

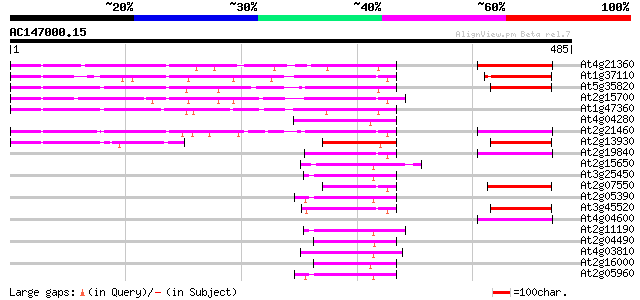
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147000.15 - phase: 0 /pseudo
(485 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g21360 putative transposable element 90 2e-18
At1g37110 79 4e-15
At5g35820 copia-like retrotransposable element 75 6e-14
At2g15700 copia-like retroelement pol polyprotein 75 6e-14
At1g47360 polyprotein, putative 68 1e-11
At4g04280 putative transposon protein 62 9e-10
At2g21460 putative retroelement pol polyprotein 57 3e-08
At2g13930 putative retroelement pol polyprotein 54 1e-07
At2g19840 copia-like retroelement pol polyprotein 53 3e-07
At2g15650 putative retroelement pol polyprotein 52 7e-07
At3g25450 hypothetical protein 50 3e-06
At2g07550 putative retroelement pol polyprotein 49 5e-06
At2g05390 putative retroelement pol polyprotein 49 5e-06
At3g45520 copia-like polyprotein 48 1e-05
At4g04600 putative polyprotein 47 2e-05
At2g11190 putative retroelement pol polyprotein 47 2e-05
At2g04490 putative retroelement pol polyprotein 47 2e-05
At4g03810 putative retrotransposon protein 45 1e-04
At2g16000 putative retroelement pol polyprotein 44 3e-04
At2g05960 putative retroelement pol polyprotein 44 3e-04
>At4g21360 putative transposable element
Length = 1308
Score = 90.1 bits (222), Expect = 2e-18
Identities = 96/353 (27%), Positives = 165/353 (46%), Gaps = 38/353 (10%)
Query: 1 FNQICKDSGIVRHKQIRGTPRENCLDH*RMDRKIMERFICLMSNANNSTLFWDETV**FF 60
F++ CK +GI RH+ TP++N + RM+R +ME+ CL++ + +FW E
Sbjct: 567 FDEFCKQNGIERHRTCTYTPQQNGVAK-RMNRTLMEKVRCLLNESGLEEVFWAEAAATAA 625
Query: 61 LIS*IDSLTLNLV*RH*KRCGWSCSKP*-KYESFWLCPLYAHIKRTILN-QGLKLYVFEV 118
+ ++ + V + W KP K+ + C Y H+ + L + LK
Sbjct: 626 YL--VNRSPASAVDHNVPEELWLDKKPGYKHLRRFGCIAYVHLDQGKLKPRALKGVFLGY 683
Query: 119 SLRCKRVQIWCMEKGET*VLINKDVVFKESEMFYSKETSS*H--QP*LDCEERVQFKVE- 175
K ++W +++ + +I++++VF E++++ SS + D E +F+V
Sbjct: 684 PQGTKGYKVWLLDEEKC--VISRNIVFNENQVYKDIRESSEQSVKDISDLEGYNEFQVSV 741
Query: 176 ------SLHGSTNLDAKVKDKD*NDDTTPLFFLMENVAIRF*ISFSSSFRFKLRTSTS-- 227
S G ++ ++ D + T E + +S S R + R + +
Sbjct: 742 KEHGECSKTGGVTIEEIDQESDSENSVT-----QEPLIASIDLSNYQSARDRERRAPNPP 796
Query: 228 LSLSSNTTLEN*L**KK*VVVSGEIDGDEPVN*DGFVNASKSREWI---ASMKEELSSLH 284
L+ T L V++ EI+ +EP + +A K + WI MKEE+ SL
Sbjct: 797 QKLADYTHFALAL------VMAEEIESEEP---QCYHDAKKDKHWIKWNGGMKEEIDSLL 847
Query: 285 KNGTWLLVNKPKRMKNMSCI*LFKLKEGVNYMD---*NSRFKARGFT*REGID 334
KNGTW +V PK K +SC LFKLK G+ ++ +R ARGFT ++GID
Sbjct: 848 KNGTWDIVEWPKEQKVISCRWLFKLKPGIPGVEAQRYKARLVARGFTQQKGID 900
Score = 52.0 bits (123), Expect = 7e-07
Identities = 28/65 (43%), Positives = 40/65 (61%)
Query: 405 YLNETMHKYVDYMILKIDDMLIACNIKEEFDKVKVML*IEFQMKDMRAAS*ILAKKILRD 464
Y+ E Y++L +DDML+A K E K+K L ++F+MKDM AAS IL I+R+
