

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147007.4 - phase: 0
(419 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
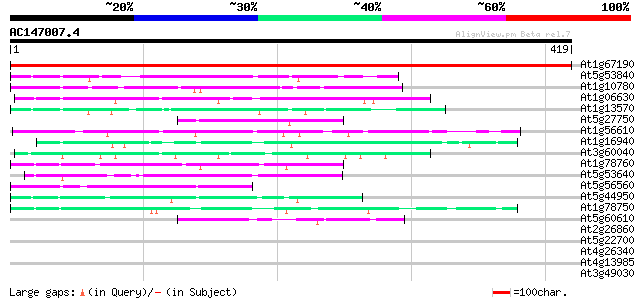
Score E
Sequences producing significant alignments: (bits) Value
At1g67190 unknown protein 540 e-154
At5g53840 putative protein 69 4e-12
At1g10780 grr1 gi|2407790 like 69 4e-12
At1g06630 unknown protein 54 2e-07
At1g13570 hypothetical protein 50 2e-06
At5g27750 unknown protein 46 3e-05
At1g56610 unknown protein 46 3e-05
At1g16940 hypothetical protein 46 4e-05
At3g60040 putative protein 45 6e-05
At1g78760 hypothetical protein 45 7e-05
At5g53640 heat shock transcription factor HSF30-like protein 44 1e-04
At5g56560 putative protein 43 3e-04
At5g44950 putative protein 43 3e-04
At1g78750 hypothetical protein 43 3e-04
At5g60610 unknown protein 42 8e-04
At2g26860 unknown protein 41 0.001
At5g22700 unknown protein 40 0.002
At4g26340 unknown protein 40 0.002
At4g13985 unknown protein 40 0.002
At3g49030 putative protein 40 0.002
>At1g67190 unknown protein
Length = 419
Score = 540 bits (1390), Expect = e-154
Identities = 263/419 (62%), Positives = 327/419 (77%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
M+ LPVEV+G ILS LG ARDV+IAS TC+KWREA + HLQTLSFN++DWP YR++ ++
Sbjct: 1 MDYLPVEVIGNILSRLGGARDVVIASATCRKWREACRKHLQTLSFNSADWPFYRDLTTNR 60
Query: 61 LEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRDSLRELRYNVRTSPNFNII 120
LE+LIT T+FQT GLQ L+I MDD ++FS VIAWLMYTRD+LR L YNVRT+PN NI+
Sbjct: 61 LEILITQTIFQTMGLQGLSIMMDDANKFSAATVIAWLMYTRDTLRRLSYNVRTTPNVNIL 120
Query: 121 EKCSRKTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSISALDLSLLLSACPKLETLS 180
E C R+ LE L LA N I+GVEP + +FPCLKSLSLS+VSISALDL+LLLSACP +E+L
Sbjct: 121 EICGRQKLEALVLAHNSITGVEPSFQRFPCLKSLSLSYVSISALDLNLLLSACPMIESLE 180
Query: 181 IVCPEIAMSDSEASIELSSSSLKDFFVESYSFDKLILVADMLENLHLKDCSFDAFELINK 240
+V EIAMSD++ +IELSS +LK + + S DK IL AD +E LH+KDC + FELI
Sbjct: 181 LVSLEIAMSDAQVTIELSSPTLKSVYFDGISLDKFILEADSIEFLHMKDCVLELFELIGN 240
Query: 241 GTLKVLKLDDVSVIHLDIGDNTENLEIVDVCNFIFMWQNFYNMISKASKLKKLRLWSVVF 300
GTLK KLDDVSVIHLDI + +E+LE+VDV +F +W FY MIS++ KLKKLRLW VVF
Sbjct: 241 GTLKHFKLDDVSVIHLDIMETSESLEVVDVNHFTMVWPKFYQMISRSQKLKKLRLWDVVF 300
Query: 301 DDEDEVVDIETISVCFPRLTHLSLSYDLKDGVLHYGLQGLSFLMNVVVLELGWTTISDLF 360
DD+DE++D+E+I+ F LTHLSLSYDLKDG HY LQG + L NV VLELGWT I+D+F
Sbjct: 301 DDDDEIIDVESIAAGFSHLTHLSLSYDLKDGAAHYSLQGTTQLENVTVLELGWTVINDVF 360
Query: 361 SVWVAGLLEGCPNLKKMVIYGYVAEIKTHEECQTFTKFTEFMIQLGRKYSHIKFEFEYE 419
S+WV LL CPNLKK++IYG V+E KT +CQ FT ++QL RKY H++ +FEYE
Sbjct: 361 SIWVEELLRRCPNLKKLIIYGVVSETKTQGDCQILATFTWSIVQLMRKYIHVEVQFEYE 419
>At5g53840 putative protein
Length = 444
Score = 69.3 bits (168), Expect = 4e-12
Identities = 78/306 (25%), Positives = 135/306 (43%), Gaps = 51/306 (16%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICS-- 58
+ LP ++ +ILSHL S +D + S+ +WR WQ + L F++ + + E S
Sbjct: 20 LSQLPDHLICVILSHL-STKDAVRTSILSTRWRNLWQL-VPVLDFDSRELRSFSEFVSFA 77
Query: 59 ---------SHLEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRDSLRELRY 109
S+++ L C ++ G LT ++D + TR ++ +
