

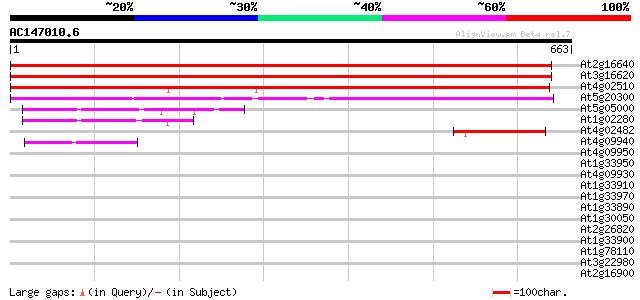
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147010.6 + phase: 0
(663 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g16640 putative chloroplast outer membrane protein 1039 0.0
At3g16620 putative GTP-binding protein 1024 0.0
At4g02510 chloroplast protein import component Toc159-like 646 0.0
At5g20300 putative protein 486 e-137
At5g05000 GTP-binding protein (gb|AAD09203.1) 130 2e-30
At1g02280 unknown protein 117 2e-26
At4g02482 putative protein 74 3e-13
At4g09940 AIG1-like protein 43 5e-04
At4g09950 AIG1-like protein 41 0.002
At1g33950 AIG1-like protein 41 0.002
At4g09930 AIG1-like protein 40 0.003
At1g33910 AIG1-like protein 39 0.007
At1g33970 unknown protein 38 0.016
At1g33890 AIG1-like protein 37 0.027
At1g30050 hypothetical protein 37 0.027
At2g26820 similar to avrRpt2-induced protein 1 37 0.046
At1g33900 AIG1-like protein 37 0.046
At1g78110 unknown protein 36 0.079
At3g22980 eukaryotic translation elongation factor 2, putative 34 0.23
At2g16900 unknown protein 34 0.23
>At2g16640 putative chloroplast outer membrane protein
Length = 1206
Score = 1039 bits (2686), Expect = 0.0
Identities = 511/640 (79%), Positives = 577/640 (89%), Gaps = 1/640 (0%)
Query: 1 MAEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGM 60
MAEQLE+AGQ+PLDFSCTIMVLGKSGVGKS+TINSIFDEVKF TDAF MGTK+VQDV G+
Sbjct: 559 MAEQLEAAGQDPLDFSCTIMVLGKSGVGKSATINSIFDEVKFCTDAFQMGTKRVQDVEGL 618
Query: 61 VQGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMP 120
VQGIKVRVIDTPGLLPSWSDQ NEKIL+SVK FIKK PPDIVLYLDRLDMQSRD DMP
Sbjct: 619 VQGIKVRVIDTPGLLPSWSDQAKNEKILNSVKAFIKKNPPDIVLYLDRLDMQSRDSGDMP 678
Query: 121 LLRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGD 180
LLRTI+D+FGP IWFNAIV LTHAAS PPDGPNGT SSYDMFVTQRSHV+QQAIRQAAGD
Sbjct: 679 LLRTISDVFGPSIWFNAIVGLTHAASVPPDGPNGTASSYDMFVTQRSHVIQQAIRQAAGD 738
Query: 181 MRLMNPVSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQDNPR 240
MRLMNPVSLVENHSACRTN AGQRVLPNGQVWKP LLLLSFASKILAEANALLKLQDN
Sbjct: 739 MRLMNPVSLVENHSACRTNRAGQRVLPNGQVWKPHLLLLSFASKILAEANALLKLQDNIP 798
Query: 241 EKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETDPDDLP 300
+P+ AR++APPLPFLLSSLLQSRPQ KLPE Q+ DE+ DDL+E SDS +E++ D LP
Sbjct: 799 GRPFAARSKAPPLPFLLSSLLQSRPQPKLPEQQYGDEED-EDDLEESSDSDEESEYDQLP 857
Query: 301 PFKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDLPSD 360
PFK LTKAQ+ LS++QKK YLDE+EYREKL MKKQ+K E+K+RKM K+ A +KDLP
Sbjct: 858 PFKSLTKAQMATLSKSQKKQYLDEMEYREKLLMKKQMKEERKRRKMFKKFAAEIKDLPDG 917
Query: 361 YVENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVLETHGWDHD 420
Y ENVEEESGG ASVPVPMPD+SLPASFDSD PTHRYR+LDSSNQWLVRPVLETHGWDHD