Sbjct: 1003 YVKEVNPGEFVYLLLYVDDMLLAAKSKSEISKLKEALSLKFEMKDMGAASRILGIDIIRN 1062
Query: 465 ISKGT 469
+GT
Sbjct: 1063 RKEGT 1067
>At1g37110
Length = 1356
Score = 79.3 bits (194), Expect = 4e-15
Identities = 92/370 (24%), Positives = 156/370 (41%), Gaps = 62/370 (16%)
Query: 1 FNQICKDSGIVRHKQIRGTPRENCLDH*RMDRKIMERFICLMSNANNSTLFWDETV**FF 60
F+ CK+ GI RH+ TP++N + RM+R IME+ CL++ + +FW E
Sbjct: 576 FDSYCKEHGIERHRTCTYTPQQNGVAE-RMNRTIMEKVRCLLNKSGVEEVFWAEAA---- 630
Query: 61 LIS*IDSLTLNLV*RH*KRCGWSCSKP*KYESFWLC--PLYAHIKR------------TI 106
+ L+ R S E WL P Y H+++ +
Sbjct: 631 ------ATAAYLI----NRSPASAINHNVPEEMWLNRKPGYKHLRKFGSIAYVHQDQGKL 680
Query: 107 LNQGLKLYVFEVSLRCKRVQIWCMEKGET*VLINKDVVFKESEMFYS-----KETSS*HQ 161
+ LK + K ++W +E+ + +I+++VVF+ES ++ +T + +Q
Sbjct: 681 KPRALKGFFLGYPAGTKGYKVWLLEEEKC--VISRNVVFQESVVYRDLKVKEDDTDNLNQ 738
Query: 162 P*LDCEERVQFKVESLHGSTNLDAKVKDKD*---------NDDTTPLFFLMENVAIRF*I 212
E Q K GS + D + +++ + R +
Sbjct: 739 KETTSSEVEQNKFAEASGSGGVIQLQSDSEPITEGEQSSDSEEEVEYSEKTQETPKRTGL 798
Query: 213 SFSSSFRFKLRTS----TSLSLSSNTTLEN*L**KK*VVVSGEIDGDEPVN*DGFVNASK 268
+ R ++R + T + S+ T +VV EP + + +
Sbjct: 799 TTYKLARDRVRRNINPPTRFTEESSVTFA--------LVVVENCIVQEPQSYQEAMESQD 850
Query: 269 SREWIASMKEELSSLHKNGTWLLVNKPKRMKNMSCI*LFKLKEGVNYMD*NSRFKAR--- 325
+W + +E+ SL KNGTW LV+KPK K + C LFKLK G+ ++ +RFKAR
Sbjct: 851 CEKWDMATHDEMDSLMKNGTWDLVDKPKDRKIIGCRWLFKLKSGIPGVE-PTRFKARLVA 909
Query: 326 -GFT*REGID 334
G+T REG+D
Sbjct: 910 KGYTQREGVD 919
Score = 47.8 bits (112), Expect = 1e-05
Identities = 28/58 (48%), Positives = 36/58 (61%), Gaps = 1/58 (1%)
Query: 411 HKYVDYMILKIDDMLIACNIKEEFDKVKVML*IEFQMKDMRAAS*ILAKKILRDISKG 468
H ++ Y++L +DDMLIA K E ++VK L EF+MKDM AS IL I RD G
Sbjct: 1029 HDFI-YLLLYVDDMLIAGASKAEINRVKEQLSTEFEMKDMGGASRILGIDIYRDRKGG 1085
>At5g35820 copia-like retrotransposable element
Length = 1342
Score = 75.5 bits (184), Expect = 6e-14
Identities = 86/363 (23%), Positives = 157/363 (42%), Gaps = 52/363 (14%)
Query: 1 FNQICKDSGIVRHKQIRGTPRENCLDH*RMDRKIMERFICLMSNANNSTLFWDETV**-F 59
F+ C+ G++RH+ TP++N + RM+R IM + C++S + FW E
Sbjct: 564 FDSFCRKEGVIRHRTCAYTPQQNGVAE-RMNRTIMNKVRCMLSESGLGKQFWAEAASTAV 622
Query: 60 FLIS*IDSLTLNLV*RH*KRCGWSCSKP*-KYESFWLCPLYAHIKRTILNQGLKLYVF-E 117
FLI+ S ++ K W+ P K + Y H + LN K +F
Sbjct: 623 FLINKSPSSSIEFDIPEEK---WTGHPPDYKILKKFGSVAYIHSDQGKLNPRAKKGIFLG 679
Query: 118 VSLRCKRVQIWCMEKGET*VLINKDVVFKESEMF--YSKETSS*HQP*LDCEERVQFKVE 175
KR ++W +E + ++++D+VF+E++M+ K S L ER +++
Sbjct: 680 YPDGVKRFKVWLLEDRKC--VVSRDIVFQENQMYKELQKNDMSEEDKQLTEVERTLIELK 737
Query: 176 SLHG---------------------STNLDAKVKDKD*NDDTTPLFFLMENVAIRF*ISF 214
+L S + D +V++ D +DD + L + R
Sbjct: 738 NLSADDENQSEGGDNSNQEQASTTRSASKDKQVEETDSDDDCLENYLLARDRIRR---QI 794
Query: 215 SSSFRFKLRTSTSLSLSSNTTLEN*L**KK*VVVSGEIDGDEPVN*DGFVNASKSREWIA 274
