 26 8.7
At4g25520 putative protein 26 8.7
>At3g16630 kinesin-like protein
Length = 794
Score = 199 bits (505), Expect = 6e-52
Identities = 107/148 (72%), Positives = 119/148 (80%), Gaps = 14/148 (9%)
Query: 1 MGGQ---SNAAAAAALYDHAGGAVPLHPAPAGTAPDAGDAVMARWLQSAGLQHLASPLAN 57
MGGQ +NAAAA ALYD GA+P + DAGDAVMARWLQSAGLQHLASP+A+
Sbjct: 1 MGGQMQQNNAAAATALYD---GALPTN--------DAGDAVMARWLQSAGLQHLASPVAS 49
Query: 58 TAIDQRLLPNLLMQGYGAQSAEEKQRLFKLMRSLNFNGESGSELYTPTSQTLGGAAVSDG 117
T DQR LPNLLMQGYGAQ+AEEKQRLF+LMR+LNFNGES SE YTPT+ T S+G
Sbjct: 50 TGNDQRHLPNLLMQGYGAQTAEEKQRLFQLMRNLNFNGESTSESYTPTAHTSAAMPSSEG 109
Query: 118 FYSPDFRGDFGAGLLDLHAMDDTELLPE 145
F+SP+FRGDFGAGLLDLHAMDDTELL E
Sbjct: 110 FFSPEFRGDFGAGLLDLHAMDDTELLSE 137
>At3g16060 kinesin like protein
Length = 684
Score = 45.1 bits (105), Expect = 1e-05
Identities = 45/146 (30%), Positives = 62/146 (41%), Gaps = 29/146 (19%)
Query: 1 MGGQSNAAAAAALYDHAGGAVPLHPAPAGTAPDAGDAVMARWLQSAGLQHLASPLANTAI 60
M G+ + AAA + PL + + RWLQS GLQH S +A
Sbjct: 1 MSGRQRSVAAAVHHQRQLSDNPLDMSSSN----------GRWLQSTGLQHFQS----SAN 46
Query: 61 DQRLLPNLLMQGYGAQSAEEKQRLFKLMRSLNFNGESGSELYTPTSQTLGGAAVSDGFYS 120
D QG G Q+A Q N + G+E + + GA ++ +
Sbjct: 47 DYGYYAG--GQGGGGQAARGYQ-----------NAQRGNEFFGEPTTPQYGARPTNQRKN 93
Query: 121 PDFRGDFGAGLLDLHAMDDTELLPEV 146
D +F GLLDLH+ DTELLPE+
Sbjct: 94 ND-ESEFSPGLLDLHSF-DTELLPEI 117
>At4g23340 unknown protein
Length = 277
Score = 32.0 bits (71), Expect = 0.12
Identities = 14/33 (42%), Positives = 20/33 (60%)
Query: 70 MQGYGAQSAEEKQRLFKLMRSLNFNGESGSELY 102
MQ YGA+ AE +RL K++ + E+G LY
Sbjct: 72 MQEYGAKMAELSKRLIKILLMMTLGDETGKRLY 104
>At3g10760 unknown protein
Length = 335
Score = 28.9 bits (63), Expect = 1.0
Identities = 33/131 (25%), Positives = 46/131 (34%), Gaps = 14/131 (10%)
Query: 2 GGQSNAAAAAALYDHAGGAVPLHPAPAGTAPDAGDAVMARWLQSAGLQHLASPLANTAID 61
GG + A L+ A VP H P + D M ++ A LQ A A
Sbjct: 170 GGAGSDPATDRLF--ASSPVPAHFLHPNRVPSS-DHFMPSFVPIATLQQQQQMAAAAAAA 226
Query: 62 QRLLPNLLMQGYGAQSAEEKQRLFKLMRSLNFNGESGSELYTPTSQTLGGAAVSDGFYSP 121
P+L + Q A + +F + +PTS G+ S+GF SP
Sbjct: 227 AAANPHLQPPQFHRQIA-----------AAHFGSPTNGGFSSPTSNGQFGSPTSNGFGSP 275
Query: 122 DFRGDFGAGLL 132
G F L
Sbjct: 276 TTNGKFDPSFL 286
>At1g55160 unknown protein
Length = 188
Score = 27.7 bits (60), Expect = 2.3
Identities = 14/23 (60%), Positives = 15/23 (64%)
Query: 5 SNAAAAAALYDHAGGAVPLHPAP 27
S AAAAAA Y GG+VP P P
Sbjct: 41 SPAAAAAASYTMGGGSVPPPPPP 63
>At5g23680 unknown protein
Length = 295
Score = 27.3 bits (59), Expect = 3.0
Identities = 12/41 (29%), Positives = 23/41 (55%)
Query: 41 RWLQSAGLQHLASPLANTAIDQRLLPNLLMQGYGAQSAEEK 81
+W++S+G + A+ NTA +R+ N + G + +EK
Sbjct: 136 KWVKSSGGETAATTTTNTASAKRVRSNWATRNDGVEQGDEK 176
>At5g64390 HEN4
Length = 857
Score = 26.9 bits (58), Expect = 3.9
Identities = 18/63 (28%), Positives = 28/63 (43%)
Query: 20 AVPLHPAPAGTAPDAGDAVMARWLQSAGLQHLASPLANTAIDQRLLPNLLMQGYGAQSAE 79
A P AP G + +G AR G H ++ + NT ++ R+ N + YG Q
Sbjct: 738 ASPPPAAPRGLSDASGGLSSARAGHVLGSGHKSAIVTNTTVEIRVPANAMSFVYGEQGYN 797
Query: 80 EKQ 82
+Q
Sbjct: 798 LEQ 800
>At4g02660 putative protein
Length = 3449
