

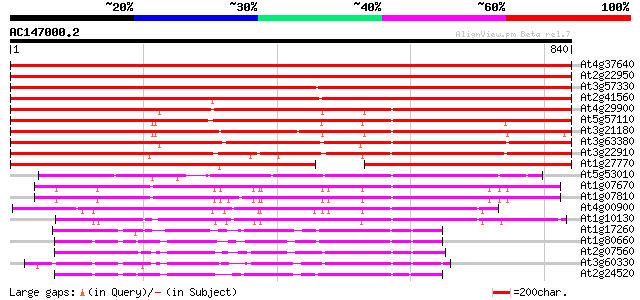
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147000.2 - phase: 0
(840 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g37640 plasma membrane-type calcium ATPase (ACA2) 1376 0.0
At2g22950 pseudogene 1367 0.0
At3g57330 Ca2+-transporting ATPase-like protein 1104 0.0
At2g41560 putative Ca2+-ATPase 1083 0.0
At4g29900 Ca2+-transporting ATPase - like protein 860 0.0
At5g57110 Ca2+-transporting ATPase-like protein (emb|CAB79748.1) 824 0.0
At3g21180 putative Ca2+-transporting ATPase 790 0.0
At3g63380 Ca2+-transporting ATPase -like protein 757 0.0
At3g22910 calmodulin-stimulated calcium-ATPase, putative 744 0.0
At1g27770 envelope Ca2+-ATPase 694 0.0
At5g53010 Ca2+-transporting ATPase-like protein 396 e-110
At1g07670 endoplasmic reticulum-type calcium-transporting ATPase... 350 2e-96
At1g07810 ER-type Ca2+-pump protein 348 7e-96
At4g00900 Ca2+-transporting ATPase - like protein 343 3e-94
At1g10130 putative calcium ATPase 310 2e-84
At1g17260 H+-transporting ATPase AHA10 189 4e-48
At1g80660 H+-transporting ATPase 9 (aha9) 185 8e-47
At2g07560 pseudogene 183 3e-46
At3g60330 plasma membrane H+-ATPase - like 179 4e-45
At2g24520 putative plasma membrane proton ATPase 174 2e-43
>At4g37640 plasma membrane-type calcium ATPase (ACA2)
Length = 1014
Score = 1376 bits (3561), Expect = 0.0
Identities = 689/843 (81%), Positives = 766/843 (90%), Gaps = 3/843 (0%)
Query: 1 MTLMILGVCAFVSLIVGVLTEGWPKGAHDGLGIVASILLVVFVTATSDYRQSLQFKDLDK 60
MTLMILGVCAFVSLIVG+ TEGWPKG+HDGLGI ASILLVVFVTATSDYRQSLQF+DLDK
Sbjct: 172 MTLMILGVCAFVSLIVGIATEGWPKGSHDGLGIAASILLVVFVTATSDYRQSLQFRDLDK 231
Query: 61 EKKKISIQVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGES 120
EKKKI++QVTRNG+RQK+SIY+LLPGDIVHL IGDQVPADGLF+SGFSV+IDESSLTGES
Sbjct: 232 EKKKITVQVTRNGFRQKLSIYDLLPGDIVHLAIGDQVPADGLFLSGFSVVIDESSLTGES 291
Query: 121 EPIMVTTQNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGV 180
EP+MV QNPFL+SGTKVQDGSC M++TTVGMRTQWGKLMATL+EGGDDETPLQVKLNGV
Sbjct: 292 EPVMVNAQNPFLMSGTKVQDGSCKMMITTVGMRTQWGKLMATLTEGGDDETPLQVKLNGV 351
Query: 181 ATIIGKIGLVFAVITFTVLVKGHLSHKIREGNFWRWTGDNAMEMLEYFAIAVTIVVVAVP 240
ATIIGKIGL FAV+TF VLV+G K+ G W W+GD A+E+LEYFAIAVTIVVVAVP
Sbjct: 352 ATIIGKIGLFFAVVTFAVLVQGMFMRKLSTGTHWVWSGDEALELLEYFAIAVTIVVVAVP 411
Query: 241 EGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVKTCI 300
EGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTN MTVVK+CI
Sbjct: 412 EGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNHMTVVKSCI 471
Query: 301 CMNSKEVSNSSSS--SDIPDSAAKLLLQSIFNNTGGEVVYNKKGKREILGTPTETAILEF 358
CMN ++V+N SS S+IP+SA KLL+QSIFNNTGGEVV NK GK E+LGTPTETAILE
Sbjct: 472 CMNVQDVANKGSSLQSEIPESAVKLLIQSIFNNTGGEVVVNKHGKTELLGTPTETAILEL 531
Query: 359 GLSLGGDSKAEREACKIVKVEPFNSEKKRMGVVVEQPDGS-VRAHCKGASEIILAACDKV 417
GLSLGG + ER++ K++KVEPFNS KKRMGVV+E P+G +RAH KGASEI+LAACDKV
Sbjct: 532 GLSLGGKFQEERKSYKVIKVEPFNSTKKRMGVVIELPEGGRMRAHTKGASEIVLAACDKV 591
Query: 418 IDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYMELENGFAAEDPIPASGYTCIGI 477
++ +G+VV LD ES YLN IN+FANEALRTLCLAYM++E GF+ +D IPASG+TC+GI
Sbjct: 592 VNSSGEVVPLDEESIKYLNVTINEFANEALRTLCLAYMDIEGGFSPDDAIPASGFTCVGI 651
Query: 478 VGIKDPVRPGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILTDDGIAIEGPDFRE 537
VGIKDPVRPGVK+SV CR AGI VRMVTGDNINTAKAIARECGILTDDGIAIEGP FRE
Sbjct: 652 VGIKDPVRPGVKESVELCRRAGITVRMVTGDNINTAKAIARECGILTDDGIAIEGPVFRE 711
Query: 538 KTQEELFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLA 597
K QEEL ELIPKIQVMARSSP+DKHTLVKQLRTTF EVVAVTGDGTNDAPALHEADIGLA
Sbjct: 712 KNQEELLELIPKIQVMARSSPMDKHTLVKQLRTTFDEVVAVTGDGTNDAPALHEADIGLA 771
Query: 598 MGIAGTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNVVALLVNFTS 657
MGIAGTEVAKESADVIILDDNFSTIVTVA+WGRSVYINIQKFVQFQLTVNVVAL+VNF+S
Sbjct: 772 MGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVALVVNFSS 831
Query: 658 ACMTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKREPVGRKGDFINNVMWRNIL 717
AC+TGSAPLTAVQLLWVNMIMDTLGALALATEPP D+LMKR PVGR+G+FI N MWRNIL
Sbjct: 832 ACLTGSAPLTAVQLLWVNMIMDTLGALALATEPPNDELMKRLPVGRRGNFITNAMWRNIL 891
Query: 718 GQALYQFVVIWFLQSVGKWVFFLRGPNADIVLNTLIFNTFVFCQVFNEINSREMEEIDVF 777
GQA+YQF+VIW LQ+ GK +F L GP++ ++LNTLIFN FVFCQVFNEI+SREMEEIDVF
Sbjct: 892 GQAVYQFIVIWILQAKGKAMFGLDGPDSTLMLNTLIFNCFVFCQVFNEISSREMEEIDVF 951
Query: 778 KGIWDNHVFVAVISATVVFQIIIVEYLGTFANTTPLSLVQWIFCLGVGYMGMPIAVRLKQ 837
KGI DN+VFV VI ATV FQIII+E+LGTFA+TTPL++ QWIF + +G++GMPIA LK
Sbjct: 952 KGILDNYVFVVVIGATVFFQIIIIEFLGTFASTTPLTITQWIFSIFIGFLGMPIAAGLKT 1011
Query: 838 IPV 840
IPV
Sbjct: 1012 IPV 1014
>At2g22950 pseudogene
Length = 1015
Score = 1367 bits (3537), Expect = 0.0
Identities = 686/843 (81%), Positives = 770/843 (90%), Gaps = 3/843 (0%)
Query: 1 MTLMILGVCAFVSLIVGVLTEGWPKGAHDGLGIVASILLVVFVTATSDYRQSLQFKDLDK 60
MTLMILGVCAFVSLIVG+ TEGWP+G+HDGLGIVASILLVVFVTATSDYRQSLQF+DLDK
Sbjct: 173 MTLMILGVCAFVSLIVGIATEGWPQGSHDGLGIVASILLVVFVTATSDYRQSLQFRDLDK 232
Query: 61 EKKKISIQVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGES 120
EKKKI++QVTRNG+RQKMSIY+LLPGD+VHL IGDQVPADGLF+SGFSV+IDESSLTGES
Sbjct: 233 EKKKITVQVTRNGFRQKMSIYDLLPGDVVHLAIGDQVPADGLFLSGFSVVIDESSLTGES 292
Query: 121 EPIMVTTQNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGV 180
EP+MVT QNPFLLSGTKVQDGSC MLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGV
Sbjct: 293 EPVMVTAQNPFLLSGTKVQDGSCKMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGV 352
Query: 181 ATIIGKIGLVFAVITFTVLVKGHLSHKIREGNFWRWTGDNAMEMLEYFAIAVTIVVVAVP 240
ATIIGKIGL FA++TF VLV+G K+ G W W+GD+A+E+LEYFAIAVTIVVVAVP
Sbjct: 353 ATIIGKIGLSFAIVTFAVLVQGMFMRKLSLGPHWWWSGDDALELLEYFAIAVTIVVVAVP 412
Query: 241 EGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVKTCI 300
EGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTN MTVVK+CI
Sbjct: 413 EGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNHMTVVKSCI 472
Query: 301 CMNSKEVSNSSSS--SDIPDSAAKLLLQSIFNNTGGEVVYNKKGKREILGTPTETAILEF 358
CMN ++V++ SSS SDIP++A KLLLQ IFNNTGGEVV N++GK EILGTPTETAILE
Sbjct: 473 CMNVQDVASKSSSLQSDIPEAALKLLLQLIFNNTGGEVVVNERGKTEILGTPTETAILEL 532
Query: 359 GLSLGGDSKAEREACKIVKVEPFNSEKKRMGVVVEQPDGS-VRAHCKGASEIILAACDKV 417
GLSLGG + ER++ K++KVEPFNS KKRMGVV+E P+G +RAH KGASEI+LAACDKV
Sbjct: 533 GLSLGGKFQEERQSNKVIKVEPFNSTKKRMGVVIELPEGGRIRAHTKGASEIVLAACDKV 592
Query: 418 IDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYMELENGFAAEDPIPASGYTCIGI 477
I+ +G+VV LD ES +LN I++FANEALRTLCLAYM++E+GF+A++ IP G+TCIGI
Sbjct: 593 INSSGEVVPLDDESIKFLNVTIDEFANEALRTLCLAYMDIESGFSADEGIPEKGFTCIGI 652
Query: 478 VGIKDPVRPGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILTDDGIAIEGPDFRE 537
VGIKDPVRPGV++SV CR AGI+VRMVTGDNINTAKAIARECGILTDDGIAIEGP FRE
Sbjct: 653 VGIKDPVRPGVRESVELCRRAGIMVRMVTGDNINTAKAIARECGILTDDGIAIEGPVFRE 712
Query: 538 KTQEELFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLA 597
K QEE+ ELIPKIQVMARSSP+DKHTLVKQLRTTF EVVAVTGDGTNDAPALHEADIGLA
Sbjct: 713 KNQEEMLELIPKIQVMARSSPMDKHTLVKQLRTTFDEVVAVTGDGTNDAPALHEADIGLA 772
Query: 598 MGIAGTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNVVALLVNFTS 657
MGIAGTEVAKE ADVIILDDNFSTIVTVA+WGRSVYINIQKFVQFQLTVNVVAL+VNF+S
Sbjct: 773 MGIAGTEVAKEIADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVALIVNFSS 832
Query: 658 ACMTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKREPVGRKGDFINNVMWRNIL 717
AC+TGSAPLTAVQLLWVNMIMDTLGALALATEPP ++LMKR PVGR+G+FI N MWRNIL
Sbjct: 833 ACLTGSAPLTAVQLLWVNMIMDTLGALALATEPPNNELMKRMPVGRRGNFITNAMWRNIL 892
Query: 718 GQALYQFVVIWFLQSVGKWVFFLRGPNADIVLNTLIFNTFVFCQVFNEINSREMEEIDVF 777
GQA+YQF++IW LQ+ GK +F L G ++ +VLNTLIFN FVFCQVFNE++SREMEEIDVF
Sbjct: 893 GQAVYQFIIIWILQAKGKSMFGLVGSDSTLVLNTLIFNCFVFCQVFNEVSSREMEEIDVF 952