Sbjct: 78 GSFFYLHKDSYIQKLRVC-IYDLAGNYYLTSWID--------------LVTRHRIQHIDI 122
Query: 110 NVRTSPNFNIIEKCSRKTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSI-SALDLSL 168
+V T F +I ++ L + ++ V ++ PCLK L L FV+ + L
Sbjct: 123 SVFTCSGFGVIPLSLYTCDTLVHLKLSRVTMVNVEFVSLPCLKILDLDFVNFTNETTLDK 182
Query: 169 LLSACPKLETLSIVCPEIAMSDSEASIELSSSSLKDFFVESYSFDK---LILVADMLENL 225
++S P LE L+IV + D+ I++ S +LK + FD+ L++ +L+ L
Sbjct: 183 IISCSPVLEELTIV---KSSEDNVKIIQVRSQTLKRVEIHR-RFDRHNGLVIDTPLLQFL 238
Query: 226 HLKDCSFDAFELINKG-TLKVLKLDDVSVIHLDIGDNTENLEIVDVCNFIFMWQNFYNMI 284
+K S + E IN G T KV D+ V LD D + M ++F+ I
Sbjct: 239 SIKAHSIKSIEFINLGFTTKV----DIDVNLLDPNDLSNR----------SMTRDFFTTI 284
Query: 285 SKASKL 290
S+ L
Sbjct: 285 SRVRSL 290
Score = 29.3 bits (64), Expect = 4.2
Identities = 24/67 (35%), Positives = 34/67 (49%), Gaps = 3/67 (4%)
Query: 150 CLKSLSLSFVSISALDLSL-LLSACPKLETLSIVCPEIAMSDSEASIELSSSSLKDFFVE 208
C S + SIS L++ L LL +CPKLE+LS+ + + E +SS+ V
Sbjct: 313 CYLSELSAVCSISNLEMLLNLLKSCPKLESLSLKLVDYEKNKKEE--VMSSTVPPPCLVS 370
Query: 209 SYSFDKL 215
S F KL
Sbjct: 371 SLKFVKL 377
>At1g10780 grr1 gi|2407790 like
Length = 418
Score = 69.3 bits (168), Expect = 4e-12
Identities = 72/302 (23%), Positives = 142/302 (46%), Gaps = 22/302 (7%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
M+ LP ++ ILS+L SARDV + K+W+E+ + ++++ F+ + + E S
Sbjct: 1 MDSLPDAILQYILSYLTSARDVAACNCVSKRWKESTDS-VKSVVFHRNSFESIMETDDS- 58
Query: 61 LEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRDSLRELRYNVRTSPNFNII 120
+ ++ + ++ L+ L ++ F+ + + +W+M+ SLR L + + ++
Sbjct: 59 -DSIVRKMISSSRRLEELVVY----SPFTSSGLASWMMHVSSSLRLLELRMDNLASEEVV 113
Query: 121 EKCSRKTLEVLTLASN----PISGV----EPKYHKFPCLKSLSLSFVSISALDLSLLLSA 172
+ K L+ + +A N + GV PK+ FP L+SL + + LS L A
Sbjct: 114 VEGPLK-LDCIGVAKNLEILKLWGVLMMSPPKWDMFPNLRSLEIVGAKMDDSSLSHALRA 172
Query: 173 CPKLETLSIVCPEIAMSDSEASIELSSSSLKDFFVESYSFDKLILVADMLENLHLKDCSF 232
CP L L ++ E S S L L DF+ + + L+L + L +L ++ CS+
Sbjct: 173 CPNLSNLLLLACEGVKSISIDLPYLEHCKL-DFYGQGNTL--LVLTSQRLVSLDVQGCSW 229
Query: 233 DAFELINKGTLKVLKLDDVS-VIHLDIGDNTENLEIVDVCNFIFMWQNFYNMISKASKLK 291
+ LK L + V+ +++ +N +LE + + + W ++ +A +K
Sbjct: 230 --IRVPETKFLKNLSISSVTGRVYMVDFNNLSSLEALSIRGVQWCWDAICMILQQARDVK 287
Query: 292 KL 293
L
Sbjct: 288 HL 289
>At1g06630 unknown protein
Length = 403
Score = 53.5 bits (127), Expect = 2e-07
Identities = 87/344 (25%), Positives = 143/344 (41%), Gaps = 42/344 (12%)
Query: 4 LPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSHLEM 63
LP E++G ILS L + + + S+ KKWR ++ + TL F+ S + + S
Sbjct: 17 LPDEILGKILSLLAT-KQAVSTSVLSKKWRTLFKL-VDTLEFDDSVSGMGEQEASYVFPE 74
Query: 64 LITCTLFQTKGLQC----------LTIFMDDEHEFS-VTPVIAWLMYTRDSLRELRYNVR 112
+ +T LQC + DDE + V I+ ++ S LR N R
Sbjct: 75 SFKDLVDRTVALQCDYPIRKLSLKCHVGRDDEQRKACVGRWISNVVGRGVSEVVLRINDR 134
Query: 113 TSPNFNIIEKCSRKTLEVLTLASNPISGVEPKYHKFPCLKSL---SLSFVSISALDLSLL 169
+F + + KTL LTL + G P Y P LK L S+ F L ++L
Sbjct: 135 -GLHFLSPQLLTCKTLVKLTLGTRLFLGKLPSYVSLPSLKFLFIHSVFFDDFGELS-NVL 192
Query: 170 LSACPKLETLSIVCPEIAMSDSEASIELSSSSLKDFFVE-SYSFDKLILV-ADMLENLHL 227
L+ CP +E L + + +SS +LK V Y F+ +I LE L