+ RF + + + T + GE+ EP + + + + +W
Sbjct: 795 RAPQRFVEEDDSLVGFALTMTED------------GEVY--EPETYEEAMRSPECEKWKQ 840
Query: 275 SMKEELSSLHKNGTWLLVNKPKRMKNMSCI*LFKLKEGVNYMD---*NSRFKARGFT*RE 331
+ EE+ S+ KN TW +++KP+ + + C +FK K G+ ++ +R A+GF+ RE
Sbjct: 841 ATIEEMDSMKKNDTWDVIDKPEGKRVIGCKWIFKRKAGIPGVEPPRYKARLVAKGFSQRE 900
Query: 332 GID 334
GID
Sbjct: 901 GID 903
Score = 44.3 bits (103), Expect = 2e-04
Identities = 23/53 (43%), Positives = 34/53 (63%)
Query: 416 YMILKIDDMLIACNIKEEFDKVKVML*IEFQMKDMRAAS*ILAKKILRDISKG 468
Y++L +DDMLIA K++ K+K L EF+MKD+ A IL +I R+ +G
Sbjct: 1017 YLLLYVDDMLIASQNKDQIQKLKESLNREFEMKDLGPARKILGMEITRNREQG 1069
>At2g15700 copia-like retroelement pol polyprotein
Length = 1166
Score = 75.5 bits (184), Expect = 6e-14
Identities = 91/387 (23%), Positives = 168/387 (42%), Gaps = 70/387 (18%)
Query: 1 FNQICKDSGIVRHKQIRGTPRENCLDH*RMDRKIMERFICLMSNANNSTLFWDETV**FF 60
F+ +CK+ GI RH+ TP++N + RM++ IM+ +++ S FW E
Sbjct: 460 FDNLCKEDGIKRHRTCTYTPQQNGVSE-RMNKTIMDIVRSMLAETGMSQEFWAEAT--ST 516
Query: 61 LIS*IDSLTLNLV*RH*KRCGWSCSKP*-KYESFWLCPLYAHIKRTILNQGLKLYVFEVS 119
+ I+ + + W+ +KP + + C Y H+ + + VF +
Sbjct: 517 AVYLINRTPNSFIGFKLPEEVWTGTKPDLSHLRRFGCSAYVHVTQDKTSPRAVKGVF-MG 575
Query: 120 LRC--KRVQIWCMEKGET*VLINKDVVFKESEMFYS---------KETSS*HQP*LDCEE 168
C K ++W ++G+ +++VVF E+E++ +E ++ +
Sbjct: 576 YPCGIKGYRVWLPKEGKC--TTSRNVVFNETELYKDTLSSADERKEEAEKEYKKLKKARK 633
Query: 169 RVQFKVESLHG-------------------------STNLDAKVKDKD*----NDDTTPL 199
RV F + L G S NL+ +++ N+ +
Sbjct: 634 RVSFSHDLLRGPSTSCCDLDDSSSQGGETSSSSSESSENLEESEMNEEVVGSENEQSLDD 693
Query: 200 FFLMENVAIRF*ISFSSSFRFKLRTSTSLSLSSNTTLEN*L**KK*VVVSGEIDGDEPVN 259
+ L ++ R I S RF+ + +L++ LE +EP +
Sbjct: 694 YLLARDMKRRSNIRPPS--RFEDEDFVAYALATAEDLEE----------------EEPKS 735
Query: 260 *DGFVNASKSREWIASMKEELSSLHKNGTWLLVNKPKRMKNMSCI*LFKLKEGVNYMD*N 319
+ + +SK ++W +MKEE+ S K+ TW L+ KP++ K + C +FKLK G+ ++
Sbjct: 736 YEEALKSSKRKQWENAMKEEMDSHKKSHTWDLIEKPEKQKLIGCKWIFKLKPGIPGVE-K 794
Query: 320 SRFKAR----GFT*REGID*CENYFLV 342
R+KAR GF+ +EGID E + LV
Sbjct: 795 QRYKARLVAKGFSQQEGIDYNEVFSLV 821
>At1g47360 polyprotein, putative
Length = 1182
Score = 68.2 bits (165), Expect = 1e-11
Identities = 92/356 (25%), Positives = 155/356 (42%), Gaps = 38/356 (10%)
Query: 1 FNQICKDSGIVRHKQIRGTPRENCLDH*RMDRKIMERFICLMSNANNSTLFWDETV-**F 59
F + CKD GIVRHK TP++N + R++R IM++ ++S + FW E
Sbjct: 514 FEKFCKDEGIVRHKTCAYTPQQNGIAE-RLNRTIMDKVRSMLSESGMDKKFWAEAASTAV 572
Query: 60 FLIS*IDSLTLNLV*RH*KRCGWSCSKP-*KYESFWLCPLYAHIKRTILNQGLKLYVFEV 118
+LI+ S L+ K W+ + P K + C +Y H + LN K VF
Sbjct: 573 YLINRSPSTALDFDLPEEK---WTGALPDLKGLRKFGCLVYIHADQGKLNPRAKKDVFTS 629
Query: 119 SLR-CKRVQIWCMEKGET*VLINKDVVFKESEMF----YSKETS---------S*HQP*L 164
L K ++W +E E +I+++V+F+E M+ + TS + P +
Sbjct: 630 YLEGVKGYKVWVLE--EKKCVISRNVIFREEIMYKDLKHDSHTSMIENDLENIRLNPPVV 687