Query: 778 KGIWDNHVFVAVISATVVFQIIIVEYLGTFANTTPLSLVQWIFCLGVGYMGMPIAVRLKQ 837
KGI DN+VFV VI ATV FQIII+E+LGTFA+TTPL++VQW F + VG++GMPIA LK+
Sbjct: 953 KGILDNYVFVVVIGATVFFQIIIIEFLGTFASTTPLTIVQWFFSIFVGFLGMPIAAGLKK 1012
Query: 838 IPV 840
IPV
Sbjct: 1013 IPV 1015
>At3g57330 Ca2+-transporting ATPase-like protein
Length = 1025
Score = 1104 bits (2855), Expect = 0.0
Identities = 557/840 (66%), Positives = 681/840 (80%), Gaps = 3/840 (0%)
Query: 1 MTLMILGVCAFVSLIVGVLTEGWPKGAHDGLGIVASILLVVFVTATSDYRQSLQFKDLDK 60
+TL+IL VCA VS+ VGV TEG+PKG +DG GI+ SI+LVV VTA SDY+QSLQF+DLD+
Sbjct: 169 ITLIILMVCAVVSIGVGVATEGFPKGMYDGTGILLSIILVVMVTAISDYKQSLQFRDLDR 228
Query: 61 EKKKISIQVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGES 120
EKKKI IQVTR+G RQ++SI++L+ GD+VHL+IGDQVPADG+F+SG+++ IDESSL+GES
Sbjct: 229 EKKKIIIQVTRDGSRQEVSIHDLVVGDVVHLSIGDQVPADGIFISGYNLEIDESSLSGES 288
Query: 121 EPIMVTTQNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGV 180
EP V + PFLLSGTKVQ+GS MLVTTVGMRT+WGKLM TLSEGG+DETPLQVKLNGV
Sbjct: 289 EPSHVNKEKPFLLSGTKVQNGSAKMLVTTVGMRTEWGKLMDTLSEGGEDETPLQVKLNGV 348
Query: 181 ATIIGKIGLVFAVITFTVLVKGHLSHKIREGNFWRWTGDNAMEMLEYFAIAVTIVVVAVP 240
ATIIGKIGL FAV+TF VL + K G+ W+ ++A+ +L+YFAIAVTI+VVAVP
Sbjct: 349 ATIIGKIGLGFAVLTFVVLCIRFVVEKATAGSITEWSSEDALTLLDYFAIAVTIIVVAVP 408
Query: 241 EGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVKTCI 300
EGLPLAVTLSLAFAMK++M+D+ALVRHLAACETMGS+T IC+DKTGTLTTN M V K I
Sbjct: 409 EGLPLAVTLSLAFAMKQLMSDRALVRHLAACETMGSSTCICTDKTGTLTTNHMVVNKVWI 468
Query: 301 CMNSKEVSNSSSSSDIPDSAAKLLLQSIFNNTGGEVVYNKKGKREILGTPTETAILEFGL 360
C N KE + ++ + +L+Q+IF NTG EVV +K+GK +ILG+PTE AILEFGL
Sbjct: 469 CENIKERQEENFQLNLSEQVKNILIQAIFQNTGSEVVKDKEGKTQILGSPTERAILEFGL 528
Query: 361 SLGGDSKAEREACKIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIILAACDKVIDL 420
LGGD +R KI+K+EPFNS+KK+M V+ G VRA CKGASEI+L C+KV+D
Sbjct: 529 LLGGDVDTQRREHKILKIEPFNSDKKKMSVLTSHSGGKVRAFCKGASEIVLKMCEKVVDS 588
Query: 421 NGDVVALDGESTNYLNSIINQFANEALRTLCLAYMELENGFAAEDPIPASGYTCIGIVGI 480
NG+ V L E ++ +I FA+EALRTLCL Y +L+ A +P GYT + +VGI
Sbjct: 589 NGESVPLSEEKIASISDVIEGFASEALRTLCLVYTDLDE--APRGDLPNGGYTLVAVVGI 646
Query: 481 KDPVRPGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILTDDGIAIEGPDFREKTQ 540
KDPVRPGV+++V C++AGI VRMVTGDNI+TAKAIA+ECGILT G+AIEG DFR
Sbjct: 647 KDPVRPGVREAVQTCQAAGITVRMVTGDNISTAKAIAKECGILTAGGVAIEGSDFRNLPP 706
Query: 541 EELFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGI 600
E+ ++PKIQVMARS PLDKHTLV LR GEVVAVTGDGTNDAPALHEADIGLAMGI
Sbjct: 707 HEMRAILPKIQVMARSLPLDKHTLVNNLR-KMGEVVAVTGDGTNDAPALHEADIGLAMGI 765
Query: 601 AGTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNVVALLVNFTSACM 660
AGTEVAKE+ADVII+DDNF+TIV VA+WGR+VYINIQKFVQFQLTVNVVAL++NF SAC+
Sbjct: 766 AGTEVAKENADVIIMDDNFATIVNVAKWGRAVYINIQKFVQFQLTVNVVALIINFVSACI 825
Query: 661 TGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKREPVGRKGDFINNVMWRNILGQA 720
TGSAPLTAVQLLWVNMIMDTLGALALATEPP + LMKR+P+GR FI MWRNI+GQ+
Sbjct: 826 TGSAPLTAVQLLWVNMIMDTLGALALATEPPNEGLMKRQPIGRTASFITRAMWRNIIGQS 885
Query: 721 LYQFVVIWFLQSVGKWVFFLRGPNADIVLNTLIFNTFVFCQVFNEINSREMEEIDVFKGI 780
+YQ +V+ L GK + L GP++ IVLNT+IFN+FVFCQVFNE+NSRE+E+I+VF+G+
Sbjct: 886 IYQLIVLGILNFAGKQILNLNGPDSTIVLNTIIFNSFVFCQVFNEVNSREIEKINVFEGM 945
Query: 781 WDNHVFVAVISATVVFQIIIVEYLGTFANTTPLSLVQWIFCLGVGYMGMPIAVRLKQIPV 840
+ + VFVAV++ATV FQ+IIVE+LG FA+T PLS W+ C+ +G + M +AV LK IPV
Sbjct: 946 FKSWVFVAVMTATVGFQVIIVEFLGAFASTVPLSWQHWLLCILIGSVSMILAVGLKCIPV 1005
>At2g41560 putative Ca2+-ATPase
Length = 1030
Score = 1083 bits (2801), Expect = 0.0
Identities = 551/843 (65%), Positives = 674/843 (79%), Gaps = 6/843 (0%)
Query: 1 MTLMILGVCAFVSLIVGVLTEGWPKGAHDGLGIVASILLVVFVTATSDYRQSLQFKDLDK 60
+TL+IL VCA VS+ VGV TEG+P+G +DG GI+ SILLVV VTA SDY+QSLQF+DLD+
Sbjct: 169 ITLIILMVCAVVSIGVGVATEGFPRGMYDGTGILLSILLVVMVTAISDYKQSLQFRDLDR 228
Query: 61 EKKKISIQVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGES 120
EKKKI +QVTR+G RQ++SI++L+ GD+VHL+IGDQVPADG+F+SG+++ IDESSL+GES
Sbjct: 229 EKKKIIVQVTRDGSRQEISIHDLVVGDVVHLSIGDQVPADGIFISGYNLEIDESSLSGES 288
Query: 121 EPIMVTTQNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGV 180
EP V + PFLLSGTKVQ+GS MLVTTVGMRT+WGKLM TL +GG+DETPLQVKLNGV
Sbjct: 289 EPSHVNKEKPFLLSGTKVQNGSAKMLVTTVGMRTEWGKLMETLVDGGEDETPLQVKLNGV 348
Query: 181 ATIIGKIGLVFAVITFTVLVKGHLSHKIREGNFWRWTGDNAMEMLEYFAIAVTIVVVAVP 240
ATIIGKIGL FAV+TF VL + K G+F W+ ++A+ +L+YFAI+VTI+VVAVP
Sbjct: 349 ATIIGKIGLSFAVLTFVVLCIRFVLDKATSGSFTNWSSEDALTLLDYFAISVTIIVVAVP 408
Query: 241 EGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVKTCI 300
EGLPLAVTLSLAFAMKK+M+D+ALVRHLAACETMGS+T IC+DKTGTLTTN M V K I
Sbjct: 409 EGLPLAVTLSLAFAMKKLMSDRALVRHLAACETMGSSTCICTDKTGTLTTNHMVVNKVWI 468
Query: 301 C---MNSKEVSNSSSSSDIPDSAAKLLLQSIFNNTGGEVVYNKKGKREILGTPTETAILE 357
C +E S S ++ + LLQ IF NTG EVV +K G +ILG+PTE AILE
Sbjct: 469 CDKVQERQEGSKESFELELSEEVQSTLLQGIFQNTGSEVVKDKDGNTQILGSPTERAILE 528
Query: 358 FGLSLGGDSKAEREACKIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIILAACDKV 417
FGL LGGD +R+ KI+K+EPFNS+KK+M V++ P G RA CKGASEI+L C+ V
Sbjct: 529 FGLLLGGDFNTQRKEHKILKIEPFNSDKKKMSVLIALPGGGARAFCKGASEIVLKMCENV 588
Query: 418 IDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYMELENGFAAEDPIPASGYTCIGI 477
+D NG+ V L E ++ II FA+EALRTLCL Y +L+ + E +P GYT + +
Sbjct: 589 VDSNGESVPLTEERITSISDIIEGFASEALRTLCLVYKDLDEAPSGE--LPDGGYTMVAV 646
Query: 478 VGIKDPVRPGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILTDDGIAIEGPDFRE 537
VGIKDPVRPGV+++V C++AGI VRMVTGDNI+TAKAIA+ECGI T+ G+AIEG +FR+
Sbjct: 647 VGIKDPVRPGVREAVQTCQAAGITVRMVTGDNISTAKAIAKECGIYTEGGLAIEGSEFRD 706
Query: 538 KTQEELFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLA 597
+ E+ +IPKIQVMARS PLDKHTLV LR GEVVAVTGDGTNDAPALHEADIGLA
Sbjct: 707 LSPHEMRAIIPKIQVMARSLPLDKHTLVSNLR-KIGEVVAVTGDGTNDAPALHEADIGLA 765
Query: 598 MGIAGTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNVVALLVNFTS 657
MGIAGTEVAKE+ADVII+DDNF TIV VARWGR+VYINIQKFVQFQLTVNVVAL++NF S
Sbjct: 766 MGIAGTEVAKENADVIIMDDNFKTIVNVARWGRAVYINIQKFVQFQLTVNVVALIINFVS 825
Query: 658 ACMTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKREPVGRKGDFINNVMWRNIL 717
AC+TGSAPLTAVQLLWVNMIMDTLGALALATEPP + LMKR P+ R FI MWRNI
Sbjct: 826 ACITGSAPLTAVQLLWVNMIMDTLGALALATEPPNEGLMKRAPIARTASFITKTMWRNIA 885
Query: 718 GQALYQFVVIWFLQSVGKWVFFLRGPNADIVLNTLIFNTFVFCQVFNEINSREMEEIDVF 777
GQ++YQ +V+ L GK + L GP++ VLNT+IFN+FVFCQVFNEINSRE+E+I+VF
Sbjct: 886 GQSVYQLIVLGILNFAGKSLLKLDGPDSTAVLNTVIFNSFVFCQVFNEINSREIEKINVF 945
Query: 778 KGIWDNHVFVAVISATVVFQIIIVEYLGTFANTTPLSLVQWIFCLGVGYMGMPIAVRLKQ 837
KG++++ VF V++ TVVFQ+IIVE+LG FA+T PLS W+ + +G + M +AV LK
Sbjct: 946 KGMFNSWVFTWVMTVTVVFQVIIVEFLGAFASTVPLSWQHWLLSILIGSLNMIVAVILKC 1005
Query: 838 IPV 840
+PV
Sbjct: 1006 VPV 1008
>At4g29900 Ca2+-transporting ATPase - like protein
Length = 1069
Score = 860 bits (2221), Expect = 0.0
Identities = 463/861 (53%), Positives = 602/861 (69%), Gaps = 24/861 (2%)
Query: 1 MTLMILGVCAFVSLIVGVLTEGWPKGAHDGLGIVASILLVVFVTATSDYRQSLQFKDLDK 60
+TL+IL V A SL +G+ TEG KG +DG+ I ++LLV+ VTATSDYRQSLQF++L++
Sbjct: 192 LTLIILIVAAVASLALGIKTEGIEKGWYDGISIAFAVLLVIVVTATSDYRQSLQFQNLNE 251
Query: 61 EKKKISIQVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGES 120
EK+ I ++VTR+G R ++SIY+++ GD++ LNIGDQVPADG+ V+G S+ +DESS+TGES
Sbjct: 252 EKRNIRLEVTRDGRRVEISIYDIVVGDVIPLNIGDQVPADGVLVAGHSLAVDESSMTGES 311
Query: 121 EPIMV-TTQNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNG 179
+ + +T++PFL+SG KV DG+ TMLVT VG+ T+WG LMA++SE ETPLQV+LNG
Sbjct: 312 KIVQKNSTKHPFLMSGCKVADGNGTMLVTGVGVNTEWGLLMASVSEDNGGETPLQVRLNG 371
Query: 180 VATIIGKIGLVFA-VITFTVLVKGHLSHKIREGNFWRWTGDNAM------EMLEYFAIAV 232
