

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147012.13 + phase: 0 /pseudo
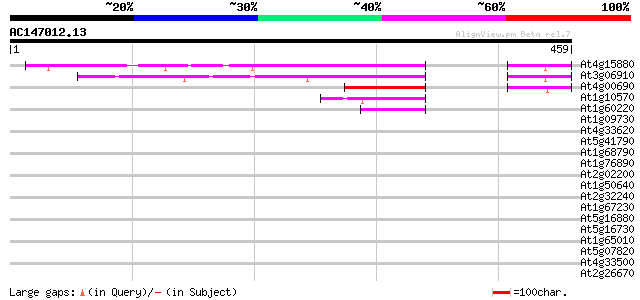
(459 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g15880 unknown protein 242 4e-64
At3g06910 unknown protein 174 7e-44
At4g00690 hypothetical protein 107 1e-23
At1g10570 protein-tyrosine phosphatase 2 like protein 56 4e-08
At1g60220 unknown protein 50 3e-06
At1g09730 unknown protein 39 0.005
At4g33620 putative protein 39 0.006
At5g41790 myosin heavy chain-like protein 37 0.023
At1g68790 putative nuclear matrix constituent protein 1 (NMCP1) 37 0.023
At1g76890 unknown protein 35 0.086
At2g02200 putative protein on transposon FARE2.10 (cds2) 34 0.15
At1g50640 ethylene responsive element binding factor 3 33 0.25
At2g32240 putative myosin heavy chain 33 0.33
At1g67230 unknown protein 32 0.56
At5g16880 TOM (target of myb1) -like protein 32 0.73
At5g16730 putative protein 32 0.73
At1g65010 hypothetical protein 32 0.73
At5g07820 putative protein 32 0.96
At4g33500 unknown protein 31 1.6
At2g26670 heme oxygenase 1 (HO1) 31 1.6
>At4g15880 unknown protein
Length = 489
Score = 242 bits (617), Expect = 4e-64
Identities = 151/339 (44%), Positives = 201/339 (58%), Gaps = 26/339 (7%)
Query: 14 HWLSVDHNRPSPNSPTF---KRQKLSTTMSNNTPPPLTTNATVFRISRYPKTKPPLRREI 70
++++ P NSP F K+++ S MS ++ P ++N T+ RISRYP K PLRREI
Sbjct: 16 NFINQQSTNPLRNSPYFQASKKRRFSFAMSEDSGKPASSNPTISRISRYPDAKAPLRREI 75
Query: 71 HAPCSSRPRKFDLSTNLKLTESSNLMASDTTDELMEKYEHAKHAALATISFREKGK---- 126
HAP R +N + +N + KY+ AK +AL + F KGK
Sbjct: 76 HAPSRGILRYGKAKSNDYCEKDANFF--------VRKYDDAKRSALEALRFVNKGKDFVD 127
Query: 127 VKDNI-KEVVVLKDSNVEDVEEFLSVVKDHKLKENGLVCLDDKVEVVNGGIQQQSTSSMV 185
+ D + KE VV DS+V+ +E + D + K+N V V G +
Sbjct: 128 LGDEVEKEEVVSDDSSVQAIE--VIDCDDDEEKKNLQPSFSSGVTDVKKG----ENFRVE 181
Query: 186 DAAKMLDNLSLS--TKNDLSGVCAYKKLLEAVDKRGDTLGRLKFEIQLNEKRKSAFDLLR 243
D + MLD+LSL ND S + AY+KL+++ +KR L L FEI LNEK+ S R
Sbjct: 182 DTSMMLDSLSLDRDVDNDASSLEAYRKLMQSAEKRNSKLEALGFEIVLNEKKLSLLRQSR 241
Query: 244 PKK-ELVEEVPREPFVPLTKEEEVEVTRAFSA-NQNKVLVNHEKSNIEISGKMFRCLRPG 301
PK E EVPREPF+PLT++EE EV RAFS N+ KVL HE SNI+I+G++ +CL P
Sbjct: 242 PKTVEKRVEVPREPFIPLTEDEEAEVYRAFSGRNRRKVLATHENSNIDITGEVLQCLTPS 301