Query: 165 DCEERVQFKVESLHGSTNLDAKVKDKD*NDDTTPLFFLMENVAIRF*ISFSSSFRFKLRT 224
C+ + + + T D +D + + T EN + S ++L
Sbjct: 688 TCDHEITDQGGATVTDTYQDDSAQDAN-SPVITQTQVSNENQSKSEEEDLSD---YQLVR 743
Query: 225 STSLSLSSNTTLEN*L**KK*VVVSGEIDGDEPVN*DGFVNASKSREW---IASMKEELS 281
++ ++ N V S + E + + A + EW +MKEE+
Sbjct: 744 DRAVKINPRYNESN------LVGFSFFTEDGEQAEPNSYQAALRDPEWDKWNEAMKEEMM 797
Query: 282 SLHKNGTWLLVNKPKRMKNMSCI*LFKLKEGVNYMD---*NSRFKARGFT*REGID 334
S+ KN T LV KP+ +K + C +F K G+ ++ +R A+GFT +EGID
Sbjct: 798 SMGKNQTLDLVEKPEGVKIIGCRWIFTRKTGIQGVEAPRYKARLVAKGFTQKEGID 853
>At4g04280 putative transposon protein
Length = 1104
Score = 61.6 bits (148), Expect = 9e-10
Identities = 37/92 (40%), Positives = 51/92 (55%), Gaps = 3/92 (3%)
Query: 246 VVVSGEIDGDEPVN*DGFVNASKSREWIASMKEELSSLHKNGTWLLVNKPKRMKNMSCI* 305
+V++ E++ +EPV +W M EE+ SL KN TW +V+KPK K +SC
Sbjct: 664 LVMAEEVESEEPVCFHDVKEDKDWEKWHGGMIEEMDSLLKNATWDIVDKPKNQKVISCHW 723
Query: 306 LFKLK---EGVNYMD*NSRFKARGFT*REGID 334
L+K K GV +R ARGF+ REGID
Sbjct: 724 LYKKKLGIPGVELPRYKARLVARGFSHREGID 755
Score = 32.7 bits (73), Expect = 0.46
Identities = 19/37 (51%), Positives = 25/37 (67%)
Query: 433 EFDKVKVML*IEFQMKDMRAAS*ILAKKILRDISKGT 469
E K+K +L EF+MKDM AAS L I+R+ S+GT
Sbjct: 850 EIKKLKKVLSREFEMKDMGAASRKLGIDIIRNRSEGT 886
Score = 32.3 bits (72), Expect = 0.60
Identities = 16/37 (43%), Positives = 23/37 (61%), Gaps = 1/37 (2%)
Query: 1 FNQICKDSGIVRHKQIRGTPRENCLDH*RMDRKIMER 37
FN C +GI RH+ TP++N + RM+R IME+
Sbjct: 481 FNDYCAKTGIERHRTCTYTPQQNGVTK-RMNRTIMEK 516
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 56.6 bits (135), Expect = 3e-08
Identities = 93/365 (25%), Positives = 157/365 (42%), Gaps = 56/365 (15%)
Query: 1 FNQICKDSGIVRHKQIRGTPRENCLDH*RMDRKIMERFICLMSNANNSTLFWDE-TV**F 59
F++ C GI+ H+ TP++N + RM+R +ME+ ++S++ FW E T
Sbjct: 555 FDEFCSQKGILWHRTCAYTPQQNGVAE-RMNRTLMEKVRSMLSDSGLPKKFWAEATHTTA 613
Query: 60 FLIS*IDSLTLNLV*RH*KRCGWSCSKP*-KYESFWLCPLYAHIKRTILNQGLKLYVFE- 117
LI+ S LN KR WS P Y + C + H LN K +
Sbjct: 614 ILINKTPSSALNYEVPD-KR--WSGKSPIYSYLRRFGCIAFVHTDDGKLNPRAKKGILVG 670
Query: 118 VSLRCKRVQIWCMEKGET*VLINKDVVFKES----EMFYSKET---------SS*HQP*L 164
+ K +IW +E+ + +++++V+F+E+ +M SK+ SS L
Sbjct: 671 YPIGVKGYKIWLLEEKKC--VVSRNVIFQENASYKDMMQSKDAEKDENEAPPSSYLDLDL 728
Query: 165 DCEERVQ-------FKVESLHGSTNLDAKVKDKD*NDDT----TPLFFLMENVAIRF*IS 213
D EE + + +S + + + N +T +PL + + V R +
Sbjct: 729 DHEEVITSGGDDPIVEAQSPFNPSPATTQTYSEGVNSETDIIQSPLSYQL--VRDRDRRT 786
Query: 214 FSSSFRFKLRTSTSLSLSSNTTLEN*L**KK*VVVSGEIDGDEPVN*DGFVNASKSREWI 273
+ RF L+ + TT + SGEI EP + + +W
Sbjct: 787 IRAPVRFD--DEDYLAEALYTTED-----------SGEI---EPADYSEAKRSMNWNKWK 830
Query: 274 ASMKEELSSLHKNGTWLLVNKPKRMKNMSCI*LFKLKEGVNYMD*NSRFKAR----GFT* 329
+M EE+ S KN TW +V +P+ K + ++K K G+ ++ RFKAR G+