VAT IG +GL A V+ F ++V+ H E ++ G +++E F +AV
Sbjct: 372 VATFIGIVGLTVAGVVLFVLVVRYFTGHTKNEQGGPQFIGGKTKFEHVLDDLVEIFTVAV 431
Query: 233 TIVVVAVPEGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNR 292
TIVVVAVPEGLPLAVTL+LA++M+KMM DKALVR L+ACETMGSATTICSDKTGTLT N
Sbjct: 432 TIVVVAVPEGLPLAVTLTLAYSMRKMMADKALVRRLSACETMGSATTICSDKTGTLTLNE 491
Query: 293 MTVVKTCICMNSKEVSNSSSSSDIPDSAAKLLLQSIFNNTGGEVVYNKKGKREILGTPTE 352
MTVV+ + +++ + SSS +P + +L++ I +NT G V ++ G+ ++ G+PTE
Sbjct: 492 MTVVECYAGL--QKMDSPDSSSKLPSAFTSILVEGIAHNTTGSVFRSESGEIQVSGSPTE 549
Query: 353 TAILEFGLSLGGDSKAEREACKIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIILA 412
AIL + + LG D A + V+ PFNSEKKR GV V+ PD SV H KGA+EI+L
Sbjct: 550 RAILNWAIKLGMDFDALKSESSAVQFFPFNSEKKRGGVAVKSPDSSVHIHWKGAAEIVLG 609
Query: 413 ACDKVIDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYMELENGFAAEDP------ 466
+C +D + V + + L I+ A +LR + +A+ E D
Sbjct: 610 SCTHYMDESESFVDMSEDKMGGLKDAIDDMAARSLRCVAIAFRTFEADKIPTDEEQLSRW 669
Query: 467 -IPASGYTCIGIVGIKDPVRPGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILTD 525
+P + IVGIKDP RPGVK SV C+ AG+ VRMVTGDNI TAKAIA ECGIL
Sbjct: 670 ELPEDDLILLAIVGIKDPCRPGVKNSVLLCQQAGVKVRMVTGDNIQTAKAIALECGILAS 729
Query: 526 DGIA-----IEGPDFREKTQEELFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTG 580
D A IEG FR ++EE + +I VM RSSP DK LV+ L+ G VVAVTG
Sbjct: 730 DSDASEPNLIEGKVFRSYSEEERDRICEEISVMGRSSPNDKLLLVQSLKRR-GHVVAVTG 788
Query: 581 DGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFV 640
DGTNDAPALHEADIGLAMGI GTEVAKE +D+IILDDNF ++V V RWGRSVY NIQKF+
Sbjct: 789 DGTNDAPALHEADIGLAMGIQGTEVAKEKSDIIILDDNFESVVKVVRWGRSVYANIQKFI 848
Query: 641 QFQLTVNVVALLVNFTSACMTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKREP 700
QFQLTVNV AL++N +A G PLTAVQLLWVN+IMDTLGALALATEPPTD LM R P
Sbjct: 849 QFQLTVNVAALVINVVAAISAGEVPLTAVQLLWVNLIMDTLGALALATEPPTDHLMDRAP 908
Query: 701 VGRKGDFINNVMWRNILGQALYQFVVIWFLQSVGKWVFFLRG-PNADIVLNTLIFNTFVF 759
VGR+ I N+MWRN+ QA+YQ V+ L G + L+ PNA+ V NT+IFN FV
Sbjct: 909 VGRREPLITNIMWRNLFIQAMYQVTVLLILNFRGISILHLKSKPNAERVKNTVIFNAFVI 968
Query: 760 CQVFNEINSREMEEIDVFKGIWDNHVFVAVISATVVFQIIIVEYLGTFANTTPLSLVQWI 819
CQVFNE N+R+ +EI++F+G+ NH+FV +IS T+V Q++IVE+LGTFA+TT L W+
Sbjct: 969 CQVFNEFNARKPDEINIFRGVLRNHLFVGIISITIVLQVVIVEFLGTFASTTKLDWEMWL 1028
Query: 820 FCLGVGYMGMPIAVRLKQIPV 840
C+G+G + P+AV K IPV
Sbjct: 1029 VCIGIGSISWPLAVIGKLIPV 1049
>At5g57110 Ca2+-transporting ATPase-like protein (emb|CAB79748.1)
Length = 1074
Score = 824 bits (2129), Expect = 0.0
Identities = 454/863 (52%), Positives = 592/863 (67%), Gaps = 30/863 (3%)
Query: 1 MTLMILGVCAFVSLIVGVLTEGWPKGAHDGLGIVASILLVVFVTATSDYRQSLQFKDLDK 60
+TL+IL V A SL +G+ TEG +G +DG I +++LV+ VTA SDY+QSLQF++L+
Sbjct: 192 LTLIILMVAAVASLALGIKTEGIKEGWYDGGSIAFAVILVIVVTAVSDYKQSLQFQNLND 251
Query: 61 EKKKISIQVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGES 120
EK+ I ++V R G R ++SIY+++ GD++ LNIG+QVPADG+ +SG S+ +DESS+TGES
Sbjct: 252 EKRNIHLEVLRGGRRVEISIYDIVVGDVIPLNIGNQVPADGVLISGHSLALDESSMTGES 311
Query: 121 EPIMV-TTQNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNG 179
+ + ++PFL+SG KV DG+ +MLVT VG+ T+WG LMA++SE +ETPLQV+LNG
Sbjct: 312 KIVNKDANKDPFLMSGCKVADGNGSMLVTGVGVNTEWGLLMASISEDNGEETPLQVRLNG 371
Query: 180 VATIIGKIGLVFAVITFTVLVKGHLSHKIREGN----FWRW---TGDNAMEMLEYFAIAV 232
VAT IG IGL A +L+ + + ++ N F + G ++++ +AV
Sbjct: 372 VATFIGSIGLAVAAAVLVILLTRYFTGHTKDNNGGPQFVKGKTKVGHVIDDVVKVLTVAV 431
Query: 233 TIVVVAVPEGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNR 292
TIVVVAVPEGLPLAVTL+LA++M+KMM DKALVR L+ACETMGSATTICSDKTGTLT N+
Sbjct: 432 TIVVVAVPEGLPLAVTLTLAYSMRKMMADKALVRRLSACETMGSATTICSDKTGTLTLNQ 491
Query: 293 MTVVKTCICMNSKEVSNSSSSSDIPDSAAKLLLQSIFNNTGGEV-VYNKKGKREILGTPT 351
MTVV+ S + + +P + L+++ I NT G + V G E G+PT
Sbjct: 492 MTVVE------SYAGGKKTDTEQLPATITSLVVEGISQNTTGSIFVPEGGGDLEYSGSPT 545
Query: 352 ETAILEFGLSLGGDSKAEREACKIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIIL 411
E AIL +G+ LG + + R I+ PFNSEKKR GV V+ DG V H KGASEI+L
Sbjct: 546 EKAILGWGVKLGMNFETARSQSSILHAFPFNSEKKRGGVAVKTADGEVHVHWKGASEIVL 605
Query: 412 AACDKVIDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYM--ELENGFAAED---- 465
A+C ID +G+V + + ++ + IN A LR + LA+ E E E+
Sbjct: 606 ASCRSYIDEDGNVAPMTDDKASFFKNGINDMAGRTLRCVALAFRTYEAEKVPTGEELSKW 665
Query: 466 PIPASGYTCIGIVGIKDPVRPGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILTD 525
+P + IVGIKDP RPGVK SV C++AG+ VRMVTGDN+ TA+AIA ECGIL+
Sbjct: 666 VLPEDDLILLAIVGIKDPCRPGVKDSVVLCQNAGVKVRMVTGDNVQTARAIALECGILSS 725
Query: 526 DG-----IAIEGPDFREKTQEELFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTG 580
D IEG FRE T E ++ KI VM RSSP DK LV+ LR G VVAVTG
Sbjct: 726 DADLSEPTLIEGKSFREMTDAERDKISDKISVMGRSSPNDKLLLVQSLRRQ-GHVVAVTG 784
Query: 581 DGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFV 640
DGTNDAPALHEADIGLAMGIAGTEVAKES+D+IILDDNF+++V V RWGRSVY NIQKF+
Sbjct: 785 DGTNDAPALHEADIGLAMGIAGTEVAKESSDIIILDDNFASVVKVVRWGRSVYANIQKFI 844
Query: 641 QFQLTVNVVALLVNFTSACMTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKREP 700
QFQLTVNV AL++N +A +G PLTAVQLLWVN+IMDTLGALALATEPPTD LM R P
Sbjct: 845 QFQLTVNVAALVINVVAAISSGDVPLTAVQLLWVNLIMDTLGALALATEPPTDHLMGRPP 904
Query: 701 VGRKGDFINNVMWRNILGQALYQFVVIWFLQSVGKWVFFLR---GPNADIVLNTLIFNTF 757
VGRK I N+MWRN+L QA+YQ V+ L G + L +A V NT+IFN F
Sbjct: 905 VGRKEPLITNIMWRNLLIQAIYQVSVLLTLNFRGISILGLEHEVHEHATRVKNTIIFNAF 964
Query: 758 VFCQVFNEINSREMEEIDVFKGIWDNHVFVAVISATVVFQIIIVEYLGTFANTTPLSLVQ 817
V CQ FNE N+R+ +E ++FKG+ N +F+ +I T+V Q+IIVE+LG FA+TT L+ Q
Sbjct: 965 VLCQAFNEFNARKPDEKNIFKGVIKNRLFMGIIVITLVLQVIIVEFLGKFASTTKLNWKQ 1024
Query: 818 WIFCLGVGYMGMPIAVRLKQIPV 840
W+ C+G+G + P+A+ K IPV
Sbjct: 1025 WLICVGIGVISWPLALVGKFIPV 1047
>At3g21180 putative Ca2+-transporting ATPase
Length = 1090
Score = 790 bits (2040), Expect = 0.0
Identities = 442/867 (50%), Positives = 586/867 (66%), Gaps = 31/867 (3%)
Query: 1 MTLMILGVCAFVSLIVGVLTEGWPKGAHDGLGIVASILLVVFVTATSDYRQSLQFKDLDK 60
+TL+IL + A SL +G+ TEG +G DG I ++LLV+ VTA SDYRQSLQF++L+
Sbjct: 206 LTLIILIIAAVTSLALGIKTEGLKEGWLDGGSIAFAVLLVIVVTAVSDYRQSLQFQNLND 265
Query: 61 EKKKISIQVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGES 120
EK+ I ++V R G K+SIY+++ GD++ L IGDQVPADG+ +SG S+ IDESS+TGES
Sbjct: 266 EKRNIQLEVMRGGRTVKISIYDVVVGDVIPLRIGDQVPADGVLISGHSLAIDESSMTGES 325
Query: 121 EPIMVTTQNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGV 180
+ + ++PFL+SG KV DG MLVT VG+ T+WG LMA++SE +ETPLQV+LNG+
Sbjct: 326 KIVHKDQKSPFLMSGCKVADGVGNMLVTGVGINTEWGLLMASISEDTGEETPLQVRLNGL 385
Query: 181 ATIIGKIGLVFAVITFTVLVKGHLSHKIREGN----FWRWT---GDNAMEMLEYFAIAVT 233
AT IG +GL A++ L+ + + ++ N F + T D + ++ F IAVT
Sbjct: 386 ATFIGIVGLSVALVVLVALLVRYFTGTTQDTNGATQFIKGTTSISDIVDDCVKIFTIAVT 445
Query: 234 IVVVAVPEGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRM 293
IVVVAVPEGLPLAVTL+LA++M+KMM DKALVR L+ACETMGSATTICSDKTGTLT N+M
Sbjct: 446 IVVVAVPEGLPLAVTLTLAYSMRKMMADKALVRRLSACETMGSATTICSDKTGTLTLNQM 505
Query: 294 TVVKTCICMNSKEVSNSSSSSDIPDSAAKLLLQSIFNNTGGEVVYNKK--GKREILGTPT 351
TVV+T + +V+++ S + L+ + + NT G + + K G+ EI G+PT
Sbjct: 506 TVVETYAGGSKMDVADNPSG--LHPKLVALISEGVAQNTTGNIFHPKVDGGEVEISGSPT 563
Query: 352 ETAILEFGLSLGGDSKAEREACKIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIIL 411
E AIL + LG R I+ PFNSEKKR GV V + D V H KGA+EI+L
Sbjct: 564 EKAILSWAYKLGMKFDTIRSESAIIHAFPFNSEKKRGGVAVLRGDSEVFIHWKGAAEIVL 623
Query: 412 AACDKVIDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYMELENGFAAEDP----- 466
A C + +D NG + +++ + + I+ A +LR + +A E ++
Sbjct: 624 ACCTQYMDSNGTLQSIESQK-EFFRVAIDSMAKNSLRCVAIACRTQELNQVPKEQEDLDK 682
Query: 467 --IPASGYTCIGIVGIKDPVRPGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILT 524