Sbjct: 918 YSENVEEESGGPASVPVPMPDLSLPASFDSDNPTHRYRYLDSSNQWLVRPVLETHGWDHD 977
Query: 421 VGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATSLGFDMQTV 480
+GYEG+N ERLFV+K+KIP+S SGQVTKDKKDANVQ+EM SSVK+GEGK+TSLGFDMQTV
Sbjct: 978 IGYEGVNAERLFVVKEKIPISVSGQVTKDKKDANVQLEMASSVKHGEGKSTSLGFDMQTV 1037
Query: 481 GKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFKLVIAGGAM 540
GK+LAYTLRSET+F NF RNKA AGLS T LGD++SAG+KVEDK IA+K F++V++GGAM
Sbjct: 1038 GKELAYTLRSETRFNNFRRNKAAAGLSVTHLGDSVSAGLKVEDKFIASKWFRIVMSGGAM 1097
Query: 541 TGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRYTNLV 600
T R D AYGG+LEAQLRDK+YPLGR L+TLGLSVMDWHGDLA+G N+QSQ+PIGR +NL+
Sbjct: 1098 TSRGDFAYGGTLEAQLRDKDYPLGRFLTTLGLSVMDWHGDLAIGGNIQSQVPIGRSSNLI 1157
Query: 601 ARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKVIGY 640
ARANLNNRGAGQ+S+R+NSSEQLQ+A++ ++PL KK++ Y
Sbjct: 1158 ARANLNNRGAGQVSVRVNSSEQLQLAMVAIVPLFKKLLSY 1197
>At3g16620 putative GTP-binding protein
Length = 1089
Score = 1024 bits (2647), Expect = 0.0
Identities = 502/640 (78%), Positives = 574/640 (89%), Gaps = 1/640 (0%)
Query: 1 MAEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGM 60
MAEQLE+A Q+PLDFSCTIMVLGKSGVGKS+TINSIFDE+K +TDAF +GTKKVQD+ G
Sbjct: 441 MAEQLEAAAQDPLDFSCTIMVLGKSGVGKSATINSIFDELKISTDAFQVGTKKVQDIEGF 500
Query: 61 VQGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMP 120
VQGIKVRVIDTPGLLPSWSDQ NEKIL SV+ FIKK+PPDIVLYLDRLDMQSRD DMP
Sbjct: 501 VQGIKVRVIDTPGLLPSWSDQHKNEKILKSVRAFIKKSPPDIVLYLDRLDMQSRDSGDMP 560
Query: 121 LLRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGD 180
LLRTITD+FGP IWFNAIV LTHAASAPPDGPNGT SSYDMFVTQRSHV+QQAIRQAAGD
Sbjct: 561 LLRTITDVFGPSIWFNAIVGLTHAASAPPDGPNGTASSYDMFVTQRSHVIQQAIRQAAGD 620
Query: 181 MRLMNPVSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQDNPR 240
MRLMNPVSLVENHSACRTN AGQRVLPNGQVWKP LLLLSFASKILAEANALLKLQDN
Sbjct: 621 MRLMNPVSLVENHSACRTNRAGQRVLPNGQVWKPHLLLLSFASKILAEANALLKLQDNIP 680
Query: 241 EKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETDPDDLP 300
+ R++APPLP LLSSLLQSRPQ KLPE Q+ DED DDLDE SDS +E++ D+LP
Sbjct: 681 GGQFATRSKAPPLPLLLSSLLQSRPQAKLPEQQYDDEDD-EDDLDESSDSEEESEYDELP 739
Query: 301 PFKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDLPSD 360
PFK LTKA++ LS++QKK YLDE+EYREKLFMK+Q+K E+K+RK++K+ A +KD+P+
Sbjct: 740 PFKRLTKAEMTKLSKSQKKEYLDEMEYREKLFMKRQMKEERKRRKLLKKFAAEIKDMPNG 799
Query: 361 YVENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVLETHGWDHD 420
Y ENVEEE ASVPVPMPD+SLPASFDSD PTHRYR+LD+SNQWLVRPVLETHGWDHD
Sbjct: 800 YSENVEEERSEPASVPVPMPDLSLPASFDSDNPTHRYRYLDTSNQWLVRPVLETHGWDHD 859
Query: 421 VGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATSLGFDMQTV 480
