

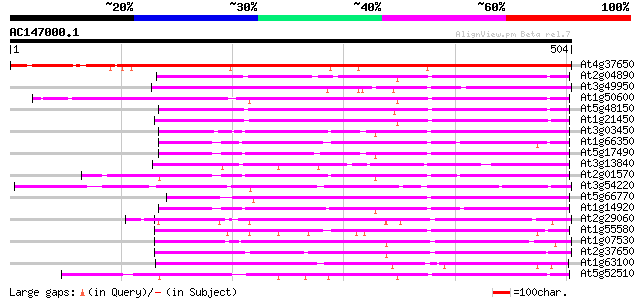
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147000.1 - phase: 0
(504 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g37650 SHORT-ROOT (SHR) 568 e-162
At2g04890 putative SCARECROW gene regulator 203 2e-52
At3g49950 putative protein 199 3e-51
At1g50600 scarecrow-like 5 (SCL5) 196 3e-50
At5g48150 SCARECROW gene regulator-like 188 5e-48
At1g21450 scarecrow-like 1 (SCL1) 178 5e-45
At3g03450 RGA1-like protein 173 2e-43
At1g66350 gibberellin regulatory protein like 170 1e-42
At5g17490 RGA-like protein 162 5e-40
At3g13840 hypothetical protein 159 3e-39
At2g01570 putative RGA1, giberellin repsonse modulation protein 159 3e-39
At3g54220 SCARECROW1 157 1e-38
At5g66770 SCARECROW gene regulator 153 2e-37
At1g14920 signal response protein (GAI) 152 3e-37
At2g29060 putative SCARECROW gene regulator 150 1e-36
At1g55580 unknown protein 148 8e-36
At1g07530 transcription factor scarecrow-like 14, putative 142 6e-34
At2g37650 scarecrow-like 9 (SCL9) 141 7e-34
At1g63100 transcription factor SCARECROW, putative 141 7e-34
At5g52510 scarecrow-like 8 (SCL8) 136 2e-32
>At4g37650 SHORT-ROOT (SHR)
Length = 531
Score = 568 bits (1463), Expect = e-162
Identities = 312/541 (57%), Positives = 390/541 (71%), Gaps = 48/541 (8%)
Query: 1 MDTLFRLVNFQQQQQQYQPDPSLNSTTTLT-TSSSSRSSRQTTYHY-YNQQEEDEECFNN 58
MDTLFRLV+ QQQQQ D + + ++L+ TS+++ S QT YHY + Q + EECFN
Sbjct: 1 MDTLFRLVSLQQQQQS---DSIITNQSSLSRTSTTTTGSPQTAYHYNFPQNDVVEECFN- 56
Query: 59 FYYMDHNNNNDEDLSSSSSKQHYYTYPYAST--TTITTPNTTYN--TINTPTTT------ 108
++MD +EDLSSSSS +++ + +T + TTP T Y+ T +TP++T
Sbjct: 57 -FFMD-----EEDLSSSSSHHNHHNHNNPNTYYSPFTTP-TQYHPATSSTPSSTAAAAAL 109
Query: 109 -----------DNYSFSPSHDYFNFEFSGHS-WSQNILLETARAFSDNNTNRIQQLMWML 156
D +FS +F+FS ++ W+ ++LLE ARAFSD +T R QQ++W L
Sbjct: 110 ASPYSSSGHHNDPSAFSIPQTPPSFDFSANAKWADSVLLEAARAFSDKDTARAQQILWTL 169
Query: 157 NELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTLTTAS--EKTCSFDSTRKMLLKFQ 214
NELS+PYGDT+QKL+SYFLQALF+RM +G+R Y+T+ TA+ EKTCSF+STRK +LKFQ
Sbjct: 170 NELSSPYGDTEQKLASYFLQALFNRMTGSGERCYRTMVTAAATEKTCSFESTRKTVLKFQ 229
Query: 215 EVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCTQWPTLLEALATRSDDTPHLRLT 274
EVSPW TFGHVAANGAILEA++G K+HI+DIS+T+CTQWPTLLEALATRSDDTPHLRLT
Sbjct: 230 EVSPWATFGHVAANGAILEAVDGEAKIHIVDISSTFCTQWPTLLEALATRSDDTPHLRLT 289
