

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147002.5 + phase: 0
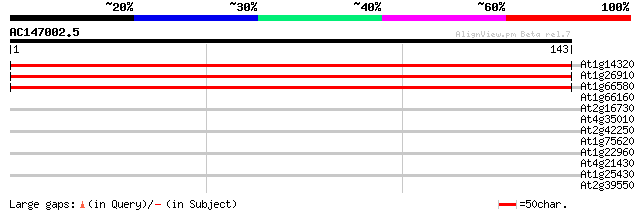
(143 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g14320 putative 60S Ribosomal Protein L10 251 7e-68
At1g26910 putative 60s ribosomal protein L10 250 2e-67
At1g66580 unknown protein 243 3e-65
At1g66160 unknown protein 28 1.3
At2g16730 putative beta-galactosidase 28 2.2
At4g35010 beta-galactosidase - like protein 27 2.9
At2g42250 putative cytochrome P450 27 3.7
At1g75620 unknown protein 27 3.7
At1g22960 putative salt-inducible protein 27 3.7
At4g21430 unknown protein 27 4.9
At1g25430 hypothetical protein 26 6.4
At2g39550 putative geranylgeranyl transferase type I beta subunit 26 8.3
>At1g14320 putative 60S Ribosomal Protein L10
Length = 220
Score = 251 bits (642), Expect = 7e-68
Identities = 120/143 (83%), Positives = 128/143 (88%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M K AGKD FHLR+RVHPFHVLRINKMLSCAGADRLQTGMRGAFGK LGTCARV+IGQVL
Sbjct: 76 MVKSAGKDAFHLRIRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKALGTCARVAIGQVL 135
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK +G HAQEALRRAKFKFPGRQKIIVSRKWGFTK + A++ KL+ E R+VPDGV
Sbjct: 136 LSVRCKDAHGHHAQEALRRAKFKFPGRQKIIVSRKWGFTKFNRADFTKLRQEKRVVPDGV 195
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
NAK L CHGPLANRQPG AFLPA
Sbjct: 196 NAKFLSCHGPLANRQPGSAFLPA 218
>At1g26910 putative 60s ribosomal protein L10
Length = 221
Score = 250 bits (639), Expect = 2e-67
Identities = 121/143 (84%), Positives = 127/143 (88%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M K AGKD FHLR+RVHPFHVLRINKMLSCAGADRLQTGMRGAFGK LGTCARV+IGQVL
Sbjct: 76 MVKSAGKDAFHLRIRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKALGTCARVAIGQVL 135
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK +G HAQEALRRAKFKFPGRQKIIVSRKWGFTK + A+Y KL+ E RIVPDGV
Sbjct: 136 LSVRCKDAHGHHAQEALRRAKFKFPGRQKIIVSRKWGFTKFNRADYTKLRQEKRIVPDGV 195
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
NAK L CHGPLANRQPG AFL A
Sbjct: 196 NAKFLSCHGPLANRQPGSAFLSA 218
>At1g66580 unknown protein
Length = 221
Score = 243 bits (620), Expect = 3e-65
Identities = 119/143 (83%), Positives = 126/143 (87%)
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
M K AGKD FHLR+RVHPFHVLRINKMLSCAGADRLQTGMRGAFGK LGTCARV+IGQVL
Sbjct: 76 MVKSAGKDAFHLRIRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKALGTCARVAIGQVL 135
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK +G HAQEALRRAKFKFPGRQKIIVSRKWGFTK + AEY KL++ RIVPDGV
Sbjct: 136 LSVRCKDNHGVHAQEALRRAKFKFPGRQKIIVSRKWGFTKFNRAEYTKLRAMKRIVPDGV 195
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
NAK L HGPLANRQPG AF+ A
Sbjct: 196 NAKFLSNHGPLANRQPGSAFISA 218
>At1g66160 unknown protein
Length = 431
Score = 28.5 bits (62), Expect = 1.3
Identities = 19/51 (37%), Positives = 27/51 (52%), Gaps = 8/51 (15%)
Query: 65 CKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGF---TKLSHAEYLKLKSE 112
CKSG+G +EALR FK K++V + G TK E LK+ ++
Sbjct: 361 CKSGDGSEVEEALRLGAFK-----KLVVMLQVGCGEGTKEKVTELLKMMNK 406
>At2g16730 putative beta-galactosidase
Length = 832
Score = 27.7 bits (60), Expect = 2.2
Identities = 10/30 (33%), Positives = 16/30 (53%)
Query: 28 LSCAGADRLQTGMRGAFGKPLGTCARVSIG 57
L C+G ++ +FG P GTC ++G
Sbjct: 746 LKCSGTKKISAVEFASFGNPNGTCGNFTLG 775