Query: 302 EWLNDEVINLYLELLKERERREPQKFLKCHFFNTFFYKK 340
WLNDEVIN+YLELLKERE REP+K+LKCH+FNTFFYKK
Sbjct: 302 AWLNDEVINVYLELLKERETREPKKYLKCHYFNTFFYKK 340
Score = 69.3 bits (168), Expect = 4e-12
Identities = 36/73 (49%), Positives = 44/73 (59%), Gaps = 21/73 (28%)
Query: 408 DEVKDKTGEDVDISSWETEFVEDLPEQMNG---------------------FVIEHMPYF 446
DE +K+G+ +D +SW+ EFVEDLP+Q NG F EHMPYF
Sbjct: 416 DEANEKSGKKIDANSWDMEFVEDLPQQKNGYDCGMFMLKYIDFFSRGLGLCFSQEHMPYF 475
Query: 447 RLRTAKEILRLKA 459
RLRTAKEILRL+A
Sbjct: 476 RLRTAKEILRLRA 488
>At3g06910 unknown protein
Length = 502
Score = 174 bits (442), Expect = 7e-44
Identities = 114/293 (38%), Positives = 164/293 (55%), Gaps = 17/293 (5%)
Query: 56 ISRYPKTKPPLRREIHAPCSSRPRKFDLSTNLKLTESSNLMASDTTDELMEKYEHAKHAA 115
I RYP+ K LRR++HAP D ST S N++ + T +A ++
Sbjct: 69 IYRYPEVKSSLRRQVHAPVRILNSGRDRSTR---QGSGNVLGTFLTRNNDMWKRNALDSS 125
Query: 116 LATISFREKGKVKDNIKEVVVLKDSN----VEDVEEFLSVVKDHKLKENGLVCLDDKVEV 171
L + RE V D + +V ++ D VE+V + V++ NGL +V
Sbjct: 126 LRYRTDREVIDVDDELGDVEMISDDTSREGVENVAMEVDEVEEKAEMGNGLF---SEVAS 182
Query: 172 VNGGIQQQSTSSMVDAAKMLDNLSLSTKNDLSGVCAYKKLLEAVDKRGDTLGRLKFEIQL 231
+ G + S +++ ++ N ++ D++ AY+K+LE+ R L F
Sbjct: 183 LKNGSLRVGECSKANSSSLVVNRPVT---DVTSFEAYRKVLESAVNRTSKLKDRGFVDFF 239
Query: 232 NEKRKSAFDLL----RPKKELVEEVPREPFVPLTKEEEVEVTRAFSANQNKVLVNHEKSN 287
E+ ++ L R +E VE V RE FVPL++EEE V RAFSAN + +LV H+ SN
Sbjct: 240 KERGRALLRSLSSFWRQDEEPVEVVQREAFVPLSREEETAVRRAFSANDSNILVTHKNSN 299
Query: 288 IEISGKMFRCLRPGEWLNDEVINLYLELLKERERREPQKFLKCHFFNTFFYKK 340
I+I+GK+ RCL+PG+WLNDEVINLY+ LLKERE REP+KFLKCHFFNTFF+ K
Sbjct: 300 IDITGKILRCLKPGKWLNDEVINLYMVLLKEREAREPKKFLKCHFFNTFFFTK 352
Score = 63.5 bits (153), Expect = 2e-10
Identities = 35/73 (47%), Positives = 41/73 (55%), Gaps = 21/73 (28%)
Query: 408 DEVKDKTGEDVDISSWETEFVEDLPEQMNG---------------------FVIEHMPYF 446
DEV+DK+ D+D+S W EFV+DLP Q NG F E MPYF
Sbjct: 429 DEVRDKSEVDLDVSRWRQEFVQDLPMQRNGFDCGMFMVKYIDFYSRGLDLCFTQEQMPYF 488
Query: 447 RLRTAKEILRLKA 459
R RTAKEIL+LKA
Sbjct: 489 RARTAKEILQLKA 501
>At4g00690 hypothetical protein
Length = 233
Score = 107 bits (267), Expect = 1e-23
Identities = 45/66 (68%), Positives = 57/66 (86%)
Query: 275 NQNKVLVNHEKSNIEISGKMFRCLRPGEWLNDEVINLYLELLKERERREPQKFLKCHFFN 334
N+ K+LV+H+ SNI+ISG+ +CLRP +WLND+V NLYLELLKER+ R+PQK+ KCHFFN
