

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147010.11 - phase: 0 /pseudo
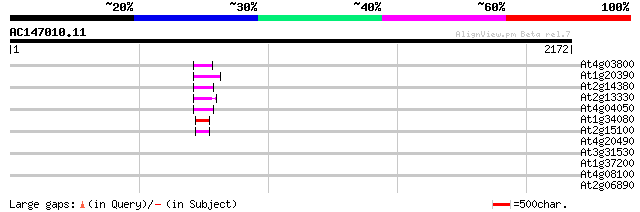
(2172 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g03800 55 4e-07
At1g20390 hypothetical protein 52 4e-06
At2g14380 putative retroelement pol polyprotein 51 6e-06
At2g13330 F14O4.9 51 6e-06
At4g04050 putative transposon protein 48 5e-05
At1g34080 putative protein 48 5e-05
At2g15100 putative retroelement pol polyprotein 46 3e-04
At4g20490 putative protein 44 0.001
At3g31530 hypothetical protein 41 0.007
At1g37200 hypothetical protein 40 0.011
At4g08100 putative polyprotein 33 1.4
At2g06890 putative retroelement integrase 31 9.1
>At4g03800
Length = 637
Score = 55.1 bits (131), Expect = 4e-07
Identities = 30/74 (40%), Positives = 40/74 (53%)
Query: 712 AFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKL 771
AF LG I LP+ G + +V F V D A+Y+ +LG PWI AV ST HQ L
Sbjct: 238 AFTSESAMSLGTIKLPVLAGKMSKIVDFVVFDKPATYNIILGTPWIFQMKAVPSTYHQWL 297
Query: 772 KFIRNGKLVTVHGE 785
KF + + T+ G+
Sbjct: 298 KFPTSNGVETIWGD 311
>At1g20390 hypothetical protein
Length = 1791
Score = 52.0 bits (123), Expect = 4e-06
Identities = 34/110 (30%), Positives = 50/110 (44%), Gaps = 7/110 (6%)
Query: 713 FDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 772
FDG +G I LPI +G V F V+ A Y+ +LG PWIH A+ ST HQ +K
Sbjct: 621 FDGDFVMTIGTIKLPIFVGGLIAWVKFVVIGKPAVYNVILGTPWIHQMQAIPSTYHQCVK 680
Query: 773 FIRNGKLVTVHG-------EEAYLVSQLSSFSCIEAGSAEGTAFQGLTIE 815
F + + T+ +Y S+L + ++ T G+ E
Sbjct: 681 FPTHNGIFTLRAPKEAKTPSRSYEESELCRTEMVNIDESDPTRCVGVGAE 730
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 51.2 bits (121), Expect = 6e-06
Identities = 27/74 (36%), Positives = 39/74 (52%)
Query: 713 FDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 772
FDG+ G I LPI +G F V+D Y+ +LG PWIHD A+ S+ HQ +K
Sbjct: 398 FDGNTMMTNGTIKLPIYLGGAATWHKFVVVDKPTIYNIILGTPWIHDMQAIPSSYHQCIK 457
Query: 773 FIRNGKLVTVHGEE 786
+ + T+ G +
Sbjct: 458 IPTSIGIETIRGNQ 471
>At2g13330 F14O4.9
Length = 889
Score = 51.2 bits (121), Expect = 6e-06
Identities = 32/86 (37%), Positives = 41/86 (47%), Gaps = 6/86 (6%)
Query: 713 FDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 772
F G +G I LP L F V+D A ++ +LGRPW+H AV ST HQ +K
Sbjct: 115 FAGDTTFSIGTIQLPTIARGVRQLTNFLVVDKKAPFNAILGRPWLHVMKAVPSTYHQCIK 174
Query: 773 FIRNGKLVTVHGEEAYLVSQLSSFSC 798
F + V+G SQ SS C
Sbjct: 175 FPSYKGIAVVYG------SQRSSRKC 194
>At4g04050 putative transposon protein
Length = 681
Score = 48.1 bits (113), Expect = 5e-05
Identities = 27/75 (36%), Positives = 37/75 (49%)
Query: 713 FDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 772
F G LG I LP L +F V++ A ++ +LGRPW+H AV ST HQ +K
Sbjct: 589 FAGDTTFSLGTIQLPTIARGVRRLTSFLVVNKKAPFNAILGRPWLHAMKAVPSTYHQCIK 648
Query: 773 FIRNGKLVTVHGEEA 787
F + + V A
Sbjct: 649 FPSDKGIAVVSAANA 663
>At1g34080 putative protein
Length = 811
Score = 48.1 bits (113), Expect = 5e-05
Identities = 23/53 (43%), Positives = 33/53 (61%)
Query: 721 LGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKF 773
LG I LP+T ++ F V D A+Y+ +LG PW+++ AV ST HQ +KF