+P + IVGIKDP RPGV+++V C SAG+ VRMVTGDN+ TAKAIA ECGIL+
Sbjct: 683 WALPEDELILLAIVGIKDPCRPGVREAVRICTSAGVKVRMVTGDNLQTAKAIALECGILS 742
Query: 525 DDGIA-----IEGPDFREKTQEELFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVT 579
D A IEG FRE +++E ++ KI VM RSSP DK LV+ LR G+VVAVT
Sbjct: 743 SDTEAVEPTIIEGKVFRELSEKEREQVAKKITVMGRSSPNDKLLLVQALRKN-GDVVAVT 801
Query: 580 GDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQKF 639
GDGTNDAPALHEADIGL+MGI+GTEVAKES+D+IILDDNF+++V V RWGRSVY NIQKF
Sbjct: 802 GDGTNDAPALHEADIGLSMGISGTEVAKESSDIIILDDNFASVVKVVRWGRSVYANIQKF 861
Query: 640 VQFQLTVNVVALLVNFTSACMTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKRE 699
+QFQLTVNV AL++N +A +G PL AVQLLWVN+IMDTLGALALATEPPTD LM R
Sbjct: 862 IQFQLTVNVAALIINVVAAMSSGDVPLKAVQLLWVNLIMDTLGALALATEPPTDHLMHRT 921
Query: 700 PVGRKGDFINNVMWRNILGQALYQFVVIWFLQSVGKWVFFLRGPN---ADIVLNTLIFNT 756
PVGR+ I N+MWRN+L Q+ YQ V+ L G + L N A V NT+IFN
Sbjct: 922 PVGRREPLITNIMWRNLLVQSFYQVAVLLVLNFAGLSILGLNHENHAHAVEVKNTMIFNA 981
Query: 757 FVFCQVFNEINSREMEEIDVFKGIWDNHVFVAVISATVVFQIIIVEYLGTFANTTPLSLV 816
FV CQ+FNE N+R+ +E++VF+G+ N +FVA++ T + QIIIV +LG FA+T L
Sbjct: 982 FVMCQIFNEFNARKPDEMNVFRGVNKNPLFVAIVGVTFILQIIIVTFLGKFAHTVRLGWQ 1041
Query: 817 QWIFCLGVGYMGM---PIAVRLKQIPV 840
W+ + +G + + P+A+ K IPV
Sbjct: 1042 LWLASIIIGLVRLVHWPLAIVGKLIPV 1068
>At3g63380 Ca2+-transporting ATPase -like protein
Length = 1033
Score = 757 bits (1954), Expect = 0.0
Identities = 419/856 (48%), Positives = 568/856 (65%), Gaps = 29/856 (3%)
Query: 1 MTLMILGVCAFVSLIVGVLTEGWPKGAHDGLGIVASILLVVFVTATSDYRQSLQFKDLDK 60
+T++IL VCA SL G+ G +G ++G I ++ LV+ V+A S++RQ QF L K
Sbjct: 164 LTILILLVCAIFSLGFGIKEHGIKEGWYEGGSIFVAVFLVIVVSALSNFRQERQFDKLSK 223
Query: 61 EKKKISIQVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGES 120
I ++V R+ RQ +SI++++ GD+V L IGDQ+PADGLF+ G S+ +DESS+TGES
Sbjct: 224 ISNNIKVEVLRDSRRQHISIFDVVVGDVVFLKIGDQIPADGLFLEGHSLQVDESSMTGES 283
Query: 121 EPIMVTTQ-NPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNG 179
+ + V + NPFL SGTK+ DG MLV +VGM T WG+ M+++++ + TPLQV+L+
Sbjct: 284 DHLEVDHKDNPFLFSGTKIVDGFAQMLVVSVGMSTTWGQTMSSINQDSSERTPLQVRLDT 343
Query: 180 VATIIGKIGLVFAVITFTVLVKGHLSHKIREGNFWRWTGDNAM------EMLEYFAIAVT 233
+ + IGKIGL A + VL+ + + + + G ++ A AVT
Sbjct: 344 LTSTIGKIGLTVAALVLVVLLVRYFTGNTEKEGKREYNGSKTPVDTVVNSVVRIVAAAVT 403
Query: 234 IVVVAVPEGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRM 293
IVVVA+PEGLPLAVTL+LA++MK+MM+D+A+VR L+ACETMGSAT IC+DKTGTLT N M
Sbjct: 404 IVVVAIPEGLPLAVTLTLAYSMKRMMSDQAMVRKLSACETMGSATVICTDKTGTLTLNEM 463
Query: 294 TVVKTCICMNS-KEVSNSSSSSDIPDSAAKLLLQSIFNNTGGEVVYNKKGKR-EILGTPT 351
V K + S E S S D+ D LL Q NT G V + G E G+PT
Sbjct: 464 KVTKFWLGQESIHEDSTKMISPDVLD----LLYQGTGLNTTGSVCVSDSGSTPEFSGSPT 519
Query: 352 ETAILEFG-LSLGGDSKAEREACKIVKVEPFNSEKKRMGVVVEQP-DGSVRAHCKGASEI 409
E A+L + L+LG D ++ ++ ++++VE F+S KKR GV+V + D +V H KGA+E+
Sbjct: 520 EKALLSWTVLNLGMDMESVKQKHEVLRVETFSSAKKRSGVLVRRKSDNTVHVHWKGAAEM 579
Query: 410 ILAACDKVIDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYMELENGFAAEDPIPA 469
+LA C G V +D + + + +II A +LR + A+ N E+
Sbjct: 580 VLAMCSHYYTSTGSVDLMDSTAKSRIQAIIQGMAASSLRCIAFAHKIASNDSVLEE---- 635
Query: 470 SGYTCIGIVGIKDPVRPGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILT----- 524
G T +GIVG+KDP RPGV ++V C+ AG+ ++M+TGDN+ TAKAIA ECGIL
Sbjct: 636 DGLTLMGIVGLKDPCRPGVSKAVETCKLAGVTIKMITGDNVFTAKAIAFECGILDHNDKD 695
Query: 525 DDGIAIEGPDFREKTQEELFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTN 584
++ +EG FR T EE + + KI+VMARSSP DK +VK LR G VVAVTGDGTN
Sbjct: 696 EEDAVVEGVQFRNYTDEERMQKVDKIRVMARSSPSDKLLMVKCLRLK-GHVVAVTGDGTN 754
Query: 585 DAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFVQFQL 644
DAPAL EADIGL+MGI GTEVAKES+D++ILDDNF+++ TV +WGR VY NIQKF+QFQL
Sbjct: 755 DAPALKEADIGLSMGIQGTEVAKESSDIVILDDNFASVATVLKWGRCVYNNIQKFIQFQL 814
Query: 645 TVNVVALLVNFTSACMTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKREPVGRK 704
TVNV AL++NF +A G PLTAVQLLWVN+IMDTLGALALATE PT++L+KR+PVGR
Sbjct: 815 TVNVAALVINFIAAISAGEVPLTAVQLLWVNLIMDTLGALALATERPTNELLKRKPVGRT 874
Query: 705 GDFINNVMWRNILGQALYQFVVIWFLQSVGKWVFFLRGPNADIVLNTLIFNTFVFCQVFN 764
I NVMWRN+L Q+LYQ V+ LQ G +F +R V +TLIFNTFV CQVFN
Sbjct: 875 EALITNVMWRNLLVQSLYQIAVLLILQFKGMSIFSVRKE----VKDTLIFNTFVLCQVFN 930
Query: 765 EINSREMEEIDVFKGIWDNHVFVAVISATVVFQIIIVEYLGTFANTTPLSLVQWIFCLGV 824
E N+REME+ +VFKG+ N +F+ +I+ T+V Q+I+VE+L FA+T L+ QW C+ +
Sbjct: 931 EFNAREMEKKNVFKGLHRNRLFIGIIAITIVLQVIMVEFLKKFADTVRLNGWQWGTCIAL 990
Query: 825 GYMGMPIAVRLKQIPV 840
+ PI K IPV
Sbjct: 991 ASLSWPIGFFTKFIPV 1006
>At3g22910 calmodulin-stimulated calcium-ATPase, putative
Length = 1017
Score = 744 bits (1922), Expect = 0.0
Identities = 421/862 (48%), Positives = 566/862 (64%), Gaps = 40/862 (4%)
Query: 1 MTLMILGVCAFVSLIVGVLTEGWPKGAHDGLGIVASILLVVFVTATSDYRQSLQFKDLDK 60
+T++IL CA +SL G+ G +G +DG I ++ LVV V+A S++RQ+ QF L K
Sbjct: 159 LTILILLGCATLSLGFGIKEHGLKEGWYDGGSIFVAVFLVVAVSAVSNFRQNRQFDKLSK 218
Query: 61 EKKKISIQVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGES 120
I I V RNG RQ++SI++++ GDIV LNIGDQVPADG+FV G + +DESS+TGES
Sbjct: 219 VSSNIKIDVVRNGRRQEISIFDIVVGDIVCLNIGDQVPADGVFVEGHLLHVDESSMTGES 278
Query: 121 EPIMVT-TQNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNG 179
+ + V+ T N FL SGTK+ DG M VT+VGM T WG++M+ +S +++TPLQ +L+
Sbjct: 279 DHVEVSLTGNTFLFSGTKIADGFGKMAVTSVGMNTAWGQMMSHISRDTNEQTPLQSRLDK 338
Query: 180 VATIIGKIGLVFAVITFTVLVKGHLSHKI------REGNFWRWTGDNAME-MLEYFAIAV 232
+ + IGK+GL+ A + VL+ + + RE N D + +++ A AV
Sbjct: 339 LTSSIGKVGLLVAFLVLLVLLIRYFTGTTKDESGNREYNGKTTKSDEIVNAVVKMVAAAV 398
Query: 233 TIVVVAVPEGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNR 292
TI+VVA+PEGLPLAVTL+LA++MK+MM D A+VR L+ACETMGSAT IC+DKTGTLT N+
Sbjct: 399 TIIVVAIPEGLPLAVTLTLAYSMKRMMKDNAMVRKLSACETMGSATVICTDKTGTLTLNQ 458
Query: 293 MTVVKTCICMNSKEVSNSSSSSDIPDSAAKLLLQSIFNNTGGEVVYNKKGKR-EILGTPT 351
M V + S +S + +L Q + NT G V K G E G+PT
Sbjct: 459 MKVTDFWFGLES------GKASSVSQRVVELFHQGVAMNTTGSVFKAKAGTEYEFSGSPT 512
Query: 352 ETAILEFG---LSLGGDSKAEREACKIVKVEPFNSEKKRMGVVVEQPDGSVR---AHCKG 405
E AIL + L +G + E +V VE FNSEKKR GV++++ + H KG
Sbjct: 513 EKAILSWAVEELEMGMEKVIEEH--DVVHVEGFNSEKKRSGVLMKKKGVNTENNVVHWKG 570
Query: 406 ASEIILAACDKVIDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYME--LENGFAA 463
A+E ILA C D +G V + + II A ++LR + AY E +N
Sbjct: 571 AAEKILAMCSTFCDGSGVVREMKEDDKIQFEKIIQSMAAKSLRCIAFAYSEDNEDNKKLK 630
Query: 464 EDPIPASGYTCIGIVGIKDPVRPGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGIL 523
E+ + + +GI+GIKDP RPGVK++V +C+ AG+ ++M+TGDNI TA+AIA ECGIL
Sbjct: 631 EEKL-----SLLGIIGIKDPCRPGVKKAVEDCQFAGVNIKMITGDNIFTARAIAVECGIL 685
Query: 524 TDDG-----IAIEGPDFREKTQEELFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAV 578
T + +EG FR TQEE E + +I+VMARSSP DK +VK L+ G VVAV
Sbjct: 686 TPEDEMNSEAVLEGEKFRNYTQEERLEKVERIKVMARSSPFDKLLMVKCLKE-LGHVVAV 744
Query: 579 TGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQK 638
TGDGTNDAPAL EADIGL+MGI GTEVAKES+D++ILDDNF+++ TV +WGR VY NIQK
Sbjct: 745 TGDGTNDAPALKEADIGLSMGIQGTEVAKESSDIVILDDNFASVATVLKWGRCVYNNIQK 804
Query: 639 FVQFQLTVNVVALLVNFTSACMTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKR 698
F+QFQLTVNV AL++NF +A G PLTAVQLLWVN+IMDTLGALALATE PT+DLMK+
Sbjct: 805 FIQFQLTVNVAALVINFVAAVSAGDVPLTAVQLLWVNLIMDTLGALALATEKPTNDLMKK 864
Query: 699 EPVGRKGDFINNVMWRNILGQALYQFVVIWFLQSVGKWVFFLRGPNADIVLNTLIFNTFV 758
+P+GR I N+MWRN+L QA YQ V+ LQ G+ +F + + V NTLIFNTFV
Sbjct: 865 KPIGRVAPLITNIMWRNLLAQAFYQISVLLVLQFRGRSIFNV----TEKVKNTLIFNTFV 920
Query: 759 FCQVFNEINSREMEEIDVFKGIWDNHVFVAVISATVVFQIIIVEYLGTFANTTPLSLVQW 818
CQVFNE N+R +E+ +VFKG+ N +F+ +I TVV Q+++VE+L FA+T L+L QW