Sbjct: 831 LAMNEEMESQIKNHTWTVVKRPQHQKVIGSRWIYKFKLGIPGVE-EGRFKARLVAKGYAQ 889
Query: 330 REGID 334
R+GID
Sbjct: 890 RKGID 894
Score = 44.3 bits (103), Expect = 2e-04
Identities = 24/65 (36%), Positives = 37/65 (56%)
Query: 405 YLNETMHKYVDYMILKIDDMLIACNIKEEFDKVKVML*IEFQMKDMRAAS*ILAKKILRD 464
Y+ E Y++L +DDML+A KE+ ++K L F MKD+ AA IL +I+R+
Sbjct: 997 YIKELSDGSRVYLLLYVDDMLVAAKNKEDISQLKEELSQRFDMKDLGAAKRILGMEIIRN 1056
Query: 465 ISKGT 469
+ T
Sbjct: 1057 REENT 1061
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 54.3 bits (129), Expect = 1e-07
Identities = 28/67 (41%), Positives = 44/67 (64%), Gaps = 3/67 (4%)
Query: 271 EWIASMKEELSSLHKNGTWLLVNKPKRMKNMSCI*LFKLKEGVNYMD*N---SRFKARGF 327
+W A+MKEE+ S+ KN TW LV KP+++K + C +F K G+ ++ +R A+GF
Sbjct: 830 KWNAAMKEEMVSMSKNHTWDLVTKPEKVKLIGCRWVFTRKAGIPGVEAPRFIARLVAKGF 889
Query: 328 T*REGID 334
T +EG+D
Sbjct: 890 TQKEGVD 896
Score = 48.1 bits (113), Expect = 1e-05
Identities = 44/155 (28%), Positives = 74/155 (47%), Gaps = 11/155 (7%)
Query: 1 FNQICKDSGIVRHKQIRGTPRENCLDH*RMDRKIMERFICLMSNANNSTLFWDETV-**F 59
F + CK+ GIVRHK TP++N + R++R IM++ ++S + FW E
Sbjct: 553 FEKFCKEEGIVRHKTCAYTPQQNGIAE-RLNRTIMDKVRSMLSRSGMEKKFWAEAASTAV 611
Query: 60 FLIS*IDSLTLNLV*RH*KRCGWSCSKP*KYESF--WLCPLYAHIKRTILNQGLKLYVF- 116
+LI+ S +N K W+ + P S + C Y H + LN K +F
Sbjct: 612 YLINRSPSTAINFDLPEEK---WTGALP-DLSSLRKFGCLAYIHADQGKLNPRSKKGIFT 667
Query: 117 EVSLRCKRVQIWCMEKGET*VLINKDVVFKESEMF 151
K ++W +E + +I+++V+F+E MF
Sbjct: 668 SYPEGVKGYKVWVLEDKK--CVISRNVIFREQVMF 700
Score = 44.3 bits (103), Expect = 2e-04
Identities = 24/53 (45%), Positives = 34/53 (63%)
Query: 416 YMILKIDDMLIACNIKEEFDKVKVML*IEFQMKDMRAAS*ILAKKILRDISKG 468
Y++L +DDMLIA K E +++K +L EF+MKD+ A IL +I RD G
Sbjct: 1010 YLLLYVDDMLIASANKSEVNELKQLLSREFEMKDLGDAKKILGMEISRDRDAG 1062
>At2g19840 copia-like retroelement pol polyprotein
Length = 1137
Score = 53.1 bits (126), Expect = 3e-07
Identities = 31/83 (37%), Positives = 49/83 (58%), Gaps = 5/83 (6%)
Query: 256 EPVN*DGFVNASKSREWIASMKEELSSLHKNGTWLLVNKPKRMKNMSCI*LFKLKEGVNY 315
EP + + S + W+ ++ EE+ SL KN TW+LVN+ + K + C +FK K G+
Sbjct: 633 EPNDYQKALQDSDYKMWLKAVDEEIESLLKNNTWVLVNRDQFQKPIGCKWVFKRKSGIVG 692
Query: 316 MD*NSRFKAR----GFT*REGID 334
++ RFKAR G++ +EGID
Sbjct: 693 VE-KPRFKARLVVKGYSQKEGID 714
Score = 43.9 bits (102), Expect = 2e-04
Identities = 25/65 (38%), Positives = 35/65 (53%)
Query: 405 YLNETMHKYVDYMILKIDDMLIACNIKEEFDKVKVML*IEFQMKDMRAAS*ILAKKILRD 464
Y E Y++L +DD+LIA K +K +L EF+MKD+ A IL +I+RD
Sbjct: 817 YFKELQSGEYIYLLLYVDDILIASRDKRTVCDLKALLNSEFEMKDLGDAKKILGMEIVRD 876
Query: 465 ISKGT 469
GT
Sbjct: 877 RKAGT 881
Score = 39.7 bits (91), Expect = 0.004
Identities = 18/52 (34%), Positives = 31/52 (59%), Gaps = 1/52 (1%)
Query: 1 FNQICKDSGIVRHKQIRGTPRENCLDH*RMDRKIMERFICLMSNANNSTLFW 52