Sbjct: 193 LAGCPVVEALYL-----NQNGESMPYTISSPTLKRLSVHYEYHFESVISFDLPNLEYLDY 247
Query: 228 KDCSFDAFELINKGTLKVLKLDDVSVIHLDIGDNTE------NLEIVDV----------- 270
D + + +N +L L+ H++ D T+ N+EI+ +
Sbjct: 248 SDYALYGYPQVNLESLVEAYLNLDKAEHVESPDVTKLIMGIRNVEILSLSPDSVGVIYSC 307
Query: 271 CNFIFMWQNFYNMISKASKLKKLRLWSVVFDDEDEVVDIETISV 314
C + + F N++S + KK R W ++ D + +ET+ +
Sbjct: 308 CKYGLLLPVFNNLVSLSFGTKKTRAWKLLADILKQSPKLETLII 351
>At1g13570 hypothetical protein
Length = 416
Score = 50.4 bits (119), Expect = 2e-06
Identities = 84/343 (24%), Positives = 138/343 (39%), Gaps = 48/343 (13%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREIC--S 58
+ DLP ++ IL+ L S RD + S+ KWR W T L L F+ + C
Sbjct: 8 ISDLPQSIIENILTRL-SIRDAIRTSVLSSKWRYKWST-LTDLVFDEKCVSPSNDRCVVE 65
Query: 59 SHLEMLITCTLFQTKG----LQCLTIFMDDEHEFSVTPVIAWLMY-TRDSLRELRYNVRT 113
++L IT L +G Q T F + WL++ +R+ ++EL +
Sbjct: 66 TNLVRFITGVLLLHQGPIHKFQLSTSFKQCRPDID-----QWLLFLSRNGIKELVLKLGE 120
Query: 114 SPNFNIIEKCSRKTLEVLTLASNPISGVEPKYHK-FPCLKSLSLSFVSISALDLSLLLSA 172
F + C L++ L P+Y K F LKSL+L + ++ + L+S
Sbjct: 121 G-EFRV-PACLFNCLKLTCLELCHCEFDPPQYFKGFSYLKSLNLHQILVAPEVIESLISG 178
Query: 173 CPKLETLSIVCPE---IAMSDSEASIELSSSSLKDFFVESYSFDKLILVA-------DML 222
CP LE LS+ + +++S KD F+E + KL+ ++ D+
Sbjct: 179 CPLLEFLSLSYFDSLVLSISAPNLMYLYLDGEFKDIFLE--NTPKLVAISVSMYMHEDVT 236
Query: 223 ENLHLKDCSFDAFELINKGTLKVLKLDDVSVIHLDIGDNTENLEIVDVCNFIFMWQNFYN 282
+ D + F L L+ L +L IGD+ L + +
Sbjct: 237 DFEQSSDYNLVKF-LGGVPLLEKLVGYIYFTKYLSIGDDPGRLPLTYI------------ 283
Query: 283 MISKASKLKKLRLWSVVFDDEDEVVDIETISVCFPRLTHLSLS 325
LK + L+ V F+D DEV+ + + P L L +S
Sbjct: 284 ------HLKTIELYQVCFEDADEVLVLLRLVTHSPNLKELKVS 320
>At5g27750 unknown protein
Length = 459
Score = 46.2 bits (108), Expect = 3e-05
Identities = 45/136 (33%), Positives = 69/136 (50%), Gaps = 14/136 (10%)
Query: 126 KTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSISA-LDLSLLLSACPKLETLSIVCP 184
KTL L L + ++ +P PCLK + L V S + L L+S CP LE L++V
Sbjct: 141 KTLVTLKLRDSALN--DPGLLSMPCLKFMILREVRWSGTMSLEKLVSGCPVLEDLTLVRT 198
Query: 185 EIA-MSDSEASI-ELSSSSLKDFFV--------ESYSFDKLILV-ADMLENLHLKDCSFD 233
I + D E + + S SLK F+V S DK++ + A LEN+ LK+ FD
Sbjct: 199 LICHIDDDELPVTRVRSRSLKTFYVPLAYGVGCRSRVPDKVLEIDAPGLENMTLKEDHFD 258
Query: 234 AFELINKGTLKVLKLD 249
+ N +L +++L+
Sbjct: 259 GIVVKNLTSLFMIELN 274
>At1g56610 unknown protein
Length = 535
Score = 46.2 bits (108), Expect = 3e-05
Identities = 96/398 (24%), Positives = 167/398 (41%), Gaps = 68/398 (17%)
Query: 3 DLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSHLE 62
DLP + ILS L + V ++ L K W + +L +L F+ SD + H
Sbjct: 77 DLPKCLAPHILSWLPTKTAVTVSLLFMKGWWRSEMKNLSSLKFSFSD-----DQEEEHFV 131
Query: 63 MLITCTLFQ--TKGLQCLTIFMDDEHEFS-VTPVIAW-LMYTRDSLRELRYNVRTSPNFN 118
+ L Q + L ++ ++DE + VT ++ + L + L+ Y+++ NF
Sbjct: 132 RFVDQVLRQRGNRKLDSFSLTLNDEIDGGFVTHLVDYPLDNGVEKLKLSIYDIK--GNFQ 189
Query: 119 IIEKC-SRKTLEVLTLASNP----ISGVEPKYHKFPCLKSLSLSFVSISALDLSL-LLSA 172
+ + S+ TL L LA+N I+G + PCLK+L L V ++ + + + LLS
Sbjct: 190 LSSRVFSQATLVTLKLATNRSLIWINGDDVAAAHLPCLKTLWLDDVLVADVKVFVRLLSR 249
Query: 173 CPKLETLSIVCPEIAMSDSEASIELSSSSLK-----DFFVESYSFDKL--ILVADMLENL 225