Sbjct: 921 LCQVFNEFNARSLEKKNVFKGLHKNRLFIGIIVVTVVLQVVMVEFLKRFADTERLNLGQW 980
Query: 819 IFCLGVGYMGMPIAVRLKQIPV 840
C+ + PI +K +PV
Sbjct: 981 GVCIAIAAASWPIGWLVKSVPV 1002
>At1g27770 envelope Ca2+-ATPase
Length = 946
Score = 694 bits (1790), Expect = 0.0
Identities = 354/460 (76%), Positives = 397/460 (85%), Gaps = 4/460 (0%)
Query: 2 TLMILGVCAFVSLIVGVLTEGWPKGAHDGLGIVASILLVVFVTATSDYRQSLQFKDLDKE 61
TLMIL CAFVSLIVG+L EGWP GAHDGLGIVASILLVVFVTATSDYRQSLQFKDLD E
Sbjct: 175 TLMILAACAFVSLIVGILMEGWPIGAHDGLGIVASILLVVFVTATSDYRQSLQFKDLDAE 234
Query: 62 KKKISIQVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGESE 121
KKKI +QVTR+ RQK+SIY+LLPGD+VHL IGDQ+PADGLF+SGFSVLI+ESSLTGESE
Sbjct: 235 KKKIVVQVTRDKLRQKISIYDLLPGDVVHLGIGDQIPADGLFISGFSVLINESSLTGESE 294
Query: 122 PIMVTTQNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVA 181
P+ V+ ++PFLLSGTKVQDGSC MLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVA
Sbjct: 295 PVSVSVEHPFLLSGTKVQDGSCKMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVA 354
Query: 182 TIIGKIGLVFAVITFTVLVKGHLSHKIREGNFWRWTGDNAMEMLEYFAIAVTIVVVAVPE 241
TIIGKIGL FAVITF VLV+G + K + + W WT D M MLEYFA+AVTIVVVAVPE
Sbjct: 355 TIIGKIGLFFAVITFAVLVQGLANQKRLDNSHWIWTADELMAMLEYFAVAVTIVVVAVPE 414
Query: 242 GLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVKTCIC 301
GLPLAVTLSLAFAMKKMMNDKALVR+LAACETMGSATTICSDKTGTLTTN MTVVK CIC
Sbjct: 415 GLPLAVTLSLAFAMKKMMNDKALVRNLAACETMGSATTICSDKTGTLTTNHMTVVKACIC 474
Query: 302 MNSKEVSNSSS----SSDIPDSAAKLLLQSIFNNTGGEVVYNKKGKREILGTPTETAILE 357
+KEV+ + +S IP+SA KLLLQSIF NTGGE+V K K EILGTPTETA+LE
Sbjct: 475 EQAKEVNGPDAAMKFASGIPESAVKLLLQSIFTNTGGEIVVGKGNKTEILGTPTETALLE 534
Query: 358 FGLSLGGDSKAEREACKIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIILAACDKV 417
FGLSLGGD + R+A +VKVEPFNS KKRMGVV+E P+ RAHCKGASEI+L +CDK
Sbjct: 535 FGLSLGGDFQEVRQASNVVKVEPFNSTKKRMGVVIELPERHFRAHCKGASEIVLDSCDKY 594
Query: 418 IDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYMEL 457
I+ +G+VV LD +ST++L +II +FA+EALRTLCLAY E+
Sbjct: 595 INKDGEVVPLDEKSTSHLKNIIEEFASEALRTLCLAYFEI 634
Score = 501 bits (1290), Expect = e-142
Identities = 247/309 (79%), Positives = 282/309 (90%)
Query: 532 GPDFREKTQEELFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHE 591
GP+FREK+ EEL +LIPK+QVMARSSP+DKHTLV+ LRT F EVVAVTGDGTNDAPALHE
Sbjct: 635 GPEFREKSDEELLKLIPKLQVMARSSPMDKHTLVRLLRTMFQEVVAVTGDGTNDAPALHE 694
Query: 592 ADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNVVAL 651
ADIGLAMGI+GTEVAKESADVIILDDNFSTIVTVA+WGRSVYINIQKFVQFQLTVNVVAL
Sbjct: 695 ADIGLAMGISGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVAL 754
Query: 652 LVNFTSACMTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKREPVGRKGDFINNV 711
+VNF SAC+TG+APLTAVQLLWVNMIMDTLGALALATEPP DDLMKR PVGRKG+FI+NV
Sbjct: 755 IVNFLSACLTGNAPLTAVQLLWVNMIMDTLGALALATEPPQDDLMKRSPVGRKGNFISNV 814
Query: 712 MWRNILGQALYQFVVIWFLQSVGKWVFFLRGPNADIVLNTLIFNTFVFCQVFNEINSREM 771
MWRNILGQ+LYQ V+IW LQ+ GK +F L GP++D+ LNTLIFN FVFCQVFNEI+SREM
Sbjct: 815 MWRNILGQSLYQLVIIWCLQTKGKTMFGLDGPDSDLTLNTLIFNIFVFCQVFNEISSREM 874
Query: 772 EEIDVFKGIWDNHVFVAVISATVVFQIIIVEYLGTFANTTPLSLVQWIFCLGVGYMGMPI 831
E+IDVFKGI N+VFVAV++ TVVFQ+II+E LGTFA+TTPL+L QW+ + +G++GMP+
Sbjct: 875 EKIDVFKGILKNYVFVAVLTCTVVFQVIIIELLGTFADTTPLNLGQWLVSIILGFLGMPV 934
Query: 832 AVRLKQIPV 840
A LK IPV
Sbjct: 935 AAALKMIPV 943
>At5g53010 Ca2+-transporting ATPase-like protein
Length = 1095
Score = 396 bits (1018), Expect = e-110
Identities = 283/795 (35%), Positives = 411/795 (51%), Gaps = 77/795 (9%)
Query: 43 VTATSDYRQSLQFKDLDKEKKKISIQVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGL 102
+ A ++Y+QS +F L +EK+ + ++V R G R ++SIY+++ GDIV L G QVPADG+
Sbjct: 278 LAAVAEYKQSCRFIKLTEEKRTVYLEVIRGGRRVRVSIYDIVVGDIVPLKNGCQVPADGV 337
Query: 103 FVSGFSVLIDESSLTGESEPIMVTTQ-NPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMA 161
S+ + E +T E + Q NPFLLSG+K+ +G TMLVT+VGM T+WG L
Sbjct: 338 LFVANSLKVAEQEVTASDEIVQKDLQTNPFLLSGSKLIEGIGTMLVTSVGMNTEWG-LKM 396
Query: 162 TLSEGGDDETPLQVKLNGVATIIGKIGLVFAVITFTVLVKGHLSHKIREGN--------- 212
+S+ D+E P Q L +A ++FA + ++ V G + + N
Sbjct: 397 EVSQKTDEEKPFQGYLKWLAISASWFVVLFASVACSIQVGGSSAPSWQGPNNRFISRYFS 456
Query: 213 -------------FWRWTGDNAME-MLEYFAIAVTIVVVAVPEGLPLAVTL--------- 249
+ T D A+E ++ + + +VVAVP GL +AV L
Sbjct: 457 GVTKKSDGTPMFIYGITTADEAIEFVITSLSFGIATIVVAVPVGLSIAVRLNSSYHFPYF 516
Query: 250 -SLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVKTCICMNSKEVS 308
S A KKM DK L M+VV + +
Sbjct: 517 ISFAKTTKKMRKDKVL----------------------------MSVVD--VWAGGIRMQ 546
Query: 309 NSSSSSDIPDSAAKLLLQSIFNNTGGEVVYNKK-GKREILGTPTETAILEFGLSLGGDSK 367
+ S +P +L+++ I NT G VV+ + E+ G+PTE AIL FG LG
Sbjct: 547 DMDDVSQLPTFLKELIIEGIAQNTNGSVVFETGVTEPEVYGSPTEQAILNFGNKLGMKFD 606
Query: 368 AEREACKIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIILAACDKVIDLNGDVVAL 427
R A + PFN +KK GV + Q H KG+++ IL++C+ +D + A+
Sbjct: 607 DARSASLVRHTIPFNPKKKYGGVAL-QLGTHAHVHWKGSAKTILSSCEGYMDGANNSRAI 665
Query: 428 DGESTNYLNSIINQFANEALRTLCLAYMELENGFAAEDPIPASGYTCIGIVGIKDPVRPG 487
+ + I + E LR LAY E G P + + IVGIKDP RPG
Sbjct: 666 NEQKRKSFEGTIENMSKEGLRCAALAYQPCELGSLPTITEPRN-LVLLAIVGIKDPCRPG 724
Query: 488 VKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILTD-DGIAIE-GPDFREKTQEELFE 545
+ ++ C S + V MVT ++ TA+AIA ECGILTD G I G FRE + E +
Sbjct: 725 TRDAIQLCNSGSVKVCMVTDNDGLTAQAIAIECGILTDASGRNIRTGAQFRELSDLEREQ 784
Query: 546 LIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEV 605
+ I V A+SSP D LV+ L+ G +VA TG G +D L EAD+ LAMG+ GT
Sbjct: 785 IAGDILVFAQSSPNDNLLLVQALKKR-GHIVAATGMGIHDPKTLREADVSLAMGVGGTAA 843
Query: 606 AKESADVIILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNVVALLVNFTSACMTGSAP 665
AKE++D IILDDNF+TIV W RS+Y N+QK + F+LTV+V AL V + + P
Sbjct: 844 AKENSDFIILDDNFATIVKCIIWSRSLYNNVQKSILFRLTVSVSALAVCVVEVVVYDAFP 903
Query: 666 LTAVQLLWVNMIMDTLGALALATEPPTD-DLMKREPVGRKGDFINNVMWRNILGQALYQF 724
L AVQ L VN+I+D LGALALA P +D LM + PVG + I MW ++ Q Y
Sbjct: 904 LNAVQFLLVNLIIDILGALALAYRPRSDHHLMGKPPVGIRDPLITKTMWSKMIIQVFYLV 963
Query: 725 VVIWFLQSVGKWVFFLRGP--NADIVLNTLIFNTFVFCQVFNEINSREMEEIDVFKGIWD 782
+ + + S K + G NA+ ++NTLIFN+FVF VFNE + +++ FK +
Sbjct: 964 LSLVLINS-EKLLKLKHGQTGNAEKMMNTLIFNSFVFYLVFNEFEIQSVDQ--TFKEVLR 1020
Query: 783 NHVFVAVISATVVFQ 797
++F+ I++T++ Q
Sbjct: 1021 ENMFLVTITSTIISQ 1035
>At1g07670 endoplasmic reticulum-type calcium-transporting ATPase 4
(ECA4)
Length = 1061
Score = 350 bits (897), Expect = 2e-96
Identities = 281/912 (30%), Positives = 433/912 (46%), Gaps = 128/912 (14%)
Query: 37 ILLVVFVTATSDYRQSLQFKDLDKEKKKISIQ---VTRNGYR-QKMSIYNLLPGDIVHLN 92
I L++ V A Q + + K+I Q V R+G + + L+PGDIV L
Sbjct: 119 IFLILIVNAIVGIWQETNAEKALEALKEIQSQQATVMRDGTKVSSLPAKELVPGDIVELR 178
Query: 93 IGDQVPADGLFVSGFS--VLIDESSLTGESEPIMVTTQNP-----------FLLSGTKVQ 139
+GD+VPAD V+ S + +++ SLTGESE + TT++ + +GT V
Sbjct: 179 VGDKVPADMRVVALISSTLRVEQGSLTGESEAVSKTTKHVDENADIQGKKCMVFAGTTVV 238
Query: 140 DGSCTMLVTTVGMRTQWGKLMATLSEGG--DDETPLQVKLNGVATIIGKIGLVFAVITFT 197
+G+C LVT GM T+ G++ + + E +++TPL+ KLN ++ I + + +
Sbjct: 239 NGNCICLVTDTGMNTEIGRVHSQIQEAAQHEEDTPLKKKLNEFGEVLTMIIGLICALVWL 298
Query: 198 VLVKGHLSHKIREGNFWRWTGDNAMEMLEY-FAIAVTIVVVAVPEGLPLAVTLSLAFAMK 256
+ VK LS + +G W + E Y F IAV + V A+PEGLP +T LA +
Sbjct: 299 INVKYFLSWEYVDG--WPRNFKFSFEKCTYYFEIAVALAVAAIPEGLPAVITTCLALGTR 356
Query: 257 KMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVKTCICMNSK-------EVSN 309
KM ALVR L + ET+G T ICSDKTGTLTTN+M V K + M S+ V
Sbjct: 357 KMAQKNALVRKLPSVETLGCTTVICSDKTGTLTTNQMAVSKL-VAMGSRIGTLRSFNVEG 415
Query: 310 SSSS------SDIPDSAAKLLLQSIFNNTG--GEVVYNKKGKREIL-GTPTETAILEFGL 360
+S D P LQ I + K ++ + G PTE A+
