

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147000.12 + phase: 0
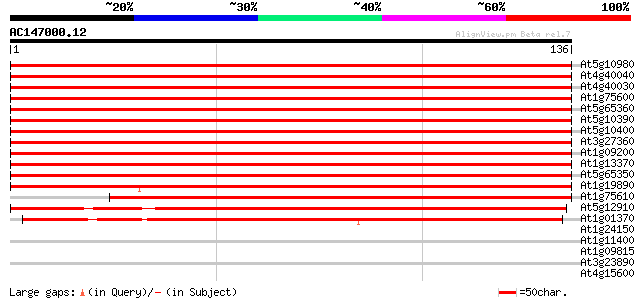
(136 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g10980 histon H3 protein 263 2e-71
At4g40040 Histon H3 263 2e-71
At4g40030 histone H3.3 263 2e-71
At1g75600 histone H3, putative 256 3e-69
At5g65360 histone H3 (sp|P05203) 254 7e-69
At5g10390 histone H3 - like protein 254 7e-69
At5g10400 histone H3 - like protein 254 7e-69
At3g27360 histone H3 like protein 254 7e-69
At1g09200 histone H3 like protein 254 7e-69
At1g13370 unknown protein 250 2e-67
At5g65350 histone H3 244 1e-65
At1g19890 hypothetical protein 237 1e-63
At1g75610 histone H3, putative 220 2e-58
At5g12910 histone H3 -like protein 187 1e-48
At1g01370 centromeric histone H3 HTR12 110 2e-25
At1g24150 unknown protein 28 1.1
At1g11400 unknown protein 28 1.9
At1g09815 unknown protein 27 2.5
At3g23890 topoisomerase II 27 3.2
At4g15600 hypothetical protein 26 5.5
>At5g10980 histon H3 protein
Length = 136
Score = 263 bits (673), Expect = 2e-71
Identities = 135/136 (99%), Positives = 135/136 (99%)
Query: 1 MARTKQTARKSTGGKAPRKPLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE 60
MARTKQTARKSTGGKAPRK LATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE
Sbjct: 1 MARTKQTARKSTGGKAPRKQLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE 60
Query: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI
Sbjct: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
Query: 121 MPKDIQLARRIRGERA 136
MPKDIQLARRIRGERA
Sbjct: 121 MPKDIQLARRIRGERA 136
>At4g40040 Histon H3
Length = 136
Score = 263 bits (673), Expect = 2e-71
Identities = 135/136 (99%), Positives = 135/136 (99%)
Query: 1 MARTKQTARKSTGGKAPRKPLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE 60
MARTKQTARKSTGGKAPRK LATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE
Sbjct: 1 MARTKQTARKSTGGKAPRKQLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE 60
Query: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI
Sbjct: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
Query: 121 MPKDIQLARRIRGERA 136
MPKDIQLARRIRGERA
Sbjct: 121 MPKDIQLARRIRGERA 136
>At4g40030 histone H3.3
Length = 136
Score = 263 bits (673), Expect = 2e-71
Identities = 135/136 (99%), Positives = 135/136 (99%)
Query: 1 MARTKQTARKSTGGKAPRKPLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE 60
MARTKQTARKSTGGKAPRK LATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE
Sbjct: 1 MARTKQTARKSTGGKAPRKQLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE 60
Query: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI
Sbjct: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
Query: 121 MPKDIQLARRIRGERA 136
MPKDIQLARRIRGERA
Sbjct: 121 MPKDIQLARRIRGERA 136
>At1g75600 histone H3, putative
Length = 136
Score = 256 bits (654), Expect = 3e-69
Identities = 131/136 (96%), Positives = 133/136 (97%)
Query: 1 MARTKQTARKSTGGKAPRKPLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE 60
MARTKQTARKS GGKAPR LATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE
Sbjct: 1 MARTKQTARKSHGGKAPRTLLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE 60
Query: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
LLIRKLPFQRLVREIAQD+KTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI
Sbjct: 61 LLIRKLPFQRLVREIAQDYKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
Query: 121 MPKDIQLARRIRGERA 136
MPKD+QLARRIRGERA
Sbjct: 121 MPKDVQLARRIRGERA 136
>At5g65360 histone H3 (sp|P05203)
Length = 136
Score = 254 bits (650), Expect = 7e-69
Identities = 131/136 (96%), Positives = 132/136 (96%)
Query: 1 MARTKQTARKSTGGKAPRKPLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE 60
MARTKQTARKSTGGKAPRK LATKAARKSAP TGGVKKPHR+RPGTVALREIRKYQKSTE
Sbjct: 1 MARTKQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPHRFRPGTVALREIRKYQKSTE 60
Query: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
LLIRKLPFQRLVREIAQDFKTDLRFQS AV ALQEAAEAYLVGLFEDTNLCAIHAKRVTI
Sbjct: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSSAVAALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
Query: 121 MPKDIQLARRIRGERA 136
MPKDIQLARRIRGERA
Sbjct: 121 MPKDIQLARRIRGERA 136
>At5g10390 histone H3 - like protein
Length = 136
Score = 254 bits (650), Expect = 7e-69
Identities = 131/136 (96%), Positives = 132/136 (96%)
Query: 1 MARTKQTARKSTGGKAPRKPLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE 60
MARTKQTARKSTGGKAPRK LATKAARKSAP TGGVKKPHR+RPGTVALREIRKYQKSTE
Sbjct: 1 MARTKQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPHRFRPGTVALREIRKYQKSTE 60
Query: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
LLIRKLPFQRLVREIAQDFKTDLRFQS AV ALQEAAEAYLVGLFEDTNLCAIHAKRVTI
Sbjct: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSSAVAALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
Query: 121 MPKDIQLARRIRGERA 136
MPKDIQLARRIRGERA
Sbjct: 121 MPKDIQLARRIRGERA 136
>At5g10400 histone H3 - like protein
Length = 136
Score = 254 bits (650), Expect = 7e-69
Identities = 131/136 (96%), Positives = 132/136 (96%)
Query: 1 MARTKQTARKSTGGKAPRKPLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE 60
MARTKQTARKSTGGKAPRK LATKAARKSAP TGGVKKPHR+RPGTVALREIRKYQKSTE
Sbjct: 1 MARTKQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPHRFRPGTVALREIRKYQKSTE 60
Query: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
LLIRKLPFQRLVREIAQDFKTDLRFQS AV ALQEAAEAYLVGLFEDTNLCAIHAKRVTI
Sbjct: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSSAVAALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
Query: 121 MPKDIQLARRIRGERA 136
MPKDIQLARRIRGERA
Sbjct: 121 MPKDIQLARRIRGERA 136
>At3g27360 histone H3 like protein
Length = 136
Score = 254 bits (650), Expect = 7e-69
Identities = 131/136 (96%), Positives = 132/136 (96%)
Query: 1 MARTKQTARKSTGGKAPRKPLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE 60
MARTKQTARKSTGGKAPRK LATKAARKSAP TGGVKKPHR+RPGTVALREIRKYQKSTE
Sbjct: 1 MARTKQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPHRFRPGTVALREIRKYQKSTE 60
Query: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
LLIRKLPFQRLVREIAQDFKTDLRFQS AV ALQEAAEAYLVGLFEDTNLCAIHAKRVTI
Sbjct: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSSAVAALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
Query: 121 MPKDIQLARRIRGERA 136
MPKDIQLARRIRGERA
Sbjct: 121 MPKDIQLARRIRGERA 136
>At1g09200 histone H3 like protein
Length = 136
Score = 254 bits (650), Expect = 7e-69
Identities = 131/136 (96%), Positives = 132/136 (96%)
Query: 1 MARTKQTARKSTGGKAPRKPLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE 60
MARTKQTARKSTGGKAPRK LATKAARKSAP TGGVKKPHR+RPGTVALREIRKYQKSTE