Query: 275 TVVTAISGGSVQ----KVMKEIGSRMEKFARLMGVPFKFKII--FSDLRELNLCDLDIKE 328
TVV A + Q ++MKEIG+RMEKFARLMGVPFKF II DL E +L +LD+K
Sbjct: 290 TVVVANKFVNDQTASHRMMKEIGNRMEKFARLMGVPFKFNIIHHVGDLSEFDLNELDVKP 349
Query: 329 DEALAINCVNSLHSISGAGNHRDLFISLLRGLEPRVLTIVEEEADL----EVCFGSDFVE 384
DE LAINCV ++H I+ G+ RD IS R L PR++T+VEEEADL E F +F+
Sbjct: 350 DEVLAINCVGAMHGIASRGSPRDAVISSFRRLRPRIVTVVEEEADLVGEEEGGFDDEFLR 409
Query: 385 GFKECLRWFRVYFEALDESFSRTSSERLMLEREAGRGIVDLVACDPYESVERRETAARWR 444
GF ECLRWFRV FE+ +ESF RTS+ERLMLER AGR IVDLVAC+P +S ERRETA +W
Sbjct: 410 GFGECLRWFRVCFESWEESFPRTSNERLMLERAAGRAIVDLVACEPSDSTERRETARKWS 469
Query: 445 RRLHGGGFNTVSFSDEVCDDVRALLRRYKEG-WSMTSSDGDTGIFLSWKDKPVVWASVWR 503
RR+ GF V +SDEV DDVRALLRRYKEG WSM GIFL W+D+PVVWAS WR
Sbjct: 470 RRMRNSGFGAVGYSDEVADDVRALLRRYKEGVWSMVQCPDAAGIFLCWRDQPVVWASAWR 529
Query: 504 P 504
P
Sbjct: 530 P 530
>At2g04890 putative SCARECROW gene regulator
Length = 413
Score = 203 bits (516), Expect = 2e-52
Identities = 118/376 (31%), Positives = 202/376 (53%), Gaps = 20/376 (5%)
Query: 133 ILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKT 192
+L+ A+A S+NN + M L + + G+ Q+L +Y L+ L +R+ +G YK+
Sbjct: 53 VLVACAKAVSENNLLMARWCMGELRGMVSISGEPIQRLGAYMLEGLVARLAASGSSIYKS 112
Query: 193 LTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCT 252
L + ++ F S +L EV P+ FG+++ANGAI EA++ ++HIID +
Sbjct: 113 LQSREPESYEFLSYVYVL---HEVCPYFKFGYMSANGAIAEAMKDEERIHIIDFQIGQGS 169
Query: 253 QWPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFKII 312
QW L++A A R P++R +T + GSV +K+ R+EK A+ VPF+F +
Sbjct: 170 QWIALIQAFAARPGGAPNIR----ITGVGDGSVLVTVKK---RLEKLAKKFDVPFRFNAV 222
Query: 313 FSDLRELNLCDLDIKEDEALAINCVNSLHSISGAG----NHRDLFISLLRGLEPRVLTIV 368
E+ + +LD+++ EAL +N LH + NHRD + +++ L P+V+T+V
Sbjct: 223 SRPSCEVEVENLDVRDGEALGVNFAYMLHHLPDESVSMENHRDRLLRMVKSLSPKVVTLV 282
Query: 369 EEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVDLVA 427
E+E + S F+ F E L ++ FE++D R ER+ +E+ R +V+++A
Sbjct: 283 EQECNTNT---SPFLPRFLETLSYYTAMFESIDVMLPRNHKERINIEQHCMARDVVNIIA 339
Query: 428 CDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDTGI 487
C+ E +ER E +W+ R GF S + +RALLR Y G+++ DG +
Sbjct: 340 CEGAERIERHELLGKWKSRFSMAGFEPYPLSSIISATIRALLRDYSNGYAIEERDG--AL 397
Query: 488 FLSWKDKPVVWASVWR 503
+L W D+ +V + W+
Sbjct: 398 YLGWMDRILVSSCAWK 413
>At3g49950 putative protein
Length = 410
Score = 199 bits (506), Expect = 3e-51