F+++CK GIVRH+ TP++N + R++R IM + ++S + FW
Sbjct: 371 FDEVCKKEGIVRHRTCTYTPQQNGVAE-RLNRTIMNKVRSMLSESGLDKKFW 421
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 52.0 bits (123), Expect = 7e-07
Identities = 35/107 (32%), Positives = 55/107 (50%), Gaps = 11/107 (10%)
Query: 252 IDGDEPVN*DGFVNASKSREWIASMKEELSSLHKNGTWLLVNKPKRMKNMSCI*LFKLKE 311
+ +EP D A +EW +M EE+ + KN TW LV+KP++ +S ++K+K
Sbjct: 832 VANEEPQTYD---EARGDKEWEEAMNEEIKVIEKNRTWKLVDKPEKKNVISVKWIYKIKT 888
Query: 312 GV--NYMD*NSRFKARGFT*REGID*CENYFLVWTA*GTNSYDSTRA 356
N++ +R ARGF+ GID E + V + YD+ RA
Sbjct: 889 DASGNHVKHKARLVARGFSQEYGIDYLETFAPV------SRYDTIRA 929
>At3g25450 hypothetical protein
Length = 1343
Score = 50.1 bits (118), Expect = 3e-06
Identities = 31/82 (37%), Positives = 45/82 (54%), Gaps = 5/82 (6%)
Query: 255 DEPVN*DGFVNASKSREWIASMKEELSSLHKNGTWLLVNKPKRMKNMSCI*LFKLKEGV- 313
DEP + F A+KS+EW + KEE+ S+ KN TW LV+ P K + +FKLK
Sbjct: 818 DEPWD---FKEANKSKEWRDACKEEIQSIEKNRTWSLVDLPVGSKAIGVKWVFKLKHNSD 874
Query: 314 -NYMD*NSRFKARGFT*REGID 334
+ +R A+G+ R G+D
Sbjct: 875 GSINKYKARLVAKGYVQRHGVD 896
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 49.3 bits (116), Expect = 5e-06
Identities = 26/68 (38%), Positives = 40/68 (58%), Gaps = 5/68 (7%)
Query: 271 EWIASMKEELSSLHKNGTWLLVNKPKRMKNMSCI*LFKLKEGVNYMD*NSRFKAR----G 326
+W +M EE+ S KN TW +V +P+ + + C +FK K G+ ++ RFKAR G
Sbjct: 851 KWKLAMDEEIDSQEKNNTWTIVTRPENQRIIGCRWIFKYKLGILGVE-EPRFKARLVAKG 909
Query: 327 FT*REGID 334
+ +EGID
Sbjct: 910 YAQKEGID 917
Score = 43.9 bits (102), Expect = 2e-04
Identities = 22/55 (40%), Positives = 35/55 (63%)
Query: 414 VDYMILKIDDMLIACNIKEEFDKVKVML*IEFQMKDMRAAS*ILAKKILRDISKG 468
V Y+++ +DD+L+A KE +K L + F+MKD+ AA IL +I+RD + G
Sbjct: 1029 VMYLLIYVDDILVASKNKEAITALKANLGMRFEMKDLGAAKKILGMEIIRDRTLG 1083
Score = 38.5 bits (88), Expect = 0.008
Identities = 39/154 (25%), Positives = 73/154 (47%), Gaps = 9/154 (5%)
Query: 1 FNQICKDSGIVRHKQIRGTPRENCLDH*RMDRKIMERFICLMSNANNSTLFWDE-TV**F 59
F+ C+ GI RH+ TP++N + RM+R IME+ ++S++ FW E T
Sbjct: 577 FDGFCESIGIHRHRTCAYTPQQNGVAE-RMNRTIMEKVRSMLSDSGLPKRFWAEATHTTV 635
Query: 60 FLIS*IDSLTLNLV*RH*KRCGWSCSKP-*KYESFWLCPLYAHIKRTILN-QGLKLYVFE 117
LI+ S LN K WS + P Y + C + H L + K +
Sbjct: 636 LLINKTPSSALNFEIPDKK---WSGNPPVYSYLRRYGCVAFVHTDDGKLEPRAKKGVLIG 692
Query: 118 VSLRCKRVQIWCMEKGET*VLINKDVVFKESEMF 151
+ K ++W ++ E +++++++F+E+ ++
Sbjct: 693 YPVGVKGYKVWILD--ERKCVVSRNIIFQENAVY 724
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 49.3 bits (116), Expect = 5e-06
Identities = 34/98 (34%), Positives = 51/98 (51%), Gaps = 13/98 (13%)
Query: 247 VVSGEIDG--------DEPVN*DGFVNASKSREWIASMKEELSSLHKNGTWLLVNKPKRM 298
V+ EI+G DEP + F A+K +EW + KEE+ S+ KN TW L++ P R
Sbjct: 763 VLMAEIEGEQVLLAINDEPWD---FKEANKLKEWRDACKEEILSIEKNKTWSLIDLPVRR 819