CP LE L ++ ++ + EA +S+S + +VE +D+ ++ D L
Sbjct: 250 CPILEELVMI--DMKWHNWEACFVVSASLRRLKIVWTDYVEMDEYDRCPQSVLFDTPNVL 307
Query: 226 HLKDCSFDAFELINKGTLKVLKLDDV--SVIHLDIGDNTENLEIVDVCNFIFMWQNFYNM 283
+L+ A G +LK + + I L++ D E D + + N
Sbjct: 308 YLEYTDHIA------GQYPLLKFSSLIEAKIRLEMIDEKEE---EDEGQEVIVGDNATAF 358
Query: 284 ISKASKLKKLRLWSVVFDDEDEVVDIETISVCFPRLTHLSLSYDLKDGVLHYGLQGLSFL 343
I+ + ++KL L++ D F LTHL++ D
Sbjct: 359 ITGITSVRKLYLYANTIQVLHHYFDPPIPE--FVYLTHLTIQSD---------------- 400
Query: 344 MNVVVLELGWTTISDLFSVWVAGLLEGCPNLKKMVIYG 381
ELGW I + LL CP+L+ +V+ G
Sbjct: 401 -----KELGWDAIPE--------LLSKCPHLETLVLEG 425
>At1g16940 hypothetical protein
Length = 434
Score = 45.8 bits (107), Expect = 4e-05
Identities = 83/378 (21%), Positives = 150/378 (38%), Gaps = 45/378 (11%)
Query: 21 DVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSHLEMLITCTLFQTKGL----- 75
+V+ SL C++WR WQ+ L L + Y + M + + G
Sbjct: 32 EVVKTSLICRRWRYVWQS-LPGLDLVINGSKNYDKFDFLERFMFLQRVKLRYVGYGHNCR 90
Query: 76 QCLTIFMDD--EHEFSVTPVIAWLMYTRDSLRELRYNVRTSPNFNIIEKCSRKTLEVLTL 133
++ M++ +H+ V + Y D + E+ + TS C R L
Sbjct: 91 NMTSMMMNNVIKHKIQHLDVGSNRRYVYDRV-EIPPTIYTS--------CERLVFLKLHR 141
Query: 134 ASNPISGVEPKYHKFPCLKSLSLSFVS-ISALDLSLLLSACPKLETLSIVCPEIAMSDSE 192
A+ P S P PCLK + L ++ + +LD+ L+S CP LETL++ D
Sbjct: 142 ANLPKS---PDSVSLPCLKIMDLQKINFVDSLDMEKLVSVCPALETLTM--------DKM 190
Query: 193 ASIELSSSSLKDFFVES----YSFDKLILVADMLENLHLKDCSFDAFELINKGTLKVLKL 248
++SS SL F + + Y ++++ L+ L L + + ++ +L L
Sbjct: 191 YGAKVSSQSLLSFCLTNNETGYLKTQVVMQTPKLKYLKLNRQFIQRIVINDLSSIVMLNL 250
Query: 249 DDVSVIHLDIGDNTENLEIVDVCNFIFMWQNFYNMISKASKLKKLRLWSVVFDDEDEVVD 308
DDV+ + + + V F Y SK+ L K SV+ + V
Sbjct: 251 DDVAYFGETLLSILKLISCVRDLTISFDILQDYRHFSKSKSLPKFHKLSVLSVKDMAVGS 310
Query: 309 IETISVCFPRLTHLSLSYDLKDGVLHYGLQGLSF-------LMNVVVLELGWTTISDLFS 361
E++ + +L L G Y G++F L ++ +E+ ++D+
Sbjct: 311 WESLLIFLESCQNLK---SLVMGFRDYN-WGINFSDVPQCVLSSLEFVEVKAREVADMKK 366
Query: 362 VWVAGLLEGCPNLKKMVI 379
+W + +E LKK +
Sbjct: 367 LW-SYFMENSTVLKKFTL 383
>At3g60040 putative protein
Length = 838
Score = 45.4 bits (106), Expect = 6e-05
Identities = 88/353 (24%), Positives = 144/353 (39%), Gaps = 49/353 (13%)
Query: 4 LPVEVVGIILSHLGSARDVLIASLTCKKWREAWQT--HLQTL-SFNTSDWPLYREICSSH 60
LP EV+G ILS L + + SL KKWR ++ HL+ SF+ H
Sbjct: 237 LPDEVLGKILS-LIPTKQAVSTSLLAKKWRTIFRLVDHLELDDSFSLQAVKDQTPGLRKH 295
Query: 61 LEMLIT----CTLFQTKGLQC--------LTIFMDDEHEFSVTPVIAWLM-YTRDSLREL 107
+ + T + +T LQC L + E V W+ + EL
Sbjct: 296 VRFVFTEDFKIFVDRTLALQCDYPIKNFSLKCHVSKYDERQKACVGRWISNVVGRGVFEL 355
Query: 108 RYNVRTSPNFNIIEK--CSRKTLEVLTLASNPISGVEPKYHKFPCLKSL---SLSFVSIS 162
++ P + + + KTL LTL + G P Y P LKSL ++ F I
Sbjct: 356 DLRMK-DPGIHFLPPHLVASKTLVKLTLGTQLCLGQLPSYVSLPSLKSLFIDTIVFYDIE 414
Query: 163 ALDLSLLLSACPKLETLSIVCPEIAMSDSEASIELSSSSLKDFFVESYSFDKLILVADM- 221
L +LL+ CP LE LS+ + + +SS +LK V+ + D + + M
Sbjct: 415 DL-CCVLLAGCPVLEELSVHHHDFIATPH----TISSPTLKRLSVDYHCPDDVDSASHMS 469
Query: 222 -----LENLHLKDCSFDAFELINKGTLKVLKLD---DVSVIHLDIGD------NTENLEI 267
L L C+ + IN +L KLD + V+ +D+ D N ++L +
Sbjct: 470 FDLPKLVYLEYSHCALGEYWQINLESLVEAKLDLGLERKVLRMDVTDLIIGIRNVQSLHL 529