Sbjct: 1 MARTKQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPHRFRPGTVALREIRKYQKSTE 60
Query: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
LLIRKLPFQRLVREIAQDFKTDLRFQS AV ALQEAAEAYLVGLFEDTNLCAIHAKRVTI
Sbjct: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSSAVAALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
Query: 121 MPKDIQLARRIRGERA 136
MPKDIQLARRIRGERA
Sbjct: 121 MPKDIQLARRIRGERA 136
>At1g13370 unknown protein
Length = 136
Score = 250 bits (638), Expect = 2e-67
Identities = 128/136 (94%), Positives = 131/136 (96%)
Query: 1 MARTKQTARKSTGGKAPRKPLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE 60
MARTKQ+ARKS GGKAP K LATKAARKSAPTTGGVKKPHR+RPGTVALREIRKYQKSTE
Sbjct: 1 MARTKQSARKSHGGKAPTKQLATKAARKSAPTTGGVKKPHRFRPGTVALREIRKYQKSTE 60
Query: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
LL RKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI
Sbjct: 61 LLNRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
Query: 121 MPKDIQLARRIRGERA 136
MPKD+QLARRIR ERA
Sbjct: 121 MPKDVQLARRIRAERA 136
>At5g65350 histone H3
Length = 139
Score = 244 bits (623), Expect = 1e-65
Identities = 125/136 (91%), Positives = 130/136 (94%)
Query: 1 MARTKQTARKSTGGKAPRKPLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE 60
MARTKQTAR STGGKAPRK LA KAAR+SAP TGGVKKPHR+RPGTVALR+IRKYQKSTE
Sbjct: 1 MARTKQTARISTGGKAPRKQLAPKAARQSAPATGGVKKPHRFRPGTVALRDIRKYQKSTE 60
Query: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
+LIRKLPFQRLVREIAQDFKTDLRFQS AV ALQEAAEAYLVGLFEDTNLCAIHAKRVTI
Sbjct: 61 ILIRKLPFQRLVREIAQDFKTDLRFQSSAVAALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
Query: 121 MPKDIQLARRIRGERA 136
MPK+IQLARRIRGERA
Sbjct: 121 MPKEIQLARRIRGERA 136
>At1g19890 hypothetical protein
Length = 137
Score = 237 bits (605), Expect = 1e-63
Identities = 124/137 (90%), Positives = 128/137 (92%), Gaps = 1/137 (0%)
Query: 1 MARTKQTARKSTGGKAPRKPLATKAARKSA-PTTGGVKKPHRYRPGTVALREIRKYQKST 59
MARTKQTARKSTGGK PRK LATKAARK+ P GGVK+ HR+RPGTVALREIRKYQKST
Sbjct: 1 MARTKQTARKSTGGKGPRKELATKAARKTRRPYRGGVKRAHRFRPGTVALREIRKYQKST 60
Query: 60 ELLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVT 119
+LLIRKLPFQRLVREIAQDFK DLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVT
Sbjct: 61 DLLIRKLPFQRLVREIAQDFKVDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVT 120
Query: 120 IMPKDIQLARRIRGERA 136
IM KDIQLARRIRGERA
Sbjct: 121 IMSKDIQLARRIRGERA 137
>At1g75610 histone H3, putative
Length = 115
Score = 220 bits (560), Expect = 2e-58
Identities = 111/112 (99%), Positives = 112/112 (99%)
Query: 25 AARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR 84
AARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR
Sbjct: 4 AARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR 63
Query: 85 FQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA 136
FQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKD+QLARRIRGERA
Sbjct: 64 FQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDVQLARRIRGERA 115
>At5g12910 histone H3 -like protein
Length = 131
Score = 187 bits (475), Expect = 1e-48
Identities = 96/135 (71%), Positives = 115/135 (85%), Gaps = 5/135 (3%)
Query: 1 MARTKQTARKSTGGKAPRKPLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTE 60
MAR+ QTARK+TGGKAP A + + S P +KKP+RY+PGTVALREIRKYQK+T+
Sbjct: 1 MARSNQTARKATGGKAPH--FAMRVWQHSTPP---LKKPYRYKPGTVALREIRKYQKTTD 55