>At4g35010 beta-galactosidase - like protein
Length = 831
Score = 27.3 bits (59), Expect = 2.9
Identities = 9/30 (30%), Positives = 16/30 (53%)
Query: 28 LSCAGADRLQTGMRGAFGKPLGTCARVSIG 57
L C+G ++ +FG P+G C ++G
Sbjct: 745 LKCSGTKKIAAVEFASFGNPIGVCGNFTLG 774
>At2g42250 putative cytochrome P450
Length = 514
Score = 26.9 bits (58), Expect = 3.7
Identities = 17/52 (32%), Positives = 24/52 (45%), Gaps = 2/52 (3%)
Query: 21 VLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL--LSVRCKSGNG 70
+ R+ C+G D +R K L ++S+G VL L V SGNG
Sbjct: 192 ICRMAMSTRCSGTDNEAEEIRELVKKSLELAGKISVGDVLGPLKVMDFSGNG 243
>At1g75620 unknown protein
Length = 547
Score = 26.9 bits (58), Expect = 3.7
Identities = 14/36 (38%), Positives = 19/36 (51%), Gaps = 2/36 (5%)
Query: 26 KMLSCAGADR--LQTGMRGAFGKPLGTCARVSIGQV 59
++L C GA + R F K L TCAR++I V
Sbjct: 280 EVLVCGGAPKGSYNLSWRNTFVKALDTCARININDV 315
>At1g22960 putative salt-inducible protein
Length = 718
Score = 26.9 bits (58), Expect = 3.7
Identities = 34/129 (26%), Positives = 52/129 (39%), Gaps = 18/129 (13%)
Query: 12 LRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFG-KPLGTCARVSIGQVLLSVRCKSGNG 70
LR R++P + + A A RL+ + + K G V LL CK+GN
Sbjct: 577 LRKRLYPSVITYFVLIYGHAKAGRLEQAFQYSTEMKKRGVRPNVMTHNALLYGMCKAGNI 636
Query: 71 DHA--------QEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSE---NRIVPDG 119
D A +E + K+ + +++S+ F K E +KL E I PDG
Sbjct: 637 DEAYRYLCKMEEEGIPPNKYSY----TMLISKNCDFEKWE--EVVKLYKEMLDKEIEPDG 690
Query: 120 VNAKVLGCH 128
+ L H
Sbjct: 691 YTHRALFKH 699
>At4g21430 unknown protein
Length = 927
Score = 26.6 bits (57), Expect = 4.9
Identities = 11/34 (32%), Positives = 23/34 (67%)
Query: 72 HAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAE 105
H+Q++L+R+K K K++ SR+ G +++ +E
Sbjct: 38 HSQQSLKRSKQKVAESSKLVRSRRGGGDEVASSE 71
>At1g25430 hypothetical protein
Length = 1213
Score = 26.2 bits (56), Expect = 6.4
Identities = 23/87 (26%), Positives = 43/87 (48%), Gaps = 12/87 (13%)
Query: 22 LRINK---MLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVLLSVRCKSGNGDHAQEALR 78
L++NK L AG ++L++ A+G P+GT +G L++ + + + E +
Sbjct: 728 LKVNKDKSHLYLAGLNQLESNANAAYGFPIGTLPIRYLGLPLMNRKLRIAEYEPLLEKI- 786
Query: 79 RAKFK--------FPGRQKIIVSRKWG 97
A+F+ F GR ++I S +G
Sbjct: 787 TARFRSWVNKCLSFAGRIQLISSVIFG 813
>At2g39550 putative geranylgeranyl transferase type I beta subunit
Length = 375
Score = 25.8 bits (55), Expect = 8.3
Identities = 23/79 (29%), Positives = 30/79 (37%), Gaps = 17/79 (21%)
Query: 39 GMRGAFGKPLGTCARVSIGQVLLSVRCKSGNGDHAQEALRRAKF------------KFPG 86
G +G KP TC IG VL K GD + + KF KFPG
Sbjct: 281 GFQGRTNKPSDTCYAFWIGAVL-----KLIGGDALIDKMALRKFLMSCQSKYGGFSKFPG 335
Query: 87 RQKIIVSRKWGFTKLSHAE 105
+ + +G+T S E
Sbjct: 336 QLPDLYHSYYGYTAFSLLE 354
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.140 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,080,663
Number of Sequences: 26719
Number of extensions: 113489
Number of successful extensions: 303
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 296
Number of HSP's gapped (non-prelim): 12
length of query: 143
length of database: 11,318,596
effective HSP length: 89
effective length of query: 54
effective length of database: 8,940,605
effective search space: 482792670
effective search space used: 482792670
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC147002.5