Query: 268 -VDVCNFIFMWQN-----FYNMISKASKLKKLRLWSVVFDDEDEVVDIETISV 314
D + I+ + F N+++ + K R W ++ + + +ET+ V
Sbjct: 530 SPDSVHVIYSYCRHGLPVFKNLVNLSFGSKNKRGWRLLANLLKQSTKLETLIV 582
>At1g78760 hypothetical protein
Length = 452
Score = 45.1 bits (105), Expect = 7e-05
Identities = 66/260 (25%), Positives = 116/260 (44%), Gaps = 21/260 (8%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
+ +LP ++ I+ ++ + +DV+ +S+ K+WR W+ ++ L+ + + Y S
Sbjct: 18 LSNLPESLICQIMLNIPT-KDVVKSSVLSKRWRNLWR-YVPGLNVEYNQFLDYNAFVSFV 75
Query: 61 LEMLITCTLFQTKGLQCLTIFMD-DEHEFSVTPVIAWLMYTRD-SLRELRYNVRTSPNFN 118
L L + Q + D DE E +++ V W+ D L+ L T N
Sbjct: 76 DRFL---ALDRESCFQSFRLRYDCDEEERTISNVKRWINIVVDQKLKVLDVLDYTWGNEE 132
Query: 119 IIEKCSRKTLEVLTLASNPISGV---EPKYHKFPCLKSLSLSFVSIS-ALDLSLLLSACP 174
+ S T E +L S + V PK P +K + L V S AL L ++S+C
Sbjct: 133 VQIPPSVYTCE--SLVSLKLCNVILPNPKVISLPLVKVIELDIVKFSNALVLEKIISSCS 190
Query: 175 KLETLSIVCPEIAMSDSEASIELSSSSLKDF-----FVESYSFDKLILVADMLENLHLKD 229
LE+L I + D + +SS SL F + + ++ + A LE L++ D
Sbjct: 191 ALESLIISRSSV---DDINVLRVSSRSLLSFKHIGNCSDGWDELEVAIDAPKLEYLNISD 247
Query: 230 CSFDAFELINKGTLKVLKLD 249
S F++ N G+L K++
Sbjct: 248 HSTAKFKMKNSGSLVEAKIN 267
>At5g53640 heat shock transcription factor HSF30-like protein
Length = 454
Score = 44.3 bits (103), Expect = 1e-04
Identities = 57/242 (23%), Positives = 111/242 (45%), Gaps = 19/242 (7%)
Query: 12 ILSHLGSARDVLIASLTCKKWREAWQT--HLQTLSFNTSDWPLYREICSSHLEMLITCTL 69
IL++L + +DV+ S+ +WR W L+ S + SD+ + C + + +
Sbjct: 35 ILNYLPT-KDVVKTSVLSTRWRSLWLLVPSLELDSRDFSDFNTFVSFCDRYFDSNRVLCI 93
Query: 70 FQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRDSLRELRY-NVRTSPNFNIIEKCSRKTL 128
+ K LTI ++E F + +W+ + R++++ +V+ P F+ I K
Sbjct: 94 NKLK----LTISENEEDGFYLK---SWI--DAAAKRKIQHLDVQFLPQFHKIHFNLYKCE 144
Query: 129 EVLTLASNPISGVEPKYHKFPCLKSLSL-SFVSISALDLSLLLSACPKLETLSIVCPEIA 187
+++L +S + + FPC+K++ L V + L+S CP LE L++ I
Sbjct: 145 ALVSLRLFEVSLDKGRIFSFPCMKTMHLEDNVYPNEATFKELISCCPVLEDLTV----II 200
Query: 188 MSDSEASIELSSSSLK-DFFVESYSFDKLILVADMLENLHLKDCSFDAFELINKGTLKVL 246
++ S SLK F+ + +++ A +L +L + D +F + N G+ L
Sbjct: 201 YGMDRKVFQVHSQSLKSSSFLHEVALSGVVIDAPLLCSLRINDNVSKSFIVNNLGSNDKL 260
Query: 247 KL 248
L
Sbjct: 261 DL 262
>At5g56560 putative protein
Length = 477
Score = 43.1 bits (100), Expect = 3e-04
Identities = 50/184 (27%), Positives = 83/184 (44%), Gaps = 9/184 (4%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
+ DLP E++ ILS L + L L ++W+ W+ + ++F+ D P S
Sbjct: 7 LSDLPDELLLKILSALPMFKVTLATRLISRRWKGPWKL-VPDVTFDDDDIPFK----SFE 61
Query: 61 LEMLITCTLFQTKGLQCL-TIFMDDEHEFSVTPVIAWL-MYTRDSLRELRYNVRTSPNFN 118
M F + Q L + + ++S + + W+ + S+RELR ++
Sbjct: 62 TFMSFVYGSFLSNDAQILDRLHLKLNQKYSASDINFWVQVAVNRSVRELRIDLFGKTLEL 121
Query: 119 IIEKCSRKTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSISALD-LSLLLSACPKLE 177
CS TL+ LTL I V P + + P LK+L L V S+ ++ +L CP LE
Sbjct: 122 PCCLCSCITLKELTLHDLCIK-VVPAWFRLPSLKTLHLLSVKFSSDGFVASILRICPVLE 180
Query: 178 TLSI 181
L +
Sbjct: 181 RLVV 184
>At5g44950 putative protein
Length = 438
Score = 43.1 bits (100), Expect = 3e-04