Query: 299 KNMSCI*LFKLKEGV--NYMD*NSRFKARGFT*REGID 334
K + +FK+K + +R A+G+ R GID
Sbjct: 820 KVIGLKWVFKIKRNSDGSINKYKARLVAKGYVQRHGID 857
>At3g45520 copia-like polyprotein
Length = 1363
Score = 48.1 bits (113), Expect = 1e-05
Identities = 30/88 (34%), Positives = 47/88 (53%), Gaps = 7/88 (7%)
Query: 253 DGD--EPVN*DGFVNASKSREWIASMKEELSSLHKNGTWLLVNKPKRMKNMSCI*LFKLK 310
DGD EP + V +W +M EE+ S KN TW V +P++ + + ++K K
Sbjct: 838 DGDAVEPADYKEAVRDENWDKWRLAMNEEIESQLKNDTWTTVTRPEKQRIIGSRWIYKYK 897
Query: 311 EGVNYMD*NSRFKAR----GFT*REGID 334
+G+ ++ RFKAR G+ REG+D
Sbjct: 898 QGIPGVE-EPRFKARLVAKGYAQREGVD 924
Score = 43.1 bits (100), Expect = 3e-04
Identities = 22/53 (41%), Positives = 33/53 (61%)
Query: 416 YMILKIDDMLIACNIKEEFDKVKVML*IEFQMKDMRAAS*ILAKKILRDISKG 468
Y++ +DDML+A N + D +K L I+F+MKD+ AA IL +I+ D G
Sbjct: 1038 YLLFYVDDMLVAANNMQAIDALKKELSIKFEMKDLGAAKKILGIEIIIDREAG 1090
Score = 34.7 bits (78), Expect = 0.12
Identities = 17/54 (31%), Positives = 30/54 (55%), Gaps = 1/54 (1%)
Query: 1 FNQICKDSGIVRHKQIRGTPRENCLDH*RMDRKIMERFICLMSNANNSTLFWDE 54
F+ C++ G RH+ TP++N + RM+R IME+ ++ ++ FW E
Sbjct: 584 FDGFCEEKGFQRHRTCAYTPQQNGVVE-RMNRTIMEKVRSMLCDSGLPKRFWAE 636
>At4g04600 putative polyprotein
Length = 922
Score = 47.0 bits (110), Expect = 2e-05
Identities = 28/65 (43%), Positives = 38/65 (58%)
Query: 405 YLNETMHKYVDYMILKIDDMLIACNIKEEFDKVKVML*IEFQMKDMRAAS*ILAKKILRD 464
Y+ E Y+++ +DDMLIA E +KVK L + F+MKDM AAS IL I R+
Sbjct: 669 YVKEVSPDQFVYLLVYVDDMLIAGKSMAEVNKVKEGLSLNFEMKDMGAASRILGIDIERN 728
Query: 465 ISKGT 469
+GT
Sbjct: 729 REEGT 733
Score = 32.7 bits (73), Expect = 0.46
Identities = 15/37 (40%), Positives = 25/37 (67%), Gaps = 1/37 (2%)
Query: 1 FNQICKDSGIVRHKQIRGTPRENCLDH*RMDRKIMER 37
F++ CK +GI RH+ TP++N + RM+R +ME+
Sbjct: 398 FDEYCKKNGIERHRTCTYTPQQNGVAE-RMNRTLMEK 433
>At2g11190 putative retroelement pol polyprotein
Length = 469
Score = 47.0 bits (110), Expect = 2e-05
Identities = 30/90 (33%), Positives = 49/90 (54%), Gaps = 5/90 (5%)
Query: 255 DEPVN*DGFVNASKSREWIASMKEELSSLHKNGTWLLVNKPKRMKNMSCI*LFKLKEGV- 313
+EP N F +A +S+EW + EE+SS+ KN TW LV P K + +FK+++
Sbjct: 155 EEPWN---FNDAKESKEWTQACVEEISSIEKNHTWDLVGLPTGAKPIGLKWVFKIQQNSD 211
Query: 314 -NYMD*NSRFKARGFT*REGID*CENYFLV 342
+ +R A+G+ R G+D E + +V
Sbjct: 212 GSINKHKARLVAKGYVQRHGVDFDEVFAIV 241
>At2g04490 putative retroelement pol polyprotein
Length = 1015
Score = 47.0 bits (110), Expect = 2e-05
Identities = 25/74 (33%), Positives = 40/74 (53%), Gaps = 2/74 (2%)
Query: 263 FVNASKSREWIASMKEELSSLHKNGTWLLVNKPKRMKNMSCI*LFKLKEGVN--YMD*NS 320
F A KS +WI +M EL +L TW + + P K + C ++K+K V+ +
Sbjct: 633 FFQAKKSDDWIKAMNAELQALEGTATWEICSLPSNKKAIGCKWVYKVKLNVDGTLERYKA 692
Query: 321 RFKARGFT*REGID 334
R A+G+T +EG+D
Sbjct: 693 RLVAKGYTQQEGVD 706