+GYEG+N ERLFV+KDKIPVSFSGQVTKDKKDA+VQ+E+ SSVK+GEG++TSLGFDMQ
Sbjct: 860 IGYEGVNAERLFVVKDKIPVSFSGQVTKDKKDAHVQLELASSVKHGEGRSTSLGFDMQNA 919
Query: 481 GKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFKLVIAGGAM 540
GK+LAYT+RSET+F F +NKA AGLS TLLGD++SAG+KVEDKLIANKRF++V++GGAM
Sbjct: 920 GKELAYTIRSETRFNKFRKNKAAAGLSVTLLGDSVSAGLKVEDKLIANKRFRMVMSGGAM 979
Query: 541 TGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRYTNLV 600
T R DVAYGG+LEAQ RDK+YPLGR LSTLGLSVMDWHGDLA+G N+QSQ+PIGR +NL+
Sbjct: 980 TSRGDVAYGGTLEAQFRDKDYPLGRFLSTLGLSVMDWHGDLAIGGNIQSQVPIGRSSNLI 1039
Query: 601 ARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKVIGY 640
ARANLNNRGAGQ+SIR+NSSEQLQ+A++ L+PL KK++ Y
Sbjct: 1040 ARANLNNRGAGQVSIRVNSSEQLQLAVVALVPLFKKLLTY 1079
>At4g02510 chloroplast protein import component Toc159-like
Length = 1503
Score = 646 bits (1667), Expect = 0.0
Identities = 332/647 (51%), Positives = 445/647 (68%), Gaps = 11/647 (1%)
Query: 2 AEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMV 61
A + E+ G E L FS I+VLGK+GVGKS+TINSI + DAF + T V+++ G V
Sbjct: 841 AVESEAEGNEELIFSLNILVLGKAGVGKSATINSILGNQIASIDAFGLSTTSVREISGTV 900
Query: 62 QGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPL 121
G+K+ IDTPGL + DQ N K+L SVK+ +KK PPDIVLY+DRLD Q+RD +++PL
Sbjct: 901 NGVKITFIDTPGLKSAAMDQSTNAKMLSSVKKVMKKCPPDIVLYVDRLDTQTRDLNNLPL 960
Query: 122 LRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGDM 181
LRTIT G IW NAIV LTHAASAPPDGP+GTP SYD+FV Q SH+VQQ+I QA GD+
Sbjct: 961 LRTITASLGTSIWKNAIVTLTHAASAPPDGPSGTPLSYDVFVAQCSHIVQQSIGQAVGDL 1020
Query: 182 RLMNP-----VSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQ 236
RLMNP VSLVENH CR N G +VLPNGQ W+ QLLLL ++ K+L+E N+LL+ Q
Sbjct: 1021 RLMNPSLMNPVSLVENHPLCRKNREGVKVLPNGQTWRSQLLLLCYSLKVLSETNSLLRPQ 1080
Query: 237 DN-PREKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDS----G 291
+ K + R R+PPLP+LLS LLQSR KLP DQ D + ++D+ SDS G
Sbjct: 1081 EPLDHRKVFGFRVRSPPLPYLLSWLLQSRAHPKLPGDQGGDSVDSDIEIDDVSDSEQEDG 1140
Query: 292 DETDPDDLPPFKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMA 351
++ + D LPPFKPL K Q+ LS Q+KAY +E +YR KL KKQ + E K+ K MK+
Sbjct: 1141 EDDEYDQLPPFKPLRKTQLAKLSNEQRKAYFEEYDYRVKLLQKKQWREELKRMKEMKKNG 1200
Query: 352 ESVKDLPSDYV-ENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRP 410
+ + + Y E + E+G A+VPVP+PDM LP SFDSD +RYR+L+ ++Q L RP
Sbjct: 1201 KKLGESEFGYPGEEDDPENGAPAAVPVPLPDMVLPPSFDSDNSAYRYRYLEPTSQLLTRP 1260
Query: 411 VLETHGWDHDVGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKA 470
VL+THGWDHD GY+G+N E L + P + + QVTKDKK+ N+ ++ + S K+GE +
Sbjct: 1261 VLDTHGWDHDCGYDGVNAEHSLALASRFPATATVQVTKDKKEFNIHLDSSVSAKHGENGS 1320