Identities = 62/277 (22%), Positives = 110/277 (39%), Gaps = 27/277 (9%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
+ +LP ++ IL +L + + S+ +WR+ W + L N D+P + S
Sbjct: 6 ISELPDGLLNHILMYL-HIEESIRTSVLSSRWRKLW-LKVPGLDVNVHDFPADGNLFESL 63
Query: 61 LEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRD-SLRELRYNVRTSPNFNI 119
++ + + LQ + + + + + W+ D ++ L P + I
Sbjct: 64 MDKFLEVNRGR---LQNFKLNYESNLYYLMDRFVPWIATVVDRGIQHLDVTATDCPPWTI 120
Query: 120 ----IEKCSRKTLEVLTLASNPISGVEPKYH-KFPCLKSLSLSFVSISALDLSLLLSACP 174
C KTL L L + + PK+ PCLK + L + S L L+S CP
Sbjct: 121 DFMPANICKSKTLVSLKLVNVGLD--TPKFVVSLPCLKIMHLEDIFYSPLIAENLISGCP 178
Query: 175 KLETLSIVCPEIAMSDSEASIELSSSSLKDFFVESYSFD--------KLILVADMLENLH 226
LE L+IV D + + S +LK+F +FD + + A L+ +
Sbjct: 179 VLEDLTIVRNH---EDFLNFLRVMSQTLKNF---RLTFDWGMGSNDFSVEIDAPGLKYMS 232
Query: 227 LKDCSFDAFELINKGTLKVLKLDDVSVIHLDIGDNTE 263
+D D + N +L + LD + +G E
Sbjct: 233 FRDSQSDRIVVKNLSSLVKIDLDTEFNLKFGLGSPLE 269
>At1g78750 hypothetical protein
Length = 458
Score = 43.1 bits (100), Expect = 3e-04
Identities = 93/401 (23%), Positives = 156/401 (38%), Gaps = 74/401 (18%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
+ +LP ++ +L +L + +DV+ +S+ +WR W+ ++ + + D+ + H
Sbjct: 20 ISNLPETLLCQVLFYLPT-KDVVKSSVLSSRWRNLWK-YVPGFNLSYCDFHVRNTFSYDH 77
Query: 61 LEMLITCTLFQTKGLQ-CLTIF-MDDEHEFSVTPVIAWLMYTRDSL--RELR-------- 108
L F Q CL F ++ + P +A + +S+ R+++
Sbjct: 78 NTFLRFVDSFMGFNSQSCLQSFRLEYDSSGYGEPKLALIRRWINSVVSRKVKYLGVLDDS 137
Query: 109 ---YNVRTSPNFNIIEKCSRKTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSIS-AL 164
Y P E TL+ L+LAS PK+ P LK L LS V + +
Sbjct: 138 CDNYEFEMPPTLYTCETLVYLTLDGLSLAS-------PKFVSLPSLKELHLSIVKFADHM 190
Query: 165 DLSLLLSACPKLETLSIVCPEIAMSDSEASIELSSSSLKDF---FVESYSFDKLILVADM 221
L L+S CP LE L+I + S DF V S S VAD
Sbjct: 191 ALETLISQCPVLENLNI----------------NRSFCDDFEFTCVRSQSLLSFTHVADT 234
Query: 222 LENLHLKDCSFDAFELINKGTLKVLKLDDVSVIHLDIGDNTENLE--IVDVCNFIFMWQN 279
E L + D I+ LK L+L D + + D +E I V N I
Sbjct: 235 DEML-----NEDLVVAIDAPKLKYLRLSDHRIASFILNDLASLVEADIDTVFNLICKKMF 289
Query: 280 FYNMISKASKLKKLRLWSVVFDDEDEVVDIETISVCFPRLTHLSLSYDLKDGVLHYGLQG 339
N ++K + ++ D +V I +I + L + YD + +
Sbjct: 290 NPNDLNKRNMIR------------DFLVGISSIKTLIIASSTLEVIYD------YSRCEP 331
Query: 340 LSFLMNVVVLELGWTTIS-DLFSVWVAGLLEGCPNLKKMVI 379
L N+ L + + ++ ++ LE CPNLK +V+
Sbjct: 332 LPLFRNLSSLRVDFYGYKWEMLPIF----LESCPNLKSLVV 368
>At5g60610 unknown protein
Length = 388
Score = 41.6 bits (96), Expect = 8e-04
Identities = 49/174 (28%), Positives = 77/174 (44%), Gaps = 31/174 (17%)
Query: 126 KTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSISALD-LSLLLSACPKLETLSIVCP 184
K+L L L I P+ P LK+L L V+ S D L LLLS CP LE L
Sbjct: 123 KSLVTLKLLGLGIRVDVPRDVCLPSLKTLLLQCVAYSEEDPLRLLLSCCPVLEDL----- 177
Query: 185 EIAMSDSEASIELSSSSLKDFFVESYSFDKLILVADMLENLHLK---DCSFDAFELINKG 241
I + D+ +++ L+++ L+ L LK CS + + LI
Sbjct: 178 VIELDDANQNVK-----------------ALVVIVPTLQCLSLKIPASCSDERY-LIVTP 219
Query: 242 TLKVLKLDDVSVIHLDIGDNTENLEIVDVCNFIFMWQNFYNMISKASKLKKLRL 295
+LK K++D I + +N LE D I++ Q+ ++ + +K+L L
Sbjct: 220 SLKYFKVEDDREIFNALIENMPELEEAD----IYVTQHIETLLESVTSVKRLTL 269
>At2g26860 unknown protein
Length = 405