Sbjct: 20 NRKKILVSHKNSNIDISGETLQCLRPNQWLNDDVTNLYLELLKERQTRDPQKYFKCHFFN 79
Query: 335 TFFYKK 340
TFFY K
Sbjct: 80 TFFYVK 85
Score = 59.3 bits (142), Expect = 4e-09
Identities = 32/80 (40%), Positives = 44/80 (55%), Gaps = 28/80 (35%)
Query: 408 DEVKDKTGEDVDISSWETEFVEDLPEQMNGF----------------------------V 439
DEVK K+ +++D+SSW E+VE+ P+Q NG+ +
Sbjct: 153 DEVKQKSQKNIDVSSWGMEYVEERPQQQNGYDCGMFMLKYIDFYSRGLSLQFSQVIRDVI 212
Query: 440 IEHMPYFRLRTAKEILRLKA 459
+ MPYFRLRTAKEILRL+A
Sbjct: 213 KKDMPYFRLRTAKEILRLRA 232
>At1g10570 protein-tyrosine phosphatase 2 like protein
Length = 571
Score = 56.2 bits (134), Expect = 4e-08
Identities = 30/90 (33%), Positives = 47/90 (51%), Gaps = 6/90 (6%)
Query: 255 EPFVPLTKEEEVEVTRAFSANQNKVLVNHEKSN----IEISGKMFRCLRPGEWLNDEVIN 310
EP P+ EE E+ ++ + ++S+ +++S K +CL PGE+L VIN
Sbjct: 296 EPLSPMVVEEACELPEGLP--EDIYYPSSDQSDGRDLVQVSLKDLKCLSPGEYLTSPVIN 353
Query: 311 LYLELLKERERREPQKFLKCHFFNTFFYKK 340
Y+ ++ + CHFFNTFFYKK
Sbjct: 354 FYIRYVQHHVFSADKTAANCHFFNTFFYKK 383
>At1g60220 unknown protein
Length = 604
Score = 49.7 bits (117), Expect = 3e-06
Identities = 21/53 (39%), Positives = 31/53 (57%)
Query: 288 IEISGKMFRCLRPGEWLNDEVINLYLELLKERERREPQKFLKCHFFNTFFYKK 340
+++ K CL P E+L V+N Y+ L+++ Q CHFFNT+FYKK
Sbjct: 363 VQVCLKDLECLAPREYLTSPVMNFYMRFLQQQISSSNQISADCHFFNTYFYKK 415
>At1g09730 unknown protein
Length = 865
Score = 39.3 bits (90), Expect = 0.005
Identities = 25/91 (27%), Positives = 46/91 (50%), Gaps = 16/91 (17%)
Query: 263 EEEVEVTRAFSANQNK-------------VLVNHEKSNIEISGKMFRCLRPGEWLNDEVI 309
E++VEV+ ++ NQ K V + + I + L+P ++ND +I
Sbjct: 341 EDDVEVS-GYNLNQQKRYFPSFDEPFEDVVYPKGDPDAVSICKRDVELLQPETFVNDTII 399
Query: 310 NLYLELLKERERREPQKFLKCHFFNTFFYKK 340
+ Y+ LK + + E + + HFFN+FF++K
Sbjct: 400 DFYINYLKNQIQTEEKH--RFHFFNSFFFRK 428
>At4g33620 putative protein
Length = 710
Score = 38.9 bits (89), Expect = 0.006
Identities = 17/43 (39%), Positives = 29/43 (66%), Gaps = 2/43 (4%)
Query: 298 LRPGEWLNDEVINLYLELLKERERREPQKFLKCHFFNTFFYKK 340
L+P ++ND +I+ Y++ LK R P++ + HFFN FF++K
Sbjct: 196 LKPRRFINDTIIDFYIKYLK--NRISPKERGRFHFFNCFFFRK 236
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 37.0 bits (84), Expect = 0.023
Identities = 30/117 (25%), Positives = 56/117 (47%), Gaps = 10/117 (8%)