Query: 471 TSLGFDMQTVGKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKR 530
T GFD+Q VGK LAY +R ETKF N +NK T G S T LG+ ++ GVK+ED++ KR
Sbjct: 1321 TMAGFDIQNVGKQLAYVVRGETKFKNLRKNKTTVGGSVTFLGENIATGVKLEDQIALGKR 1380
Query: 531 FKLVIAGGAMTGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQ 590
LV + G M + D AYG +LE +LR+ ++P+G+ S+ GLS++ W GDLA+G NLQSQ
Sbjct: 1381 LVLVGSTGTMRSQGDSAYGANLEVRLREADFPIGQDQSSFGLSLVKWRGDLALGANLQSQ 1440
Query: 591 IPIGRYTNLVARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKV 637
+ +GR + + RA LNN+ +GQI++R +SS+QLQIAL ++P+ +
Sbjct: 1441 VSVGRNSKIALRAGLNNKMSGQITVRTSSSDQLQIALTAILPIAMSI 1487
>At5g20300 putative protein
Length = 787
Score = 486 bits (1252), Expect = e-137
Identities = 274/645 (42%), Positives = 390/645 (59%), Gaps = 23/645 (3%)
Query: 1 MAEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGM 60
+A + ES+G LDFS I+VLGK+GVGKS+TINSIF + K TDAF GT ++++V+G
Sbjct: 145 LAREQESSGIPELDFSLRILVLGKTGVGKSATINSIFGQPKSETDAFRPGTDRIEEVMGT 204
Query: 61 VQGIKVRVIDTPGLLP-SWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDM 119
V G+KV IDTPG P S S N KIL S+KR++KK PPD+VLYLDRLDM +SD
Sbjct: 205 VSGVKVTFIDTPGFHPLSSSSTRKNRKILLSIKRYVKKRPPDVVLYLDRLDMIDMRYSDF 264
Query: 120 PLLRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAG 179
LL+ IT+IFG IW N I+V+TH+A A +G NG +Y+ +V QR VVQ I QA
Sbjct: 265 SLLQLITEIFGAAIWLNTILVMTHSA-ATTEGRNGQSVNYESYVGQRMDVVQHYIHQAVS 323
Query: 180 DMRLMNPVSLVENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQDN- 238
D +L NPV LVENH +C+ N AG+ VLPNG VWKPQ + L +K+L + +LL+ +D+
Sbjct: 324 DTKLENPVLLVENHPSCKKNLAGEYVLPNGVVWKPQFMFLCVCTKVLGDVQSLLRFRDSI 383
Query: 239 PREKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETDPDD 298
+P + R + LP LLS L+ R E + + LN DL+E E + D
Sbjct: 384 GLGQPSSTRTAS--LPHLLSVFLRRRLSSGADETEKEIDKLLNLDLEE------EDEYDQ 435
Query: 299 LPPFKPLTKAQIRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDLP 358
LP + L K++ LS++QKK YLDE++YRE L++KKQLK E ++R+ K + E
Sbjct: 436 LPTIRILGKSRFEKLSKSQKKEYLDELDYRETLYLKKQLKEECRRRRDEKLVEE------ 489
Query: 359 SDYVENVEEESGGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVLETHGWD 418
+ +E+ E+ A VP+PDM+ P SFDSD P HRYR + + +QWLVRPV + GWD
Sbjct: 490 -ENLEDTEQRDQAA----VPLPDMAGPDSFDSDFPAHRYRCVSAGDQWLVRPVYDPQGWD 544
Query: 419 HDVGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKAT-SLGFDM 477
DVG++G+N+E + + S +GQV++DK+ +Q E ++ + T S+ D+
Sbjct: 545 RDVGFDGINIETAAKINRNLFASATGQVSRDKQRFTIQSETNAAYTRNFREQTFSVAVDL 604
Query: 478 QTVGKDLAYTLRSETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFKLVIAG 537
Q+ G+DL Y+ + TK F N G+ T G G K+ED L+ KR KL
Sbjct: 605 QSSGEDLVYSFQGGTKLQTFKHNTTDVGVGLTSFGGKYYVGGKLEDTLLVGKRVKLTANA 664