Sbjct: 535 LGTITLPVTAQGVVKMIEFTVFDRPAAYNIILGTPWLYEMKAVPSTYHQCVKF 587
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 45.8 bits (107), Expect = 3e-04
Identities = 22/53 (41%), Positives = 32/53 (59%)
Query: 721 LGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKF 773
LG + LP+T +V F V D A+Y+ +LG PW+++ V ST HQ +KF
Sbjct: 353 LGTVVLPVTAQGVVKMVEFTVFDRPAAYNVILGTPWLYEMKVVPSTYHQCVKF 405
>At4g20490 putative protein
Length = 853
Score = 43.9 bits (102), Expect = 0.001
Identities = 25/89 (28%), Positives = 39/89 (43%), Gaps = 14/89 (15%)
Query: 713 FDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTST------ 766
+DG K +GEI L + +G F V+D Y+ +LG PWI+ A+ ST
Sbjct: 531 YDGEAKMSIGEIKLQVQVGGITQKTKFVVIDSEPIYNAILGSPWIYSMKAIPSTNLHTIR 590
Query: 767 --------LHQKLKFIRNGKLVTVHGEEA 787
L + + + G L+ + G A
Sbjct: 591 QPEGLTDMLRDREEAAKEGNLIAIKGRRA 619
>At3g31530 hypothetical protein
Length = 831
Score = 41.2 bits (95), Expect = 0.007
Identities = 20/53 (37%), Positives = 28/53 (52%)
Query: 721 LGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKF 773
LG I LP+ +V F + D Y+ ++G PW++ AV ST H LKF
Sbjct: 3 LGTIKLPVRAKGVTKIVDFSITDQPTVYNAIIGTPWLNQFRAVASTYHLCLKF 55
>At1g37200 hypothetical protein
Length = 1564
Score = 40.4 bits (93), Expect = 0.011
Identities = 21/53 (39%), Positives = 29/53 (54%), Gaps = 6/53 (11%)
Query: 746 ASYSCLLGRPWIHDAGAVTSTLHQKLKFIRNGKLVTVHGEEAYLVSQLSSFSC 798
A ++ +LGRPW+H AV ST HQ +KF + ++G SQ SS C
Sbjct: 530 APFNAILGRPWLHAMKAVPSTYHQCIKFPSEKGIAVIYG------SQRSSRRC 576
>At4g08100 putative polyprotein
Length = 1054
Score = 33.5 bits (75), Expect = 1.4
Identities = 22/84 (26%), Positives = 40/84 (47%), Gaps = 7/84 (8%)
Query: 705 EVLSWVKAFDGSRKNVLGE------IDLPITIGPENFLVTFQVMDINASYSCLLGRPWIH 758
EVL + + +N GE + +P++IG + + ++ ++AS+ LLGRPW
Sbjct: 395 EVLKHPRPYSLQWRNETGEMSVKEQVKVPLSIGKYHDEIMCDILHMDASH-ILLGRPWQS 453
Query: 759 DAGAVTSTLHQKLKFIRNGKLVTV 782
D + + F NG+ T+
Sbjct: 454 DRKVLQDGFTNRQTFEHNGRKTTL 477
>At2g06890 putative retroelement integrase
Length = 1215
Score = 30.8 bits (68), Expect = 9.1
Identities = 21/77 (27%), Positives = 34/77 (43%), Gaps = 4/77 (5%)
Query: 707 LSWVKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTST 766
L W+ D + V ++ +PI IG + V+ + A + LLGRPW D +
Sbjct: 268 LKWLN--DSGKMRVKNQVVVPIVIGKYEDEILCDVLPMEAGH-ILLGRPWQSDRKVMHDG 324
Query: 767 LHQKLKF-IRNGKLVTV 782
+ F + GK + V
Sbjct: 325 FTNRHSFEFKGGKTILV 341
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.354 0.154 0.541
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,628,142
Number of Sequences: 26719
Number of extensions: 1437319
Number of successful extensions: 6373
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 6355
Number of HSP's gapped (non-prelim): 20
length of query: 2172
length of database: 11,318,596
effective HSP length: 115
effective length of query: 2057
effective length of database: 8,245,911
effective search space: 16961838927
effective search space used: 16961838927
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 68 (30.8 bits)
Medicago: description of AC147010.11