Query: 75 SSRPRKFDLSTNLKLTESSNLMASDTTDELMEKYEHAKHAALATISFREKGKVKDNIKE- 133
SS R DL+ +LK E N S E+M+K E A++ + E G++KD KE
Sbjct: 341 SSEQRISDLTVDLKDAEEENKAISSKNLEIMDKLEQAQNTIKELMD--ELGELKDRHKEK 398
Query: 134 ------VVVLKDSNVEDVEEFL-SVVKDHKLKENGLVCLDDKVEVVNGGIQQQSTSS 183
+V D V D+++ L + ++ K+ ++ + ++++ IQ+ + S
Sbjct: 399 ESELSSLVKSADQQVADMKQSLDNAEEEKKMLSQRILDISNEIQEAQKTIQEHMSES 455
Score = 31.2 bits (69), Expect = 1.2
Identities = 38/170 (22%), Positives = 71/170 (41%), Gaps = 21/170 (12%)
Query: 75 SSRPRKFDLSTNLKLTESSNLMASDTTDELMEKYEHAKHAALATISFREKGKVKDNIKEV 134
SS R +LS +LK E + S E ++ E + I +E +KE
Sbjct: 672 SSEHRVLELSESLKAAEEESRTMSTKISETSDELERTQ------IMVQELTADSSKLKEQ 725
Query: 135 VVLKDSNVEDVEEFLSVVKDHKLK------ENGLVCLDDKVEVVNGGIQQQSTSSMVDAA 188
+ K+S + FL KD K + E + L+ ++E V I T +
Sbjct: 726 LAEKESKL-----FLLTEKDSKSQVQIKELEATVATLELELESVRARIIDLETE-IASKT 779
Query: 189 KMLDNLSLSTKNDLSGVCAYKKLLEAVDKRGDTLGRLKFEIQLNEKRKSA 238
+++ L + ++ + +K +E +RG L L +++ N+K+ S+
Sbjct: 780 TVVEQLEAQNREMVARISELEKTME---ERGTELSALTQKLEDNDKQSSS 826
>At1g68790 putative nuclear matrix constituent protein 1 (NMCP1)
Length = 1085
Score = 37.0 bits (84), Expect = 0.023
Identities = 51/283 (18%), Positives = 111/283 (39%), Gaps = 58/283 (20%)
Query: 68 REIHAPCSSRPRKFDLSTNLKLTESSNLMASDTTDELMEKYEHAKHAALATI-SFREKGK 126
RE H + R+ +L+ E + D E+ H + + + +K K
Sbjct: 229 REAHEAVFYKQRE-----DLQEWEKKLTLEEDRLSEVKRSINHREERVMENERTIEKKEK 283
Query: 127 VKDNIKEVVVLKDSNVEDVEEFLSV-VKDHKLKENGLVCLDDKVEVVNGGIQQQSTS--- 182
+ +N+++ + + S + + EE + + + D LKE + KV++ + + +
Sbjct: 284 ILENLQQKISVAKSELTEKEESIKIKLNDISLKEKDFEAMKAKVDIKEKELHEFEENLIE 343
Query: 183 -SMVDAAKMLDNLSLSTKNDLSGVCAYKKLLEAVDKRGDTLGRLKFEIQLNEKRKSAFDL 241
++ K+LD+ +D R R +FE++L + R+S +
Sbjct: 344 REQMEIGKLLDDQKA-----------------VLDSR-----RREFEMELEQMRRSLDEE 381
Query: 242 LRPKKELVEEVPREPFVPLTKEEEVEVTRAFSANQNKVLVNHEKSNIEISGKMFRCLRPG 301
L KK +E++ V ++ +EE R + + + V ++ +++
Sbjct: 382 LEGKKAEIEQLQ----VEISHKEEKLAKREAALEKKEEGVKKKEKDLDAR---------- 427
Query: 302 EWLNDEVINLYLELLKERERREPQKFLKCHFFNTFFYKKQECL 344
L+ +KE+E+ + K H N + +ECL
Sbjct: 428 -----------LKTVKEKEKALKAEEKKLHMENERLLEDKECL 459
>At1g76890 unknown protein
Length = 575
Score = 35.0 bits (79), Expect = 0.086