Query: 538 GAMTGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRYT 597
G M G A GGS EA +R ++YP+ L ++ + + +L + LQ+Q R T
Sbjct: 665 GQMRGSGQTANGGSFEACIRGRDYPVRNEQIGLTMTALSFKRELVLNYGLQTQFRPARGT 724
Query: 598 NLVARANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKVIGYSQ 642
N+ N+NNR G+I+++LNSSE +IALI + + K ++ S+
Sbjct: 725 NIDVNINMNNRKMGKINVKLNSSEHWEIALISALTMFKALVRRSK 769
>At5g05000 GTP-binding protein (gb|AAD09203.1)
Length = 313
Score = 130 bits (328), Expect = 2e-30
Identities = 90/271 (33%), Positives = 136/271 (49%), Gaps = 21/271 (7%)
Query: 16 SCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMVQGIKVRVIDTPGLL 75
S T++V+GK GVGKSST+NS+ E F + V G + +IDTPGL+
Sbjct: 38 SLTVLVMGKGGVGKSSTVNSVIGEKAAAVSTFQSEGLRPTLVSRTRSGFTLNIIDTPGLI 97
Query: 76 PSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPLLRTITDIFGPPIWF 135
N++ ++ +KRF+ D++LY+DRLD+ D D ++ ITD FG IW
Sbjct: 98 EGGYV---NDQAINIIKRFLLNMTIDVLLYVDRLDVYRVDDLDRQVVGAITDAFGKEIWK 154
Query: 136 NAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAA----GDMRLMN-PVSLV 190
+ +VLTHA +PPDG N Y+ FV++RS+ + + I+ A D++ + PV LV
Sbjct: 155 KSALVLTHAQFSPPDGLN-----YNHFVSKRSNALLKVIQTGAQLKKQDLQGFSIPVILV 209
Query: 191 ENHSACRTNTAGQRVLPNGQVWKPQLL----LLSFASKILAEANALLKLQDNPREKPYTA 246
EN C N + +++LP G W P L +SF + L NP E+
Sbjct: 210 ENSGRCHKNESDEKILPCGTSWIPNLFNKITEISFNGNKAIHVDKKLVEGPNPNER---- 265
Query: 247 RARAPPLPFLLSSLLQSRPQLKLPEDQFSDE 277
+ PL F LL +P ++ + S E
Sbjct: 266 GKKLIPLMFAFQYLLVMKPLVRAIKSDVSRE 296
>At1g02280 unknown protein
Length = 297
Score = 117 bits (293), Expect = 2e-26
Identities = 74/208 (35%), Positives = 105/208 (49%), Gaps = 15/208 (7%)
Query: 16 SCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMVQGIKVRVIDTPGLL 75
S T++VLGK GVGKSST+NS+ E F + V + G + +IDTPGL+
Sbjct: 36 SMTVLVLGKGGVGKSSTVNSLIGEQVVRVSPFQAEGLRPVMVSRTMGGFTINIIDTPGLV 95
Query: 76 PSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPLLRTITDIFGPPIWF 135
+ N + L +K F+ D++LY+DRLD+ D D ++ IT FG IW
Sbjct: 96 EAGYV---NHQALELIKGFLVNRTIDVLLYVDRLDVYRVDELDKQVVIAITQTFGKEIWC 152
Query: 136 NAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGDMRLMN------PVSL 189
++VLTHA +PPD SY+ F ++RS + + IR A MR V
Sbjct: 153 KTLLVLTHAQFSPPD-----ELSYETFSSKRSDSLLKTIR-AGSKMRKQEFEDSAIAVVY 206
Query: 190 VENHSACRTNTAGQRVLPNGQVWKPQLL 217
EN C N ++ LPNG+ W P L+
Sbjct: 207 AENSGRCSKNDKDEKALPNGEAWIPNLV 234
>At4g02482 putative protein
Length = 134
Score = 73.9 bits (180), Expect = 3e-13
Identities = 38/114 (33%), Positives = 70/114 (61%), Gaps = 5/114 (4%)
Query: 525 LIANKRFKLVIAG-----GAMTGRDDVAYGGSLEAQLRDKNYPLGRSLSTLGLSVMDWHG 579