Score = 40.8 bits (94), Expect = 0.001
Identities = 83/334 (24%), Positives = 144/334 (42%), Gaps = 49/334 (14%)
Query: 4 LPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSF----NTSDWPLYREICSS 59
LP E++ IL H ++V+ S+ K+W W + L+F SD P+ ++ +
Sbjct: 7 LPDELLVEIL-HCLPTKEVVSTSILSKRWEFLW-LWVPKLTFVMNHYESDLPI-QDFITK 63
Query: 60 HLEMLITCTLFQTKGLQCLTIFM--DDEHEFSVTPV---IAWLMYTRDSLRELRYNVRTS 114
+L +L + ++ LQC + +D + VT + + L+ L L V
Sbjct: 64 NLRLLKP-QVIESFHLQCFSSSFKPEDIKHWVVTTISRRVRELIINYCDLSWLDKPVVLL 122
Query: 115 PNFNIIEKCSRKTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSISALD-LSLLLSAC 173
N + C+ +L L L + I P+ PCLK+L L V+ S + L LLLS C
Sbjct: 123 DLPNSLYTCT--SLVTLKLIGHSIIVDVPRTVSLPCLKTLELDSVAYSNEESLRLLLSYC 180
Query: 174 PKLETLSIVCPEIAMSDSEASIELSSSSLKDFFVE---SYSFDKLILVADMLENLHLKDC 230
P LE L+I M D+ ++ + SL + +YS D ++V L+ L +
Sbjct: 181 PVLEDLTI---HRDMHDNVKTLVIIVPSLLRLNLPIDGAYSCDGYVIVTPALKYLKVPGL 237
Query: 231 SFDAFELINKGTLKVLKLDDVSVIHLDIGDNTENLEIVDVCNFIFMWQNFYNMISKASKL 290
+ F + + H+ N+E D + + Q+ + + +
Sbjct: 238 YREDFSYL--------------LTHM------PNVEEAD----LSVEQDVERLFESITSV 273
Query: 291 KKLRLWSVVFDDEDEVVDIETISVCFPRLTHLSL 324
K+L L+ V+ D EDE + + F +L HL+L
Sbjct: 274 KRLSLF-VLLDIEDE--SMYHNGIYFNQLEHLNL 304
>At5g22700 unknown protein
Length = 452
Score = 40.4 bits (93), Expect = 0.002
Identities = 60/253 (23%), Positives = 111/253 (43%), Gaps = 18/253 (7%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAW-QTHLQTLSFNT-SDWPLYREICS 58
+ LP E++ ILS+L + ++ + S+ +WR W T + + + D + S
Sbjct: 23 ISSLPDELLCQILSNLPT-KNAVTTSILSTRWRSIWLSTPVLDIDIDAFDDATTFVSFAS 81
Query: 59 SHLEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRDSLRELRYNVRTSPNFN 118
LE L + K L++ DD ++ P I + R ++ L + R S +F
Sbjct: 82 RFLEFSKDSCLHKFK----LSVERDDVDMCTIMPWIQDAVNRR--IQHLEVDCRFSFHFE 135
Query: 119 IIEKCSRKTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSISALD-LSLLLSACPKLE 177
+ + +++L + ++ ++ P LK + L L+ L +S+CP LE
Sbjct: 136 AVYLTLYLSETLVSLRLHFVTLHRYEFVSLPNLKVMHLEENIYYCLETLENFISSCPVLE 195
Query: 178 TLSIV------CPEI--AMSDSEASIELSSSSLKDFFVESYSFDKLILVADMLENLHLKD 229
L++V +I S S S++L S +F++ K+I+ A L L LKD
Sbjct: 196 DLTVVRIVDIITEKILRVRSRSLNSLKLVLDSSNGWFIDDIDEWKVIIDAPRLAYLSLKD 255
Query: 230 CSFDAFELINKGT 242
+F + N G+
Sbjct: 256 DQSASFVISNLGS 268
>At4g26340 unknown protein
Length = 419
Score = 40.4 bits (93), Expect = 0.002
Identities = 61/244 (25%), Positives = 103/244 (42%), Gaps = 37/244 (15%)
Query: 2 EDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTL---SFNTSDWP-----LY 53
+DL ++++ I +DV+ SL K+W+ W + S++T D+ +Y
Sbjct: 9 DDLLLQILSFI-----PGKDVVATSLLSKRWQSLWMLVSELEYDDSYHTGDYKSFSQFVY 63
Query: 54 REICSSHLEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRDSLRELRYNVRT 113
R + S++ ++ L G C I + F++T LR+L+ N+RT
Sbjct: 64 RSLLSNNAPVI--KHLHLNLGPDCPAIDIGLWIGFALT----------RRLRQLKINIRT 111
Query: 114 SPN---FNIIEKC-SRKTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSI-SALDLSL 168
S N F++ + TLE L L + + V P P LK L L V L
Sbjct: 112 SSNDASFSLPSSLYTSDTLETLRLINFVLLDV-PSSVCLPSLKVLHLKTVDYEDDASLPS 170
Query: 169 LLSACPKLETLSIVCPEIAMSDSEASIELSSSSLKDFFVESYSF---DKLILVADMLENL 225