Identities = 39/133 (29%), Positives = 58/133 (43%), Gaps = 32/133 (24%)
Query: 13 HHWLSVDHNRPSPNSPTFKRQKLSTTM------SNNTP----PPLT-------TNATVFR 55
HH +SV +P +PTF ++ S+T SNNT PP++ ++ +F
Sbjct: 177 HHQVSV---QPITTNPTFLAKQPSSTTPFPFYSSNNTTTVSQPPISNDLMNNVSSLNLFS 233
Query: 56 ISRYPKTKPPLRREIHAPCSSRP-RKFDLSTNLKLTESSNLMASDTTDELMEKYEHAKHA 114
S T + H SSR RK+ KLT+ ELMEK E +
Sbjct: 234 SSTSSSTASDEEEDHHQVKSSRKKRKYWKGLFTKLTK-----------ELMEKQEKMQKR 282
Query: 115 ALATISFREKGKV 127
L T+ +REK ++
Sbjct: 283 FLETLEYREKERI 295
>At2g02200 putative protein on transposon FARE2.10 (cds2)
Length = 586
Score = 34.3 bits (77), Expect = 0.15
Identities = 53/240 (22%), Positives = 100/240 (41%), Gaps = 52/240 (21%)
Query: 102 DELMEKYEHAKHAALATISFREKGK---------VKDNIKEVVVLKDSNVEDVEEFLSVV 152
+E EK E+ H + + EK + ++ KE VLK+ NVE+ +E
Sbjct: 306 EEEEEKVEYRDHHSTCNVEETEKQENPKQGDEEMEREEGKEEKVLKEENVEEHDEH---- 361
Query: 153 KDHKLKENGLVCLDDKVEVVNGGIQQQSTSSMVDAAKMLDNLSLSTKNDLSGVCAYKKLL 212
D + V L D + +++S S + ++ A ++ +
Sbjct: 362 -DETEDQEAYVILSDDEDNGTAPTEKESESQKEETTEV----------------AKEENV 404
Query: 213 EAVDKRGDTLGRLKFEIQLNEKRKSAFDLLR---PKKELVEEVPREP--------FVPLT 261
E D+ +T + + I +++ + P+KE ++EVPRE PL+
Sbjct: 405 EEHDEHDETEDQEAYVILSDDEDNGTAPTEKESQPQKEEIKEVPRETKKDDEDVNQTPLS 464
Query: 262 KEEEVEVTRAFSANQ---NKVLVNHEKSNIEISGKMFRCLRPGEWLNDEVINLYLELLKE 318
+EE E+T+ S+ Q ++++HE EI L+ +W +++I L L+E
Sbjct: 465 TQEE-EITQGQSSLQTPLTPIMLSHEVME-EID------LKVKKWAKNKLIRDLLSSLEE 516
>At1g50640 ethylene responsive element binding factor 3
Length = 225
Score = 33.5 bits (75), Expect = 0.25
Identities = 23/71 (32%), Positives = 30/71 (41%), Gaps = 5/71 (7%)
Query: 14 HWLSVDHNRPSP--NSPTFKRQKLSTTMSNNTPPPLTTNATVFRISRYPKTKPPLRREIH 71
H L DH + P N PT ST S + P P T + RYP+T P + + H
Sbjct: 114 HRLFTDHQQQFPIVNRPTSSSMS-STVESFSGPRPTTMKPATTK--RYPRTPPVVPEDCH 170
Query: 72 APCSSRPRKFD 82
+ C S D
Sbjct: 171 SDCDSSSSVID 181
>At2g32240 putative myosin heavy chain
Length = 1333
Score = 33.1 bits (74), Expect = 0.33
Identities = 41/177 (23%), Positives = 73/177 (41%), Gaps = 33/177 (18%)
Query: 86 NLKLTESSNLMAS-----DTTDELMEKYEHAKHAA------------LATISFREKGKVK 128
++K++ES NL+ S + T +E E+ AA A S +KG+
Sbjct: 724 SVKISESENLLESIRNELNVTQGKLESIENDLKAAGLQESEVMEKLKSAEESLEQKGREI 783