Sbjct: 416 TSFDPRDGKIEDWPTGRMDANLQMIAKIAAICNDANVEKSDQQFVSRGMPTEAALKVLVE 475
Query: 361 SLG---GDSKAEREA-----CKI-------VKVEPFNSEKKRMGVVVEQPDGSVRAHCKG 405
+G G ++A + C++ + F+ ++K MGV+V+ G KG
Sbjct: 476 KMGFPEGLNEASSDGNVLRCCRLWSELEQRIATLEFDRDRKSMGVMVDSSSGKKLLLVKG 535
Query: 406 ASEIILAACDKVIDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYMELENGFAAED 465
A E +L + L+G LD S + + ++ + ALR L AY ++ + FA D
Sbjct: 536 AVENVLERSTHIQLLDGSTRELDQYSRDLILQSLHDMSLSALRCLGFAYSDVPSDFATYD 595
Query: 466 P----------IPASGYTCI-------GIVGIKDPVRPGVKQSVAECRSAGIVVRMVTGD 508
+ S Y+ I G VG++DP R V+Q++A+CR+AGI V ++TGD
Sbjct: 596 GSEDHPAHQQLLNPSNYSSIESNLVFVGFVGLRDPPRKEVRQAIADCRTAGIRVMVITGD 655
Query: 509 NINTAKAIARECGILTDD----GIAIEGPDFREKTQEELFELIPKIQVMARSSPLDKHTL 564
N +TA+AI RE G+ D ++ G +F + ++ + +R+ P K +
Sbjct: 656 NKSTAEAICREIGVFEADEDISSRSLTGKEFMDVKDQKNHLRQTGGLLFSRAEPKHKQEI 715
Query: 565 VKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVT 624
V+ L+ GEVVA+TGDG NDAPAL ADIG+AMGI+GTEVAKE++D+++ DDNFSTIV
Sbjct: 716 VRLLKED-GEVVAMTGDGVNDAPALKLADIGVAMGISGTEVAKEASDLVLADDNFSTIVA 774
Query: 625 VARWGRSVYINIQKFVQFQLTVNVVALLVNFTSACMTGSAPLTAVQLLWVNMIMDTLGAL 684
GRS+Y N++ F+++ ++ N+ + F +A + + VQLLWVN++ D A
Sbjct: 775 AVGEGRSIYNNMKAFIRYMISSNIGEVASIFLTAALGIPEGMIPVQLLWVNLVTDGPPAT 834
Query: 685 ALATEPPTDDLMKREPVGRKGDFINN-VMWRNI-----LGQALYQFVVIWFLQS------ 732
AL PP D+MK+ P I +++R + +G A +IW+ +
Sbjct: 835 ALGFNPPDKDIMKKPPRRSDDSLITAWILFRYMVIGLYVGVATVGVFIIWYTHNSFMGID 894
Query: 733 -------------------VGKWVFFLRGP-------------------NADIVLNTLIF 754
W F P I +TL
Sbjct: 895 LSQDGHSLVSYSQLAHWGQCSSWEGFKVSPFTAGSQTFSFDSNPCDYFQQGKIKASTLSL 954
Query: 755 NTFVFCQVFNEINSREMEEIDVFKGIWDNH--VFVAVISATVVFQIIIVEYLGTFANTTP 812
+ V ++FN +N+ + V W N + +S + F I+ V +L P
Sbjct: 955 SVLVAIEMFNSLNALSEDGSLVTMPPWVNPWLLLAMAVSFGLHFVILYVPFLAQVFGIVP 1014
Query: 813 LSLVQWIFCLGV 824
LSL +W+ L V
Sbjct: 1015 LSLNEWLLVLAV 1026
>At1g07810 ER-type Ca2+-pump protein
Length = 1061
Score = 348 bits (893), Expect = 7e-96
Identities = 284/915 (31%), Positives = 434/915 (47%), Gaps = 134/915 (14%)
Query: 37 ILLVVFVTATSDYRQSLQFKDLDKEKKKISIQ---VTRNGYR-QKMSIYNLLPGDIVHLN 92
I L++ V A Q + + K+I Q V R+G + + L+PGDIV L
Sbjct: 119 IFLILIVNAIVGIWQETNAEKALEALKEIQSQQATVMRDGTKVSSLPAKELVPGDIVELR 178
Query: 93 IGDQVPADGLFVSGFS--VLIDESSLTGESEPIMVTTQNP-----------FLLSGTKVQ 139
+GD+VPAD V+ S + +++ SLTGESE + TT++ + +GT V
Sbjct: 179 VGDKVPADMRVVALISSTLRVEQGSLTGESEAVSKTTKHVDENADIQGKKCMVFAGTTVV 238
Query: 140 DGSCTMLVTTVGMRTQWGKLMATLSEGG--DDETPLQVKLNGVATIIGKIGLVFAVITFT 197
+G+C LVT GM T+ G++ + + E +++TPL+ KLN ++ I + + +
Sbjct: 239 NGNCICLVTDTGMNTEIGRVHSQIQEAAQHEEDTPLKKKLNEFGEVLTMIIGLICALVWL 298
Query: 198 VLVKGHLSHKIREGNFWRWTGDNAMEMLEY-FAIAVTIVVVAVPEGLPLAVTLSLAFAMK 256
+ VK LS + +G W + E Y F IAV + V A+PEGLP +T LA +
Sbjct: 299 INVKYFLSWEYVDG--WPRNFKFSFEKCTYYFEIAVALAVAAIPEGLPAVITTCLALGTR 356
Query: 257 KMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVKTCICMNSK-------EVSN 309
KM ALVR L + ET+G T ICSDKTGTLTTN+M V K + M S+ V
Sbjct: 357 KMAQKNALVRKLPSVETLGCTTVICSDKTGTLTTNQMAVSKL-VAMGSRIGTLRSFNVEG 415
Query: 310 SSSS------SDIPDSAAKLLLQ------SIFNNTGGEVVYNKKGKREILGTPTETAILE 357
+S D P LQ +I N+ E + R G PTE A+
Sbjct: 416 TSFDPRDGKIEDWPMGRMDANLQMIAKIAAICNDANVEQSDQQFVSR---GMPTEAALKV 472
Query: 358 FGLSLG---GDSKAEREA-----CKI-------VKVEPFNSEKKRMGVVVEQPDGSVRAH 402
+G G ++A + C++ + F+ ++K MGV+V+ G+
Sbjct: 473 LVEKMGFPEGLNEASSDGDVLRCCRLWSELEQRIATLEFDRDRKSMGVMVDSSSGNKLLL 532
Query: 403 CKGASEIILAACDKVIDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYMELENGFA 462
KGA E +L + L+G LD S + + + + ALR L AY ++ + FA
Sbjct: 533 VKGAVENVLERSTHIQLLDGSKRELDQYSRDLILQSLRDMSLSALRCLGFAYSDVPSDFA 592
Query: 463 AEDP----------IPASGYTCI-------GIVGIKDPVRPGVKQSVAECRSAGIVVRMV 505
D + S Y+ I G VG++DP R V+Q++A+CR+AGI V ++
Sbjct: 593 TYDGSEDHPAHQQLLNPSNYSSIESNLIFVGFVGLRDPPRKEVRQAIADCRTAGIRVMVI 652
Query: 506 TGDNINTAKAIARECGILTDD----GIAIEGPDFREKTQEELFELIPKIQVMARSSPLDK 561
TGDN +TA+AI RE G+ D ++ G +F + ++ + +R+ P K
Sbjct: 653 TGDNKSTAEAICREIGVFEADEDISSRSLTGIEFMDVQDQKNHLRQTGGLLFSRAEPKHK 712
Query: 562 HTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFST 621
+V+ L+ GEVVA+TGDG NDAPAL ADIG+AMGI+GTEVAKE++D+++ DDNFST
Sbjct: 713 QEIVRLLKED-GEVVAMTGDGVNDAPALKLADIGVAMGISGTEVAKEASDMVLADDNFST 771
Query: 622 IVTVARWGRSVYINIQKFVQFQLTVNVVALLVNFTSACMTGSAPLTAVQLLWVNMIMDTL 681
IV GRS+Y N++ F+++ ++ N+ + F +A + + VQLLWVN++ D
Sbjct: 772 IVAAVGEGRSIYNNMKAFIRYMISSNIGEVASIFLTAALGIPEGMIPVQLLWVNLVTDGP 831
Query: 682 GALALATEPPTDDLMKREPVGRKGDFINN-VMWRNI-----LGQALYQFVVIWFLQS--- 732
A AL PP D+MK+ P I +++R + +G A +IW+ S
Sbjct: 832 PATALGFNPPDKDIMKKPPRRSDDSLITAWILFRYMVIGLYVGVATVGVFIIWYTHSSFM 891
Query: 733 ----------------------VGKWVFFLRGP-------------------NADIVLNT 751
W F P I +T
Sbjct: 892 GIDLSQDGHSLVSYSQLAHWGQCSSWEGFKVSPFTAGSQTFSFDSNPCDYFQQGKIKAST 951
Query: 752 LIFNTFVFCQVFNEINSREMEEIDVFKGIWDNH--VFVAVISATVVFQIIIVEYLGTFAN 809
L + V ++FN +N+ + V W N + +S + F I+ V +L
Sbjct: 952 LSLSVLVAIEMFNSLNALSEDGSLVTMPPWVNPWLLLAMAVSFGLHFVILYVPFLAQVFG 1011
Query: 810 TTPLSLVQWIFCLGV 824
PLSL +W+ L V
Sbjct: 1012 IVPLSLNEWLLVLAV 1026
>At4g00900 Ca2+-transporting ATPase - like protein
Length = 1054
Score = 343 bits (879), Expect = 3e-94
Identities = 267/823 (32%), Positives = 413/823 (49%), Gaps = 102/823 (12%)
Query: 5 ILGVCAFVSLIVGVLTE--GWPKGAHDGLGIVASILLVVFVTATSDYRQSLQFKDLD--K 60
IL AF+S ++ L E G G + +L+++ +++S K L+ K
Sbjct: 69 ILLGAAFISFVLAFLGEEHGSGSGFEAFVEPFVIVLILILNAVVGVWQESNAEKALEALK 128
Query: 61 EKKKISIQVTRNG-YRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGF---SVLIDESSL 116
E + S +V R+G + L+PGDIV LN+GD+VPAD + VSG ++ +++SSL
Sbjct: 129 EMQCESAKVLRDGNVLPNLPARELVPGDIVELNVGDKVPAD-MRVSGLKTSTLRVEQSSL 187
Query: 117 TGESEPIM------------VTTQNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLS 164
TGE+ P++ + + + +GT V +GSC +VT++GM T+ GK+ +
Sbjct: 188 TGEAMPVLKGANLVVMDDCELQGKENMVFAGTTVVNGSCVCIVTSIGMDTEIGKIQRQIH 247
Query: 165 EGG--DDETPLQVKLNGVATIIGKIGLVFAVITFTVLVKGHLSHKIREGNFWRWTGDNAM 222
E + ETPL+ KL+ + + + V+ + + K +S + +G +
Sbjct: 248 EASLEESETPLKKKLDEFGSRLTTAICIVCVLVWMINYKNFVSWDVVDGYKPVNIKFSFE 307
Query: 223 EMLEYFAIAVTIVVVAVPEGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICS 282
+ YF IAV + V A+PEGLP +T LA +KM A+VR L + ET+G T ICS
Sbjct: 308 KCTYYFKIAVALAVAAIPEGLPAVITTCLALGTRKMAQKNAIVRKLPSVETLGCTTVICS 367
Query: 283 DKTGTLTTNRMTVVKTCIC----MNSKEVSNSSSSSDIPDSA--------------AKLL 324
DKTGTLTTN+M+ + ++ S S ++ D D A
Sbjct: 368 DKTGTLTTNQMSATEFFTLGGKTTTTRVFSVSGTTYDPKDGGIVDWGCNNMDANLQAVAE 427
Query: 325 LQSIFNNTGGEVVYNKKGKREILGTPTETAILEFGLSLGGDSKAERE------------- 371
+ SI N+ G V Y K R G PTE A+ +G K E
Sbjct: 428 ICSICNDAG--VFYEGKLFRAT-GLPTEAALKVLVEKMGIPEKKNSENIEEVTNFSDNGS 484
Query: 372 ----AC--------KIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIILAACDKVID 419
AC K V F+ +K M V+V +P+G R KGA+E IL
Sbjct: 485 SVKLACCDWWNKRSKKVATLEFDRVRKSMSVIVSEPNGQNRLLVKGAAESILERSSFAQL 544
Query: 420 LNGDVVALDGESTNYLNSIINQFANEALRTLCLAYM----ELENGFAAEDP-----IPAS 470
+G +VALD S + ++ ++ LR L LAY E + + E P + S
Sbjct: 545 ADGSLVALDESSREVILKKHSEMTSKGLRCLGLAYKDELGEFSDYSSEEHPSHKKLLDPS 604
Query: 471 GYTCI-------GIVGIKDPVRPGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGIL 523
Y+ I G+VG++DP R V +++ +CR AGI V ++TGDN +TA+AI E +
Sbjct: 605 SYSNIETNLIFVGVVGLRDPPREEVGRAIEDCRDAGIRVMVITGDNKSTAEAICCEIRLF 664
Query: 524 TDDG----IAIEGPDFREKTQEELFELIPKI--QVMARSSPLDKHTLVKQLRTTFGEVVA 577
+++ + G +F E++ K +V +R+ P K +V+ L+ GE+VA
Sbjct: 665 SENEDLSQSSFTGKEFMSLPASRRSEILSKSGGKVFSRAEPRHKQEIVRMLKE-MGEIVA 723
Query: 578 VTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQ 637
+TGDG NDAPAL ADIG+AMGI GTEVAKE++D+++ DDNFSTIV+ GRS+Y N++