Score = 26.9 bits (58), Expect = 3.9
Identities = 25/73 (34%), Positives = 32/73 (43%), Gaps = 9/73 (12%)
Query: 70 MQGYGAQSAEEKQRLFKLMRSLNF-----NGESGSELYTPTSQTLGGAAVSDGFYSPDF- 123
+ G Q ++E RLFKLM + +F NGE + Y TL G SD P F
Sbjct: 2863 ISGSAKQESKEGSRLFKLM-AKSFTKRWQNGEISNFQYLMHLNTLAGRGYSDLTQYPVFP 2921
Query: 124 --RGDFGAGLLDL 134
D+ LDL
Sbjct: 2922 WILADYDGESLDL 2934
>At2g23350 putative poly(A) binding protein
Length = 662
Score = 26.9 bits (58), Expect = 3.9
Identities = 19/53 (35%), Positives = 23/53 (42%), Gaps = 1/53 (1%)
Query: 14 YDHAGGAVPLHPAPAGTAPDAGDAVMARWLQSAGLQHLASPLAN-TAIDQRLL 65
Y G +P P P G P A D + + Q LA+ LAN T QR L
Sbjct: 525 YPSGGRNMPDGPMPGGMVPVAYDMNVMPYSQPMSAGQLATSLANATPAQQRTL 577
>At5g41920 SCARECROW gene regulator-like protein
Length = 405
Score = 26.2 bits (56), Expect = 6.7
Identities = 14/43 (32%), Positives = 21/43 (48%)
Query: 27 PAGTAPDAGDAVMARWLQSAGLQHLASPLANTAIDQRLLPNLL 69
P+ A G+AV+ W+Q + L I +RL PNL+
Sbjct: 227 PSQLATRQGEAVVVHWMQHRLYDVTGNNLETLEILRRLKPNLI 269
>At5g27290 unknown protein
Length = 341
Score = 26.2 bits (56), Expect = 6.7
Identities = 18/43 (41%), Positives = 22/43 (50%), Gaps = 8/43 (18%)
Query: 82 QRLFKLMRSLNFNGESGSELYTPTSQTLGG-------AAVSDG 117
QRL+ L L NG + + L +PT TLG AAVS G
Sbjct: 86 QRLYTL-EELKLNGINAASLLSPTDTTLGSIERNLQIAAVSGG 127
>At2g31190 unknown protein
Length = 433
Score = 26.2 bits (56), Expect = 6.7
Identities = 17/77 (22%), Positives = 33/77 (42%), Gaps = 15/77 (19%)
Query: 28 AGTAPDAGDAVMARWLQSAGLQHLASPLANTAIDQRLLPNLLMQGYGAQSAEEKQRLFKL 87
AG P A + W+ G+QH+ L+ GA+ E +R +++
Sbjct: 113 AGLRPTPAQATVVSWILKDGMQHVG--------------KLICSNLGARMDSEPKR-WRI 157
Query: 88 MRSLNFNGESGSELYTP 104
+ + ++ +G EL +P
Sbjct: 158 LADVLYDLGTGLELVSP 174
>At4g31795 WRKY like transcription factor
Length = 310
Score = 25.8 bits (55), Expect = 8.7
Identities = 15/45 (33%), Positives = 21/45 (46%), Gaps = 2/45 (4%)
Query: 76 QSAEEKQRLFKLMRSL--NFNGESGSELYTPTSQTLGGAAVSDGF 118
Q E+K +L R + +FN S +Y PT + V DGF
Sbjct: 135 QQHEQKNQLLSCKRPVTDSFNKAKVSTVYVPTETSDTSLTVKDGF 179
>At4g25520 putative protein
Length = 748
Score = 25.8 bits (55), Expect = 8.7
Identities = 20/89 (22%), Positives = 38/89 (42%), Gaps = 12/89 (13%)
Query: 66 PNLLMQGYGAQSAEEKQRLFKLMRSLN-----------FNGESGSELYTPTSQTLGGAAV 114
P+ L + + E+Q L +L++ ++ F+G+SGS + T + +
Sbjct: 640 PHQLQSPHSHGNTPEQQMLHQLLQEMSENGGSVQQQQAFSGQSGSNSNAERNTTASTSNI 699
Query: 115 SDGFYSPDFRGDF-GAGLLDLHAMDDTEL 142
S G +P F A +LH +D +
Sbjct: 700 SGGGRAPSRNNSFKAASNNNLHFSEDISI 728
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,459,517
Number of Sequences: 26719
Number of extensions: 136931
Number of successful extensions: 224
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 212
Number of HSP's gapped (non-prelim): 15
length of query: 147
length of database: 11,318,596
effective HSP length: 90
effective length of query: 57
effective length of database: 8,913,886
effective search space: 508091502
effective search space used: 508091502
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 55 (25.8 bits)
Medicago: description of AC147012.14