Query: 129 DNIKEVVVLKDSNVEDVEEFLSVVKDHKLKENGLVCLDDKVEVVNGGIQQQSTSSMVDAA 188
D K +E + + LS+ +H+L++ +++ + SS+ +
Sbjct: 784 DE----ATTKRMELEALHQSLSIDSEHRLQK----AMEEFTS------RDSEASSLTEKL 829
Query: 189 KMLDNLSLSTKNDLSGVCAYKKLLEAVDKRGDTLGRLKFEIQLNEKRKSAFDLLRPK 245
+ L+ S + L+ L+ +K TLGRL +NEK K FD + K
Sbjct: 830 RDLEGKIKSYEEQLAEASGKSSSLK--EKLEQTLGRLAAAESVNEKLKQEFDQAQEK 884
>At1g67230 unknown protein
Length = 1132
Score = 32.3 bits (72), Expect = 0.56
Identities = 39/167 (23%), Positives = 73/167 (43%), Gaps = 24/167 (14%)
Query: 122 REKGKVKDNIKEVVVLKDSNVEDVEEFLSV-VKDHKLKENGLVCLDDKVEVVNGGIQQQS 180
++KGK + ++ + + V+ +E+ +S +KD L+E L +E +Q
Sbjct: 268 KQKGKELEEAQKKIDAANLAVKKLEDDVSSRIKDLALREQETDVLKKSIETKARELQ--- 324
Query: 181 TSSMVDAAKMLDNLSLSTKNDLSGVCAYKKLLEAVDKRGDTLGRLKFEIQLNEKRKSAFD 240
+L K + A ++L++ + D+ R +FE+++ +KRKS D
Sbjct: 325 --------------ALQEKLEAREKMAVQQLVDEHQAKLDSTQR-EFELEMEQKRKSIDD 369
Query: 241 LLRPKKELVEEVPREPFVPLTKEEEVEVTRAFSANQNKVLVNHEKSN 287
L+ K VE+ E K E +V + A K+ + EK N
Sbjct: 370 SLKSKVAEVEKREAE-----WKHMEEKVAKREQALDRKLEKHKEKEN 411
>At5g16880 TOM (target of myb1) -like protein
Length = 407
Score = 32.0 bits (71), Expect = 0.73
Identities = 38/188 (20%), Positives = 84/188 (44%), Gaps = 18/188 (9%)
Query: 135 VVLKDSNVEDVEEFLSVVKDHKLKENGLVCLDDKVEVVNGGIQQQSTSSMVDAAKMLDNL 194
+VL ++ V++ E+ S V ++ + + +DD VVN + A ML
Sbjct: 105 LVLLETCVKNCEKAFSEVAAERVLDEMVKLIDDPQTVVNNR----------NKALMLIEA 154
Query: 195 SLSTKNDLSGVCAYKKLLEAVDKRGDTLGRLKFEIQLNEKRKSAFDLLR--PKKELVEEV 252
+ ++L + +++ +++ RG ++F + NE F R P EL ++
Sbjct: 155 WGESTSELRYLPVFEETYKSLKARG-----IRFPGRDNESLAPIFTPARSTPAPELNADL 209
Query: 253 PREPFVPLTKEEEVEVTRAFSANQNKVLVNHEKSNIEISGKMFRCLRPGEWLNDEVINLY 312
P+ P + +V V R+F+A Q K + +++IE+ + + L D++
Sbjct: 210 PQHVHEPAHIQYDVPV-RSFTAEQTKEAFDIARNSIELLSTVLSSSPQHDALQDDLTTTL 268
Query: 313 LELLKERE 320
++ ++ +
Sbjct: 269 VQQCRQSQ 276
>At5g16730 putative protein
Length = 853
Score = 32.0 bits (71), Expect = 0.73
Identities = 66/295 (22%), Positives = 116/295 (38%), Gaps = 55/295 (18%)
Query: 12 PHHWLSVDHNRPSPNSPTFKRQ-KLSTTMSNNTPPPLTTNATVFRI--SRYPKTKPPLRR 68
PH LS+D + P+ S +R KL T PP + A V + + P+T L +
Sbjct: 50 PHSRLSLDRSSPNSKSSVERRSPKLPT-------PPEKSQARVAAVKGTESPQTTTRLSQ 102