>At4g03810 putative retrotransposon protein
Length = 964
Score = 44.7 bits (104), Expect = 1e-04
Identities = 27/90 (30%), Positives = 45/90 (50%), Gaps = 2/90 (2%)
Query: 252 IDGDEPVN*DGFVNASKSREWIASMKEELSSLHKNGTWLLVNKPKRMKNMSCI*LFKLKE 311
I+ DEP + + + S +W+ + K E+ S+ +N W LV+ P +K + C +FK K
Sbjct: 437 IESDEPTSYEEALMGPDSDKWLEAAKSEMESMSQNKVWTLVDLPDGVKPIECKWIFKKKI 496
Query: 312 GV--NYMD*NSRFKARGFT*REGID*CENY 339
+ N + A+G+ GID E Y
Sbjct: 497 DMDGNIQIYKAGLVAKGYKQVHGIDYDETY 526
Score = 37.7 bits (86), Expect = 0.014
Identities = 25/76 (32%), Positives = 38/76 (49%), Gaps = 7/76 (9%)
Query: 404 CYLNETMHKYVDYMILKIDDMLIACNIKEEFDKVKVML*IEFQMKDMRAAS*ILAKKILR 463
C +T V +++L +DD+L+ N VK L F MKDM A+ IL +I R
Sbjct: 622 CVYKKTSGSAVAFLVLYVDDILLLGNDIPLLQSVKTWLGSCFSMKDMGEAAYILGIRIYR 681
Query: 464 D-------ISKGTFLD 472
D +S+ T++D
Sbjct: 682 DRLNKIIGLSQDTYID 697
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 43.5 bits (101), Expect = 3e-04
Identities = 22/74 (29%), Positives = 38/74 (50%), Gaps = 2/74 (2%)
Query: 263 FVNASKSREWIASMKEELSSLHKNGTWLLVNKPKRMKNMSCI*LFKLK--EGVNYMD*NS 320
+ A ++EW ++ E+ ++ K TW + PK K + C +F LK N +
Sbjct: 943 YAEAQDTKEWCEAVDAEIGAMEKTNTWEITTLPKGKKAVGCKWVFTLKFLADGNLERYKA 1002
Query: 321 RFKARGFT*REGID 334
R A+G+T +EG+D
Sbjct: 1003 RLVAKGYTQKEGLD 1016
>At2g05960 putative retroelement pol polyprotein
Length = 1200
Score = 43.5 bits (101), Expect = 3e-04
Identities = 32/98 (32%), Positives = 49/98 (49%), Gaps = 13/98 (13%)
Query: 247 VVSGEIDG--------DEPVN*DGFVNASKSREWIASMKEELSSLHKNGTWLLVNKPKRM 298
V+ EI+G DEP + F A+K REW + EE+ S+ KN TW LV+ P +
Sbjct: 767 VLLAEIEGERLLLLINDEPWD---FKEANKYREWRDACDEEIKSIIKNRTWSLVSLPVGI 823
Query: 299 KNMSCI*LFKLKEGV--NYMD*NSRFKARGFT*REGID 334
K + +FK+K N +R A+G+ + +D
Sbjct: 824 KPIGLKWVFKIKRNSDGNITKHKARLVAKGYVQKHVVD 861
Score = 28.9 bits (63), Expect = 6.6
Identities = 14/56 (25%), Positives = 27/56 (48%), Gaps = 1/56 (1%)
Query: 1 FNQICKDSGIVRHKQIRGTPRENCLDH*RMDRKIMERFICLMSNANNSTLFWDETV 56
F C++ GI+RH TP++N + R +R ++ +M + + W E +
Sbjct: 528 FQTFCEEEGIIRHLTAPYTPQQNGVVE-RRNRTLLGMTRSIMKHMSVPNYLWGEAI 582
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.361 0.161 0.572
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,606,956
Number of Sequences: 26719
Number of extensions: 302368
Number of successful extensions: 1298
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 44
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 1136
Number of HSP's gapped (non-prelim): 134
length of query: 485
length of database: 11,318,596
effective HSP length: 103
effective length of query: 382
effective length of database: 8,566,539
effective search space: 3272417898
effective search space used: 3272417898
T: 11
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147000.15