LL CP LE L + E D E + SL+ + ++ D+ ++ L+ L
Sbjct: 171 LLFGCPNLEELFV---ERHDQDLEMDVTFVVPSLRRLSMIDKNYGQCDRYVIDVPSLKYL 227
Query: 226 HLKD 229
++ D
Sbjct: 228 NITD 231
>At4g13985 unknown protein
Length = 459
Score = 40.4 bits (93), Expect = 0.002
Identities = 35/111 (31%), Positives = 54/111 (48%), Gaps = 5/111 (4%)
Query: 144 KYHKFPCLKSLSLSFVSISA-LDLSLLLSACPKLETLSIVCPEIAMSDSEASIELSSSSL 202
++ P LK + L V +L+S CP LE+LS+ +I ++D S+++SS SL
Sbjct: 171 EFVSLPSLKVIVLDAVVFDEDFAFEMLVSGCPVLESLSV--NKINLNDISESVQVSSQSL 228
Query: 203 KDFFVESYSFDKLILVAD--MLENLHLKDCSFDAFELINKGTLKVLKLDDV 251
F + D L +V D L L L D +F + N G+L +D V
Sbjct: 229 LSFSYVADDDDYLEVVIDAPRLHYLKLNDKRTASFIMKNHGSLLKADIDFV 279
>At3g49030 putative protein
Length = 443
Score = 40.4 bits (93), Expect = 0.002
Identities = 80/335 (23%), Positives = 137/335 (40%), Gaps = 54/335 (16%)
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWP--------- 51
+ +LP +++ ILS + + +V+ S+ K+WR W+ + L+F+ + P
Sbjct: 23 ISELPEDLLLQILSDIPT-ENVIATSVLSKRWRSLWKM-VPNLTFDFTFDPKYHQTFSEN 80
Query: 52 LYREICSSHLEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRDSLRELRYNV 111
LYR + S +L + L T+G+ L I M + V V ++ + +R+ R
Sbjct: 81 LYRSLTSHEASVLESLQLNFTRGIDGLNIGMWIATAY-VRHVRKLVLVSFGDVRDKRARF 139
Query: 112 RTSP-NFNIIEKCSRKTLEVLTLASNPISGVEPKYHKFPCLKSL-SLSFVSISALD---L 166
R++ NFN TL++L + + + CLKSL L + D +
Sbjct: 140 RSALFNFN-------DTLDILEIQDYILLDLPSPV----CLKSLRELRLYEVHFKDEASV 188
Query: 167 SLLLSACPKLETLS-------------IVCP--------EIAMSDSEASIELSSSSLKDF 205
LL CP LE LS IV P + + + +++ SLK
Sbjct: 189 CNLLCGCPSLEVLSVHRERNVDVETFTIVVPSLQRLTIYDFCIGGGKGGYVINAPSLKYL 248
Query: 206 FVESYSFDKLILV--ADMLENLHLKDCSFDAFELINKG--TLKVLKLDDVSVIHLDIGDN 261
+ + L+ A L + D S A E I + ++K L L+ I G
Sbjct: 249 NIVGFEGLDFCLIENAPELVEAEISDVSHIANENILESLTSVKRLSLESPIKIKFPTGKV 308
Query: 262 TENLEIVDVCNFIFMWQNFYN-MISKASKLKKLRL 295
+ L +DV W N + M+ + KL+ L+L
Sbjct: 309 FDQLVYLDVLTKEREWWNLLSRMLESSPKLQILKL 343
Score = 31.2 bits (69), Expect = 1.1
Identities = 31/122 (25%), Positives = 49/122 (39%), Gaps = 34/122 (27%)
Query: 290 LKKLRLWSVVFDDEDEV--------------------VDIETISVCFPRLTHLSL----- 324
L++LRL+ V F DE V VD+ET ++ P L L++
Sbjct: 172 LRELRLYEVHFKDEASVCNLLCGCPSLEVLSVHRERNVDVETFTIVVPSLQRLTIYDFCI 231
Query: 325 -----SYDLKDGVLHY----GLQGLSFLMNVVVLELGWTTISDLFSVWVAGLLEGCPNLK 375
Y + L Y G +GL F + EL ISD+ + +LE ++K
Sbjct: 232 GGGKGGYVINAPSLKYLNIVGFEGLDFCLIENAPELVEAEISDVSHIANENILESLTSVK 291
Query: 376 KM 377
++
Sbjct: 292 RL 293
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,383,961
Number of Sequences: 26719
Number of extensions: 394317
Number of successful extensions: 1376
Number of sequences better than 10.0: 105
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 84
Number of HSP's that attempted gapping in prelim test: 1287
Number of HSP's gapped (non-prelim): 177
length of query: 419
length of database: 11,318,596
effective HSP length: 102
effective length of query: 317
effective length of database: 8,593,258
effective search space: 2724062786
effective search space used: 2724062786
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC147007.4