Query: 69 EIHAPCSSRPRKFDLSTNLKLTESSNLMASDTTDELMEKYEHAKHAALATISFREKGKVK 128
K DL K E + + D L E + K A T+ + K +
Sbjct: 103 ----------IKEDLK---KANERISSLEKDKAKALDELKQAKKEAEQVTLKLDDALKAQ 149
Query: 129 DNIKEVVVLK------------DSNVEDVEEFLSVVKD-HKLKENGLVCLDDKVEVVNGG 175
+++E ++ +N E++++ L VK+ H LV + ++E +N
Sbjct: 150 KHVEENSEIEKFQAVEAGIEAVQNNEEELKKELETVKNQHASDSAALVAVRQELEKIN-- 207
Query: 176 IQQQSTSSMVDAAKMLDNLSLSTKNDLSGVCAYKKLLEAVDKRGDTLGRLKFEIQLNEKR 235
+ ++ DA + +LS D S E VD L RLK + ++
Sbjct: 208 ---EELAAAFDA----KSKALSQAEDASKTAEIH--AEKVDILSSELTRLKALLDSTREK 258
Query: 236 KSAFDLLRPKKELVEEVPREPFVPLTKEEEVEVTRAFSANQNKVLVNHEKSNIEI 290
+ D E+V ++ E V + ++E R F A + + EK N+++
Sbjct: 259 TAISD-----NEMVAKLEDEIVV---LKRDLESARGFEAEVKEKEMIVEKLNVDL 305
>At1g65010 hypothetical protein
Length = 1318
Score = 32.0 bits (71), Expect = 0.73
Identities = 48/223 (21%), Positives = 91/223 (40%), Gaps = 43/223 (19%)
Query: 16 LSVDHNRPSPNSPTFKRQKLSTTMSNNTPPPLTTNATVFRISRYPKTKPPLRREIHAPCS 75
+ ++PSP P + S + SN+ P T + + R P TK +H S
Sbjct: 1 METPRSKPSPPPPRLSKLSASKSDSNSASPVPNTRLS---LDRSPPTK------VH---S 48
Query: 76 SRPRKFDLSTNLKLTESSNLMASDTTDELMEKYEHAKHAALATISFREKGKVKDNIKEVV 135
+ +L T L + +L +D EL++K +K K D++KE
Sbjct: 49 RLVKGTELQTQLNQIQ-EDLKKADEQIELLKK---------------DKAKAIDDLKESE 92
Query: 136 VLKDSNVEDVEEFLSVVKDHKLKENGLVCLDDKVEVVNGGIQQQSTSSMVDAAKMLDNLS 195
L + E ++E L+ K + +E+ V VE+ G++ +
Sbjct: 93 KLVEEANEKLKEALAAQK--RAEESFEVEKFRAVELEQAGLE------------AVQKKD 138
Query: 196 LSTKNDLSGVCAYKKL-LEAVDKRGDTLGRLKFEIQLNEKRKS 237
+++KN+L + + L + A+ + L R+K E+ + K+
Sbjct: 139 VTSKNELESIRSQHALDISALLSTTEELQRVKHELSMTADAKN 181
Score = 29.6 bits (65), Expect = 3.6
Identities = 42/189 (22%), Positives = 81/189 (42%), Gaps = 27/189 (14%)
Query: 122 REKGKVKDNIKEVVVLKDSNVEDVEEFLSVVKDHK-LKENGLVCLDDKVEVVNGGIQQQS 180
+E G +K I+E+ V +S ++V + S+V++ K LKE + L K+E ++ +
Sbjct: 687 KEAGYLK-KIEELSVANESLADNVTDLQSIVQESKDLKEREVAYL-KKIEELS-----VA 739
Query: 181 TSSMVDAAKMLDNLSLSTKNDLSGVCAYKKLLEAVDKRGDTLGRLKFEIQLNEKRKSAFD 240
S+VD L ++ EA + RG LK +L+++ ++ D
Sbjct: 740 NESLVDKETKLQHID----------------QEAEELRGREASHLKKIEELSKENENLVD 783
Query: 241 LLRPKKELVEE---VPREPFVPLTKEEEVEVTRAFSANQNKVLVNHEKSNIEISGKMFRC 297
+ + + EE + L K +E+ A+ L N + N E+ +