L ++R +L + G G +D +Y G+LE +LR+ ++P+G++ +G+S+ +
Sbjct: 19 LCNSRRNELTLGGLVTFFGTTRSEEDSSYEGNLELRLREADFPIGQNQPHMGVSLKNSKD 78
Query: 580 DLAVGCNLQSQIPIGRYTNLVARANLNNRGAGQISIRLNSSEQLQIALIGLIPL 633
DL V NL+ Q+ +GR T + +L+++ G ++R NSS+QLQIA++ L+ L
Sbjct: 79 DLTVTANLRHQVSVGRQTKVTTFVSLDSKRTGCFTVRTNSSDQLQIAVMALLLL 132
>At4g09940 AIG1-like protein
Length = 394
Score = 43.1 bits (100), Expect = 5e-04
Identities = 40/137 (29%), Positives = 60/137 (43%), Gaps = 6/137 (4%)
Query: 18 TIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVG--MVQGIKVRVIDTPGLL 75
T++++G+SG GKS+T NSI F + G ++ + G + VIDTPGL
Sbjct: 49 TLLLVGRSGNGKSATGNSILGRKAFKSKGRASGVTTACELQSSTLPNGQIINVIDTPGL- 107
Query: 76 PSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSR-DFSDMPLLRTITDIFGPPIW 134
+S P E + R T I L +++R + L + +FG I
Sbjct: 108 --FSLSPSTEFTCREILRCFSLTKEGIDAVLLVFSLKNRLTEEEKSALFALKILFGSKIV 165
Query: 135 FNAIVVLTHAASAPPDG 151
IVV T+ S DG
Sbjct: 166 DYMIVVFTNEDSLEDDG 182
>At4g09950 AIG1-like protein
Length = 336
Score = 41.2 bits (95), Expect = 0.002
Identities = 40/130 (30%), Positives = 65/130 (49%), Gaps = 8/130 (6%)
Query: 18 TIMVLGKSGVGKSSTINSIFDEVKFNTDA-FHMGTKKVQ-DVVGMVQGIKVRVIDTPGLL 75
T+++LG++G GKS+T NSI + F + A TK+ + + G+ + VIDTPGL
Sbjct: 19 TLVLLGRTGNGKSATGNSILGKTMFQSKARGKFITKECKLHKSKLPNGLTINVIDTPGL- 77
Query: 76 PSWSDQPHNEKILHSVKR--FIKKTPPDIVLYLDRLDMQSRDFSDMPLLRTITDIFGPPI 133
+S + + + R + K D VL + L + + + LRT+ +FG I
Sbjct: 78 --FSASSTTDFTIREIVRCLLLAKGGIDAVLLVFSLRNRLTE-EEQSTLRTLKILFGSQI 134
Query: 134 WFNAIVVLTH 143
IVV T+
Sbjct: 135 VDYIIVVFTN 144
>At1g33950 AIG1-like protein
Length = 311
Score = 41.2 bits (95), Expect = 0.002
Identities = 39/127 (30%), Positives = 59/127 (45%), Gaps = 6/127 (4%)
Query: 19 IMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMV--QGIKVRVIDTPGLLP 76
I+++G++G GKS+T NS+ + F + A G G+V G K+ VIDTPGL
Sbjct: 19 IVLVGRTGNGKSATGNSLIGKKVFASKAHASGVTMKCQTHGVVTKDGHKINVIDTPGL-- 76
Query: 77 SWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSR-DFSDMPLLRTITDIFGPPIWF 135
+ E I + R + I L L ++R + LRT+ +FG I
Sbjct: 77 -FDLSVSAEYISKEIVRCLTLAEGGIHAVLLVLSARTRITQEEENTLRTLQALFGSQILD 135
Query: 136 NAIVVLT 142
+VV T
Sbjct: 136 YVVVVFT 142
>At4g09930 AIG1-like protein
Length = 335
Score = 40.4 bits (93), Expect = 0.003
Identities = 42/144 (29%), Positives = 68/144 (47%), Gaps = 10/144 (6%)
Query: 12 PLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMVQ--GIKVRVI 69
P+ S T++++G SG GKS+T NSI F + K ++ + G + VI
Sbjct: 22 PMKPSRTLVLIGCSGNGKSATGNSILRSEAFKSKGQAAAVTKECELKSTKRPNGQIINVI 81
Query: 70 DTPGLLPSWSDQPHNEKILHSVKR--FIKKTPPDIVLYLDRLDMQ-SRDFSDMPLLRTIT 126
DTPGL +S P NE + + + + K D VL + L + + + +P + +