Sbjct: 724 MTGDGVNDAPALKLADIGIAMGITGTEVAKEASDMVLADDNFSTIVSAVAEGRSIYNNMK 783
Query: 638 KFVQFQLTVNVVALLVNFTSACMTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMK 697
F+++ ++ NV ++ F +A + + VQLLWVN++ D A AL P D+MK
Sbjct: 784 AFIRYMISSNVGEVISIFLTAALGIPECMIPVQLLWVNLVTDGPPATALGFNPADIDIMK 843
Query: 698 REPVGRKGD--------FINNVMWRNILGQALYQFVVIWFLQS 732
+ P RK D I ++ + +G A V+W+ Q+
Sbjct: 844 KPP--RKSDDCLIDSWVLIRYLVIGSYVGVATVGIFVLWYTQA 884
>At1g10130 putative calcium ATPase
Length = 992
Score = 310 bits (794), Expect = 2e-84
Identities = 258/857 (30%), Positives = 402/857 (46%), Gaps = 111/857 (12%)
Query: 69 VTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFV--SGFSVLIDESSLTGESEPI--- 123
V RNG + L+PGDIV + +G ++PAD + S + +D++ LTGES +
Sbjct: 122 VLRNGCFSILPATELVPGDIVEVTVGCKIPADLRMIEMSSNTFRVDQAILTGESCSVEKD 181
Query: 124 ---MVTTQNPF------LLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQ 174
+TT + L SGT V G +V VG T G + ++ + D+ TPL+
Sbjct: 182 VDCTLTTNAVYQDKKNILFSGTDVVAGRGRAVVIGVGSNTAMGSIHDSMLQTDDEATPLK 241
Query: 175 VKLNGVATIIGKIGLVFAVITFTVLVKGHLSHKIREGNFWRWTGDNAMEMLEYFAIAVTI 234
KL+ + + K+ V+ + V + GH S G F + YF IAV +
Sbjct: 242 KKLDEFGSFLAKVIAGICVLVWVVNI-GHFSDPSHGGFF--------KGAIHYFKIAVAL 292
Query: 235 VVVAVPEGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMT 294
V A+PEGLP VT LA KKM A+VR L + ET+G T ICSDKTGTLTTN M+
Sbjct: 293 AVAAIPEGLPAVVTTCLALGTKKMARLNAIVRSLPSVETLGCTTVICSDKTGTLTTNMMS 352
Query: 295 VVKTCICMNSKE--------------------VSNSSSSSDIPDSAAKL----LLQSIFN 330
V K C+ +++ ++ D+P + L + S+ N
Sbjct: 353 VSKICVVQSAEHGPMINEFTVSGTTYAPEGTVFDSNGMQLDLPAQSPCLHHLAMCSSLCN 412
Query: 331 NTGGEVVYNK-KGKREILGTPTETAILEFGLSLGGD------------SKAEREAC---- 373
++ + YN K E +G TE A+ +G SK ER +
Sbjct: 413 DS--ILQYNPDKDSYEKIGESTEVALRVLAEKVGLPGFDSMPSALNMLSKHERASYCNHY 470
Query: 374 -----KIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIILAACDKVIDLNGD--VVA 426
K V V F ++K M V+ V KGA E I+A C+K++ NGD VV
Sbjct: 471 WENQFKKVYVLEFTRDRKMMSVLCSHKQMDVM-FSKGAPESIIARCNKIL-CNGDGSVVP 528
Query: 427 LDGESTNYLNSIINQFANEALRTLCLAYMELENGFAAEDPIPASGYTCIGIVGIKDPVRP 486
L L S F +E LR L LA+ + +G + T IG+VG+ DP R
Sbjct: 529 LTAAGRAELESRFYSFGDETLRCLALAFKTVPHGQQTISYDNENDLTFIGLVGMLDPPRE 588
Query: 487 GVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILTD----DGIAIEGPDFREKTQEE 542
V+ ++ C +AGI V +VTGDN +TA+++ R+ G + G++ +F +
Sbjct: 589 EVRDAMLACMTAGIRVIVVTGDNKSTAESLCRKIGAFDNLVDFSGMSYTASEFERLPAVQ 648
Query: 543 LFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAG 602
+ ++ + +R P K LV+ L+ EVVA+TGDG NDAPAL +ADIG+AMG +G
Sbjct: 649 QTLALRRMTLFSRVEPSHKRMLVEALQKQ-NEVVAMTGDGVNDAPALKKADIGIAMG-SG 706
Query: 603 TEVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNVVALLVNFTSACMTG 662
T VAK ++D+++ DDNF++IV GR++Y N ++F+++ ++ N+ ++ F +A +
Sbjct: 707 TAVAKSASDMVLADDNFASIVAAVAEGRAIYNNTKQFIRYMISSNIGEVVCIFVAAVLGI 766
Query: 663 SAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKREPVGRKGD-------FINNVMWRN 715
L VQLLWVN++ D L A A+ D+MK +P + G+ F ++
Sbjct: 767 PDTLAPVQLLWVNLVTDGLPATAIGFNKQDSDVMKAKP-RKVGEAVVTGWLFFRYLVIGV 825
Query: 716 ILGQALYQFVVIWFLQSVG-----------------KWVFFLRGPNADIVLNTLIFNTFV 758
+G A + WF+ S G + + D +T+ V
Sbjct: 826 YVGLATVAGFIWWFVYSDGGPKLTYSELMNFETCALRETTYPCSIFEDRHPSTVAMTVLV 885
Query: 759 FCQVFNEINSREMEEIDVFKGIWDNHVFVAVISATVVFQIII--VEYLGTFANTTPLSLV 816
++FN +N+ + + N V I T++ ++I V L + TPLS
Sbjct: 886 VVEMFNALNNLSENQSLLVITPRSNLWLVGSIILTMLLHVLILYVHPLAVLFSVTPLSWA 945
Query: 817 QWIFCLGVGYMGMPIAV 833
+W L Y+ P+ +
Sbjct: 946 EWTAVL---YLSFPVII 959
>At1g17260 H+-transporting ATPase AHA10
Length = 946
Score = 189 bits (481), Expect = 4e-48
Identities = 158/593 (26%), Positives = 263/593 (43%), Gaps = 78/593 (13%)
Query: 65 ISIQVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGESEPIM 124
+ +V R+G Q+ L+PGDI+ + +GD +PAD + G + ID+S LTGES P+
Sbjct: 138 LKTRVLRDGQWQEQDASILVPGDIISIKLGDIIPADARLLEGDPLKIDQSVLTGESLPV- 196
Query: 125 VTTQNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATII 184
+ + SG+ + G +V G T +GK + D T + V T I
Sbjct: 197 TKKKGEQVFSGSTCKQGEIEAVVIATGSTTFFGKTARLV-----DSTDVTGHFQQVLTSI 251
Query: 185 GK-------IGLVFAVITFTVLVKGHLSHKIREGNFWRWTGDNAMEMLEYFAIAVTIVVV 237
G +G+V +I + H S++I N + +++
Sbjct: 252 GNFCICSIAVGMVLEIIIMFPVQ--HRSYRIGINNL------------------LVLLIG 291
Query: 238 AVPEGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVK 297
+P +P ++++LA ++ A+ + + A E M +C DKTGTLT N +TV K
Sbjct: 292 GIPIAMPTVLSVTLAIGSHRLSQQGAITKRMTAIEEMAGMDVLCCDKTGTLTLNSLTVDK 351
Query: 298 TCICMNSKEVSNSSSSSDIPDSAAKLLLQSIFNNTGGEVVYNKKGKREILGTPTETAILE 357
I D D LLL G+ L + AI
Sbjct: 352 NLI----------EVFVDYMDKDTILLLA---------------GRASRL--ENQDAIDA 384
Query: 358 FGLSLGGDSKAEREACKIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIILAACDKV 417
+S+ D + R + + PFN KR + DG KGA E +L C +
Sbjct: 385 AIVSMLADPREARANIREIHFLPFNPVDKRTAITYIDSDGKWYRATKGAPEQVLNLCQQK 444
Query: 418 IDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYMELENGFAAEDPIPASGYTCIGI 477
++ V A II++FA + LR+L +AY E+ + P + G+
Sbjct: 445 NEIAQRVYA-----------IIDRFAEKGLRSLAVAYQEIPE---KSNNSPGGPWRFCGL 490
Query: 478 VGIKDPVRPGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILTD--DGIAIEGPDF 535
+ + DP R +++ S G+ V+M+TGD + AK R G+ T+ ++ G +
Sbjct: 491 LPLFDPPRHDSGETILRALSLGVCVKMITGDQLAIAKETGRRLGMGTNMYPSSSLLGHNN 550
Query: 536 REKTQEELFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIG 595
E + ELI A P K+ +VK L+ VV +TGDG NDAPAL +ADIG
Sbjct: 551 DEHEAIPVDELIEMADGFAGVFPEHKYEIVKILQE-MKHVVGMTGDGVNDAPALKKADIG 609
Query: 596 LAMGIAGTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNV 648
+A+ A T+ A+ SAD+++ D S I++ R+++ ++ + + +++ +
Sbjct: 610 IAVADA-TDAARSSADIVLTDPGLSVIISAVLTSRAIFQRMRNYTVYAVSITI 661
>At1g80660 H+-transporting ATPase 9 (aha9)
Length = 954
Score = 185 bits (470), Expect = 8e-47
Identities = 152/586 (25%), Positives = 271/586 (45%), Gaps = 69/586 (11%)
Query: 68 QVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGESEPIMVTT 127
+V R+G + L+PGDI+ + +GD VPADG + G + ID+S+LTGES P+ T
Sbjct: 138 KVLRDGKWSEQEAAILVPGDIISIKLGDIVPADGRLLDGDPLKIDQSALTGESLPV---T 194
Query: 128 QNPF--LLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATIIG 185
++P + SG+ + G +V G+ T +GK + D T + V T IG
Sbjct: 195 KHPGQEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLV-----DSTNQEGHFQKVLTAIG 249
Query: 186 KIGLVFAVITFTVLVKGHLSHKIREGNFWRWTGDNAMEMLEYFAIAVTIVVVAVPEGLPL 245
+ I +L++ + + I++ +R DN + +L + +P +P
Sbjct: 250 NFCI--CSIAIGMLIEIVVMYPIQK-RAYRDGIDNLLVLL----------IGGIPIAMPT 296
Query: 246 AVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVKTCICMNSK 305
+++++A ++ A+ + + A E M +CSDKTGTLT N++TV K+ + + K
Sbjct: 297 VLSVTMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLTVDKSMVEVFVK 356
Query: 306 EVSNSSSSSDIPDSAAKLLLQSIFNNTGGEVVYNKKGKREILGTPTETAILEFGLSLGGD 365
++ Q + N V N + AI + + GD
Sbjct: 357 DLDKD---------------QLLVNAARASRVEN------------QDAIDACIVGMLGD 389
Query: 366 SKAEREACKIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIILAACDKVIDLNGDVV 425
+ RE V PFN KR + +G+ KGA E I+ C+
Sbjct: 390 PREAREGITEVHFFPFNPVDKRTAITYIDANGNWHRVSKGAPEQIIELCN---------- 439
Query: 426 ALDGESTNYLNSIINQFANEALRTLCLAYMELENGFAAEDPIPASGYTCIGIVGIKDPVR 485
L +++ + II++FA+ LR+L + + + P + +G++ + DP R
Sbjct: 440 -LREDASKRAHDIIDKFADRGLRSLAVGRQTVSE---KDKNSPGEPWQFLGLLPLFDPPR 495
Query: 486 PGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILTD--DGIAIEGPDFREKTQE-E 542
+++ G+ V+M+TGD + K R G+ T+ A+ G D E
Sbjct: 496 HDSAETIRRALDLGVNVKMITGDQLAIGKETGRRLGMGTNMYPSSALLGQDKDESIASLP 555
Query: 543 LFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAG 602
+ ELI K A P K+ +VK+L+ + +TGDG NDAPAL ADIG+A+ A