Sbjct: 784 NVANMQNIAEESKDLREREVAYLKKIDELSTANGTLADNVTNLQNISEENKELRERETTL 843
Query: 298 LRPGEWLND 306
L+ E L++
Sbjct: 844 LKKAEELSE 852
>At5g07820 putative protein
Length = 561
Score = 31.6 bits (70), Expect = 0.96
Identities = 35/151 (23%), Positives = 65/151 (42%), Gaps = 28/151 (18%)
Query: 31 KRQKLSTTMSNNTPPPLT---TNATVFRISRYPKTKPPLRREIHAPCSSRPRKFDLSTNL 87
K+ + +T + +P T TN V + SR + K P K DLS NL
Sbjct: 156 KKSETKSTSPSVSPVVRTVKKTNLVVNKASRISQNKSP--------------KEDLSKNL 201
Query: 88 KLTESSNLMASDTTDELMEKYE----------HAKHAALATISFREKGKVKDNIKEVVVL 137
K E ++ D+++EK + K + T+ +EK K+ + ++ VL
Sbjct: 202 KNKEKGKIVEPVRCDDVLEKTDLEVKKVSRISENKSSKEDTLKNKEKAKIDEPVRCDDVL 261
Query: 138 KDSNVEDVEEFLSVVKDHKLKENGLVCLDDK 168
+ +++ D ++ + ++ KE L L +K
Sbjct: 262 EKTSL-DAQKVSRISENKNSKEERLKNLKNK 291
>At4g33500 unknown protein
Length = 724
Score = 30.8 bits (68), Expect = 1.6
Identities = 30/131 (22%), Positives = 57/131 (42%), Gaps = 5/131 (3%)
Query: 73 PCSSRPRKFDLSTNLKLTESSNLMASDTTDELMEKYEHAKHAALATISFREKGKVKDNIK 132
P +S P KFDL ++ +L + S++ E+ EKY A+ A + K+ +
Sbjct: 63 PENSAPEKFDLVSSTQLKDGSHVFRFGDASEI-EKYLEAEEKARCVEVETQNAKIAEEAS 121
Query: 133 EVVVLKDSNVEDVEEFLSVVKDHKLKENGL---VCLDDKVEVVNGGIQQQSTSSMVDAAK 189
EV + V + E S K+ + L + + D+ V + +++ T ++ +
Sbjct: 122 EVSRKQKKLVSSIIE-TSTEKEETAAPSDLSNVIKIKDRKRVRSPTKKKKETVNVSRSED 180
Query: 190 MLDNLSLSTKN 200
+D S S N
Sbjct: 181 KIDAKSASVSN 191
>At2g26670 heme oxygenase 1 (HO1)
Length = 282
Score = 30.8 bits (68), Expect = 1.6
Identities = 12/26 (46%), Positives = 18/26 (69%), Gaps = 1/26 (3%)
Query: 312 YLELLKERERREPQKFLKCHFFNTFF 337
Y + LKE ++PQ F+ CHF+N +F
Sbjct: 180 YSQYLKELAEKDPQAFI-CHFYNIYF 204
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,365,577
Number of Sequences: 26719
Number of extensions: 455761
Number of successful extensions: 2036
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 44
Number of HSP's that attempted gapping in prelim test: 2004
Number of HSP's gapped (non-prelim): 65
length of query: 459
length of database: 11,318,596
effective HSP length: 103
effective length of query: 356
effective length of database: 8,566,539
effective search space: 3049687884
effective search space used: 3049687884
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147012.13