Sbjct: 82 DTPGL---FSLFPSNESTIREILKCSHLAKEGIDAVLMVFSLRSRLTEEEKSVPFV--LK 136
Query: 127 DIFGPPIWFNAIVVLTHAASAPPD 150
+FG I+ IVV T+ S D
Sbjct: 137 TLFGDSIFDYLIVVFTNEDSLIDD 160
>At1g33910 AIG1-like protein
Length = 301
Score = 39.3 bits (90), Expect = 0.007
Identities = 28/90 (31%), Positives = 49/90 (54%), Gaps = 8/90 (8%)
Query: 1 MAEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGM 60
M+++ + + EP+ I+++G +G GKSST NS+ + F + T K + + G
Sbjct: 1 MSDRAQPSASEPVR---NIVLVGPTGNGKSSTGNSLIGKEVFILETVECKTCKAKTLDGQ 57
Query: 61 VQGIKVRVIDTPGLLP-SWSDQPHNEKILH 89
+ + VIDTPGL S S N++I++
Sbjct: 58 I----INVIDTPGLFDLSVSTDYMNKEIIN 83
>At1g33970 unknown protein
Length = 342
Score = 38.1 bits (87), Expect = 0.016
Identities = 22/59 (37%), Positives = 35/59 (59%), Gaps = 2/59 (3%)
Query: 18 TIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMVQ--GIKVRVIDTPGL 74
T++++G++G GKS+T NSI F + A +G + +VQ G + V+DTPGL
Sbjct: 26 TLVLVGRTGNGKSATGNSILGRKAFRSRARTVGVTSTCESQRVVQEDGDIINVVDTPGL 84
>At1g33890 AIG1-like protein
Length = 334
Score = 37.4 bits (85), Expect = 0.027
Identities = 22/58 (37%), Positives = 34/58 (57%), Gaps = 2/58 (3%)
Query: 19 IMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMVQ--GIKVRVIDTPGL 74
I+++G++G GKS+T NS+ + F ++A G K V G ++ VIDTPGL
Sbjct: 16 IVLVGRTGNGKSATGNSLIGKDVFVSEAKATGVTKTCQTYKAVTPGGSRINVIDTPGL 73
>At1g30050 hypothetical protein
Length = 389
Score = 37.4 bits (85), Expect = 0.027
Identities = 38/161 (23%), Positives = 65/161 (39%), Gaps = 14/161 (8%)
Query: 236 QDNPREKPY-----TARARAPPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDS 290
Q NPRE A A A LL L + L +++ S + N L + D
Sbjct: 178 QPNPRESQLKASRDVAMATAAKAKLLLRELKTVKADLAFAKERCSQLEEENKRLRDNRDK 237
Query: 291 GDETDPDDLPPFKPLTKAQIRN-LSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRK---M 346
G+ DD L + Q+ L+ + A+ + + RE F+++ ++Y + +
Sbjct: 238 GNNNPADD-----DLIRLQLETLLAEKARLAHENSIYARENRFLREIVEYHQLTMQDVVY 292
Query: 347 MKEMAESVKDLPSDYVENVEEESGGAASVPVPMPDMSLPAS 387
+ E E V ++ + S A+ +P P S PAS
Sbjct: 293 IDEGIEEVAEVNPSITRTLSMASYSASELPSISPSPSSPAS 333
>At2g26820 similar to avrRpt2-induced protein 1
Length = 463
Score = 36.6 bits (83), Expect = 0.046
Identities = 36/128 (28%), Positives = 61/128 (47%), Gaps = 8/128 (6%)
Query: 19 IMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDV--VGMVQGIKVRVIDTPGLLP 76
I+++G++G GKSST N++ +F + G + ++ + G + VIDTPGL
Sbjct: 8 IVLVGRTGNGKSSTGNTLLGTKQFKSKNQAKGVTMICEMYRAAIQDGPIINVIDTPGLCD 67
Query: 77 SW--SDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPLLRTITDIFGPPIW 134
S+ D NE I + ++ ++L L S++ + + T+ IFG I
Sbjct: 68 SFVPGDDISNEII--NCLTMAEEGIHAVLLVLSARGRISKE--EESTVNTLQCIFGSQIL 123