Query: 61 LLIRKLPFQRLVREIAQDFKTDLRFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTI 120
L+IRKLPFQRLV+EIAQ K DLRFQ+ AV ALQEAAEA++VG+FEDTNLCA+HAKR TI
Sbjct: 56 LVIRKLPFQRLVKEIAQSLKADLRFQTGAVSALQEAAEAFMVGMFEDTNLCAMHAKRSTI 115
Query: 121 MPKDIQLARRIRGER 135
MPKDIQLA+R+RG+R
Sbjct: 116 MPKDIQLAKRLRGDR 130
>At1g01370 centromeric histone H3 HTR12
Length = 178
Score = 110 bits (276), Expect = 2e-25
Identities = 66/133 (49%), Positives = 86/133 (64%), Gaps = 5/133 (3%)
Query: 4 TKQTARKSTGGKAPRKPLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTELLI 63
T+QT ++ R+ K +R++ P G KK +RYRPGTVAL+EIR +QK T LLI
Sbjct: 45 TQQTNPTTSPATGTRR--GAKRSRQAMPR-GSQKKSYRYRPGTVALKEIRHFQKQTNLLI 101
Query: 64 RKLPFQRLVREIAQDFKTDL--RFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTIM 121
F R VR I R+ + A++ALQEAAE YLVGLF D+ LCAIHA+RVT+M
Sbjct: 102 PAASFIREVRSITHMLAPPQINRWTAEALVALQEAAEDYLVGLFSDSMLCAIHARRVTLM 161
Query: 122 PKDIQLARRIRGE 134
KD +LARR+ G+
Sbjct: 162 RKDFELARRLGGK 174
>At1g24150 unknown protein
Length = 725
Score = 28.5 bits (62), Expect = 1.1
Identities = 23/81 (28%), Positives = 37/81 (45%), Gaps = 5/81 (6%)
Query: 25 AARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTELLIRKLPFQR--LVREIAQ--DFK 80
A K +P GG KKP P + + + RK Q +T ++++ L R LV + + DF
Sbjct: 354 AVGKKSPDDGGDKKPSSASPAQIFILDPRKSQ-NTAIVLKSLGMTRDELVESLMEGHDFH 412
Query: 81 TDLRFQSHAVLALQEAAEAYL 101
D + + +E A L
Sbjct: 413 PDTLERLSRIAPTKEEQSAIL 433
>At1g11400 unknown protein
Length = 204
Score = 27.7 bits (60), Expect = 1.9
Identities = 13/36 (36%), Positives = 20/36 (55%)
Query: 34 GGVKKPHRYRPGTVALREIRKYQKSTELLIRKLPFQ 69
G ++KP R RPG E+ KYQ L+ +++ Q
Sbjct: 34 GTLRKPIRIRPGYTPEDEVVKYQSKGSLMKKEMASQ 69
>At1g09815 unknown protein
Length = 124
Score = 27.3 bits (59), Expect = 2.5
Identities = 23/60 (38%), Positives = 28/60 (46%), Gaps = 4/60 (6%)
Query: 5 KQTARKSTGGKAPRKPLATKAARKSAPTTGG-VKKPHR-YRPGTVALREIRKYQKSTELL 62
KQT TGG + KP + K A K A G V +P G+V L E Y K E+L
Sbjct: 13 KQTKSNITGGISKSKPSSRKVAPKHAAAQGSDVTQPAALISHGSVDLNE--DYDKEEEML 70
>At3g23890 topoisomerase II
Length = 1473
Score = 26.9 bits (58), Expect = 3.2
Identities = 21/59 (35%), Positives = 27/59 (45%), Gaps = 10/59 (16%)
Query: 15 KAPRKPLATKAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTELLIRKL---PFQR 70
K RKP ATKAA+ A P + TVA E+ S E +RK+ PF +
Sbjct: 1347 KGGRKPAATKAAKPPA-------APRKRGKQTVASTEVLAIGVSPEKKVRKMRSSPFNK 1398
>At4g15600 hypothetical protein
Length = 655
Score = 26.2 bits (56), Expect = 5.5
Identities = 14/36 (38%), Positives = 18/36 (49%), Gaps = 3/36 (8%)
Query: 5 KQTARKSTGGKAPR---KPLATKAARKSAPTTGGVK 37
K+ R GG R + L + RK AP TGGV+
Sbjct: 369 KENVRPQNGGNQNRSGEQSLVIQKNRKGAPATGGVR 404
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.133 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,648,692
Number of Sequences: 26719
Number of extensions: 91498
Number of successful extensions: 247
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 222
Number of HSP's gapped (non-prelim): 28
length of query: 136
length of database: 11,318,596
effective HSP length: 89
effective length of query: 47
effective length of database: 8,940,605
effective search space: 420208435
effective search space used: 420208435
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC147000.12