Sbjct: 556 VDELIEKADGFAGVFPEHKYEIVKRLQE-MKHICGMTGDGVNDAPALKRADIGIAVADA- 613
Query: 603 TEVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNV 648
T+ A+ ++D+++ + S IV+ R+++ ++ + + +++ +
Sbjct: 614 TDAARSASDIVLTEPGLSVIVSAVLTSRAIFQRMKNYTIYAVSITI 659
>At2g07560 pseudogene
Length = 949
Score = 183 bits (465), Expect = 3e-46
Identities = 155/589 (26%), Positives = 273/589 (46%), Gaps = 68/589 (11%)
Query: 68 QVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGESEPIMVTT 127
+V R+G + L+PGD++ + +GD VPAD + G + ID+S+LTGES P
Sbjct: 136 KVLRDGRWGEQEAAILVPGDLISIKLGDIVPADARLLEGDPLKIDQSALTGESLPA-TKH 194
Query: 128 QNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATI-IGK 186
Q + SG+ + G +V G+ T +GK A L + ++ Q L + I
Sbjct: 195 QGDEVFSGSTCKQGEIEAVVIATGVHTFFGKA-AHLVDSTNNVGHFQKVLTAIGNFCICS 253
Query: 187 IGLVFAVITFTVLVKGHLSH-KIREGNFWRWTGDNAMEMLEYFAIAVTIVVVAVPEGLPL 245
IG+ + +++ + H K R+G DN + +L + +P +P
Sbjct: 254 IGIGMLI---EIIIMYPIQHRKYRDGI------DNLLVLL----------IGGIPIAMPT 294
Query: 246 AVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVKTCICMNSK 305
+++++A ++ A+ + + A E M +CSDKTGTLT N++TV K I + SK
Sbjct: 295 VLSVTMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLTVDKNLIEVFSK 354
Query: 306 EVSNSSSSSDIPDSAAKLLLQSIFNNTGGEVVYNKKGKREILGTPTETAILEFGLSLGGD 365
+V D +LL + + R +T+I+ ++ GD
Sbjct: 355 DV----------DKDYVILLSA-------------RASRVENQDAIDTSIV----NMLGD 387
Query: 366 SKAEREACKIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIILAACDKVIDLNGDVV 425
K R V PFN +KR + +G KGA E I+ CD
Sbjct: 388 PKEARAGITEVHFLPFNPVEKRTAITYIDTNGEWHRCSKGAPEQIIELCD---------- 437
Query: 426 ALDGESTNYLNSIINQFANEALRTLCLAYMELENGFAAEDPIPASGYTCIGIVGIKDPVR 485
L GE+ + II++FA LR+L +A + P + +G++ + DP R
Sbjct: 438 -LKGETKRRAHEIIDKFAERGLRSLGVARQRVPEKDKESAGTP---WEFVGLLPLFDPPR 493
Query: 486 PGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILTDDGIAIEGPDFREKTQE--EL 543
+++ G+ V+M+TGD + K R G+ T+ + + ++ T +
Sbjct: 494 HDSAETIRRALDLGVNVKMITGDQLAIGKETGRRLGMGTNMYPSSSLLENKDDTTGGVPV 553
Query: 544 FELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGT 603
ELI K A P K+ +V++L+ +V +TGDG NDAPAL +ADIG+A+ A T
Sbjct: 554 DELIEKADGFAGVFPEHKYEIVRKLQER-KHIVGMTGDGVNDAPALKKADIGIAVDDA-T 611
Query: 604 EVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNVVALL 652
+ A+ ++D+++ + S IV+ R+++ ++ + + +++ + +L
Sbjct: 612 DAARSASDIVLTEPGLSVIVSAVLTSRAIFQRMKNYTIYAVSITIRIVL 660
>At3g60330 plasma membrane H+-ATPase - like
Length = 961
Score = 179 bits (455), Expect = 4e-45
Identities = 161/647 (24%), Positives = 287/647 (43%), Gaps = 80/647 (12%)
Query: 22 GWPKGAHDGLGIVASILL---VVFVTATSDYRQSLQFKDLDKEKKKISIQVTRNGYRQKM 78
G P HD +GIV +L+ + FV + + K K R+G ++
Sbjct: 91 GKPADYHDFVGIVVLLLINSTISFVEENNAGNAAAALMAQLAPKAK----AVRDGKWNEI 146
Query: 79 SIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGESEPIMVTTQNP--FLLSGT 136
L+PGDIV + +GD +PAD + G + ID+++LTGES P+ T+NP + SG+
Sbjct: 147 DAAELVPGDIVSIKLGDIIPADARLLEGDPLKIDQATLTGESLPV---TKNPGASVYSGS 203
Query: 137 KVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATIIGKIGLVFAVITF 196
+ G +V G+ T +GK + D T V T IG + +
Sbjct: 204 TCKQGEIEAVVIATGVHTFFGKAAHLV-----DSTTHVGHFQKVLTAIGNFCICSIAVGM 258
Query: 197 T---VLVKGHLSHKIREGNFWRWTGDNAMEMLEYFAIAVTIVVVAVPEGLPLAVTLSLAF 253
V++ G R G DN + +L + +P +P +++++A
Sbjct: 259 AIEIVVIYGLQKRGYRVGI------DNLLVLL----------IGGIPIAMPTVLSVTMAI 302
Query: 254 AMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVKTCICMNSKEVSNSSSS 313
++ A+ + + A E M +CSDKTGTLT N+++V K I + + + +
Sbjct: 303 GAHRLAQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFKRGIDRDMAV 362
Query: 314 SDIPDSAAKLLLQSIFNNTGGEVVYNKKGKREILGTPTETAILEFGLSLGGDSKAEREAC 373
+ AA+L Q +TAI+ S+ D K R
Sbjct: 363 L-MAARAARLENQDAI----------------------DTAIV----SMLSDPKEARAGI 395
Query: 374 KIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIILAACDKVIDLNGDVVALDGESTN 433
K + PF+ +R + +G + KGA E IL +++ V
Sbjct: 396 KELHFLPFSPANRRTALTYLDGEGKMHRVSKGAPEEILDMAHNKLEIKEKV--------- 446
Query: 434 YLNSIINQFANEALRTLCLAYMELENGFAAEDPIPASGYTCIGIVGIKDPVRPGVKQSVA 493
++ I++FA LR+L LAY E+ +G + P + + ++ + DP R Q++
Sbjct: 447 --HATIDKFAERGLRSLGLAYQEVPDGDVKGEGGP---WDFVALLPLFDPPRHDSAQTIE 501
Query: 494 ECRSAGIVVRMVTGDNINTAKAIARECGILTDDGIAIEGPDFREKTQEELFELIPKIQVM 553
G+ V+M+TGD + AK R G+ T+ + + ELI
Sbjct: 502 RALHLGVSVKMITGDQLAIAKETGRRLGMGTNMYPSSSLLSDNNTEGVSVDELIENADGF 561
Query: 554 ARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVI 613
A P K+ +VK+L++ + +TGDG NDAPAL +ADIG+A+ A T+ A+ ++D++
Sbjct: 562 AGVFPEHKYEIVKRLQSR-KHICGMTGDGVNDAPALKKADIGIAVDDA-TDAARGASDIV 619
Query: 614 ILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNVVALLVNFTSACM 660
+ + S I++ R+++ ++ + + +++ + +++ F C+
Sbjct: 620 LTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSI-TIRIVMGFMLLCV 665
>At2g24520 putative plasma membrane proton ATPase
Length = 931
Score = 174 bits (440), Expect = 2e-43
Identities = 148/587 (25%), Positives = 267/587 (45%), Gaps = 71/587 (12%)
Query: 68 QVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGESEPIMVTT 127
+V R+ + L+PGD++ + +GD +PAD + G + ID+SSLTGES P+ T
Sbjct: 115 KVLRDNQWSEQEASILVPGDVISIKLGDIIPADARLLDGDPLKIDQSSLTGESIPV---T 171
Query: 128 QNPF--LLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATIIG 185
+NP + SG+ + G +V G+ T +GK + D T V T IG
Sbjct: 172 KNPSDEVFSGSICKQGEIEAIVIATGVHTFFGKAAHLV-----DNTNQIGHFQKVLTSIG 226
Query: 186 KIGLVFAVITFTVLVKGHLSHKIREGNFWRWTGDNAMEMLEYFAIAVTIVVVAVPEGLPL 245
+ I ++V+ + + I+ + R DN + +L + +P +P
Sbjct: 227 NFCI--CSIALGIIVELLVMYPIQRRRY-RDGIDNLLVLL----------IGGIPIAMPS 273
Query: 246 AVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVKTCICMNSK 305
+++++A ++ A+ + + A E M +C DKTGTLT N++TV K + + +K
Sbjct: 274 VLSVTMATGSHRLFQQGAITKRMTAIEEMAGMDVLCCDKTGTLTLNKLTVDKNLVEVFAK 333
Query: 306 EVSNSSSSSDIPDSAAKLLLQSIFNNTGGEVVYNKKGKREILGTPTETAILEFGLSLGGD 365
V G E V+ + + + AI + + D
Sbjct: 334 GV-------------------------GKEHVFLLAARASRI--ENQDAIDAAIVGMLAD 366
Query: 366 SKAEREACKIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIILAACDKVIDLNGDVV 425
K R + V PFN KR + DG+ KGA E IL C+ D+ V
Sbjct: 367 PKEARAGVREVHFFPFNPVDKRTALTYVDSDGNWHRASKGAPEQILNLCNCKEDVRRKV- 425
Query: 426 ALDGESTNYLNSIINQFANEALRTLCLAYMELENGFAAEDPIPASGYTCIGIVGIKDPVR 485
+ +I++FA LR+L +A E+ + P + +G++ + DP R
Sbjct: 426 ----------HGVIDKFAERGLRSLAVARQEV---LEKKKDAPGGPWQLVGLLPLFDPPR 472
Query: 486 PGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILTD--DGIAIEGPDFREKTQEEL 543
+++ + G+ V+M+TGD + K R G+ T+ A+ G ++ + L
Sbjct: 473 HDSAETIRRALNLGVNVKMITGDQLAIGKETGRRLGMGTNMYPSSALLG-QVKDSSLGAL 531
Query: 544 --FELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIA 601
ELI K A P K+ +V +L+ + +TGDG NDAPAL +ADIG+A+ +
Sbjct: 532 PVDELIEKADGFAGVFPEHKYEIVHRLQQR-NHICGMTGDGVNDAPALKKADIGIAV-VD 589
Query: 602 GTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNV 648
T+ A+ ++D+++ + S I++ R+++ ++ + + +++ +
Sbjct: 590 ATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITI 636
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.137 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,732,021
Number of Sequences: 26719
Number of extensions: 745483
Number of successful extensions: 2004
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 47
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1730
Number of HSP's gapped (non-prelim): 124
length of query: 840
length of database: 11,318,596
effective HSP length: 108
effective length of query: 732
effective length of database: 8,432,944
effective search space: 6172915008
effective search space used: 6172915008
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC147000.2