Query: 135 FNAIVVLT 142
IVV T
Sbjct: 124 DYCIVVFT 131
>At1g33900 AIG1-like protein
Length = 326
Score = 36.6 bits (83), Expect = 0.046
Identities = 22/58 (37%), Positives = 35/58 (59%), Gaps = 2/58 (3%)
Query: 19 IMVLGKSGVGKSSTINSIFDEVKFNTDAFHMG-TKKVQDVVGMVQ-GIKVRVIDTPGL 74
I+++G++G GKS+T NS+ + F ++ G T K + V + G + VIDTPGL
Sbjct: 22 IVLVGRTGNGKSATGNSLIGKQVFRSETRATGVTMKCETCVAVTPCGTGINVIDTPGL 79
>At1g78110 unknown protein
Length = 342
Score = 35.8 bits (81), Expect = 0.079
Identities = 27/98 (27%), Positives = 46/98 (46%), Gaps = 6/98 (6%)
Query: 271 EDQFSDEDSLNDDLDEPSDSGDETDPDDLPPFKPLTKAQIRNLSRAQKKAYLDEVEYREK 330
E D+D N+ DE D D +PP L + R+ A K++L+E R K
Sbjct: 209 EQNNKDDDKNNNKCDEKRDLEDTETEPAVPPPNALLLMRCRS---APAKSWLEE---RMK 262
Query: 331 LFMKKQLKYEKKQRKMMKEMAESVKDLPSDYVENVEEE 368
+ +++ + E+K+ K ++ S+K D +EEE
Sbjct: 263 VKTEQEKREEQKEEKETEDQETSMKTKKKDLRSLMEEE 300
>At3g22980 eukaryotic translation elongation factor 2, putative
Length = 1015
Score = 34.3 bits (77), Expect = 0.23
Identities = 30/114 (26%), Positives = 47/114 (40%), Gaps = 8/114 (7%)
Query: 97 KTPPDI---VLYLDRLDMQSRDFSDMPLLRTITDIFGPPIWFNAIVVLTHAASAPPDGPN 153
+TP ++ LY + L ++S S L + P+W A + +H A A D
Sbjct: 784 ETPAEVSETALYSEALTLESSIVSGFQLATASGPLCDEPMWGLAFTIESHLAPA-EDVET 842
Query: 154 GTPSSYDMFVTQRSHVVQQAIRQAAGDMRLMNPVSLVENHSACRTNTAGQRVLP 207
P ++ +F Q V+ A R A L +VE C NTA + + P
Sbjct: 843 DKPENFGIFTGQVMTAVKDACRAAV----LQTNPRIVEAMYFCELNTAPEYLGP 892
>At2g16900 unknown protein
Length = 382
Score = 34.3 bits (77), Expect = 0.23
Identities = 25/98 (25%), Positives = 44/98 (44%), Gaps = 10/98 (10%)
Query: 561 YPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRYTNLVA--RANLNNRGAGQIS---- 614
Y + S+ST+ S++D HGD+A C L+S RY + L + GQ++
Sbjct: 213 YRVNSSVSTILQSIIDKHGDIAANCKLESASMRSRYLECLCSLMQELGSTPVGQLTELKV 272
Query: 615 ----IRLNSSEQLQIALIGLIPLLKKVIGYSQNCSLDK 648
L E + I + + +L++ Y +N +K
Sbjct: 273 KEMVAVLKDLESVNIDVGWMRSVLEEFAQYQENTDSEK 310
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.134 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,109,571
Number of Sequences: 26719
Number of extensions: 682693
Number of successful extensions: 3110
Number of sequences better than 10.0: 89
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 62
Number of HSP's that attempted gapping in prelim test: 2865
Number of HSP's gapped (non-prelim): 213
length of query: 663
length of database: 11,318,596
effective HSP length: 106
effective length of query: 557
effective length of database: 8,486,382
effective search space: 4726914774
effective search space used: 4726914774
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC147010.6