Identities = 127/393 (32%), Positives = 202/393 (51%), Gaps = 24/393 (6%)
Query: 128 SWSQNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGD 187
++ + +LL A A N+ Q++W+LN ++ P GD+ Q+L+S FL+AL SR
Sbjct: 25 NFMEQLLLHCATAIDSNDAALTHQILWVLNNIAPPDGDSTQRLTSAFLRALLSRAVSKTP 84
Query: 188 RTYKTLTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDIS 247
T++ + + L F +++PW FG +AAN AIL A+EG +HI+D+S
Sbjct: 85 TLSSTISFLPQADELHRFSVVELAAFVDLTPWHRFGFIAANAAILTAVEGYSTVHIVDLS 144
Query: 248 NTYCTQWPTLLEALATRSDDTPHLRLTTVVTAISGGS--VQKVMKEIGSRMEKFARLMGV 305
T+C Q PTL++A+A+R + P L TVV++ + +E+GS++ FA +
Sbjct: 145 LTHCMQIPTLIDAMASRLNKPPPLLKLTVVSSSDHFPPFINISYEELGSKLVNFATTRNI 204
Query: 306 PFKFKII---FSD-----LRELNLCDLDIKEDEALAINCVNSLHSI------SGAGNHRD 351
+F I+ +SD L++L + +EAL +NC L I S + + R
Sbjct: 205 TMEFTIVPSTYSDGFSSLLQQLRIYPSSF--NEALVVNCHMMLRYIPEEPLTSSSSSLRT 262
Query: 352 LFISLLRGLEPRVLTIVEEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSER 411
+F+ LR L PR++T++EE+ DL + V K +F + F+ D S +R
Sbjct: 263 VFLKQLRSLNPRIVTLIEEDVDLT---SENLVNRLKSAFNYFWIPFDTTDTFMSE---QR 316
Query: 412 LMLEREAGRGIVDLVACDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRR 471
E E I ++VA + E VER ET RW R+ F V ++ DV+A+L
Sbjct: 317 RWYEAEISWKIENVVAKEGAERVERTETKRRWIERMREAEFGGVRVKEDAVADVKAMLEE 376
Query: 472 YKEGWSMTSSDGDTGIFLSWKDKPVVWASVWRP 504
+ GW M D D + L+WK VV+A+VW P
Sbjct: 377 HAVGWGMKKEDDDESLVLTWKGHSVVFATVWVP 409
>At1g50600 scarecrow-like 5 (SCL5)
Length = 597
Score = 196 bits (497), Expect = 3e-50
Identities = 137/493 (27%), Positives = 235/493 (46%), Gaps = 25/493 (5%)
Query: 21 PSLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDEDLSSSSSKQH 80
P LN+ ++S++S SS ++ N FNN ++NN+ S++++ +
Sbjct: 120 PCLNNKNN-SSSTTSFSSNESPISQANNNNLSR--FNNHSPEENNNSPLSGSSATNTNET 176
Query: 81 YYTYPYAST-TTITTPNTTYNTINTPTTTDNYSFSPSHDYFNFEFSGHSWSQNILLETAR 139
+ T + P+ + N + S Y + E + +L E A+
Sbjct: 177 ELSLMLKDLETAMMEPDVDNSYNNQGGFGQQHGVVSSAMYRSMEMISRGDLKGVLYECAK 236
Query: 140 AFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTLTTASEK 199
A + + L+ L ++ + G+ Q+L +Y L+ L +R+ +G YK L
Sbjct: 237 AVENYDLEMTDWLISQLQQMVSVSGEPVQRLGAYMLEGLVARLASSGSSIYKALRCK--- 293
Query: 200 TCSFDSTRKMLLKFQ----EVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCTQWP 255
D T LL + E P+ FG+ +ANGAI EA++ +HIID + QW
Sbjct: 294 ----DPTGPELLTYMHILYEACPYFKFGYESANGAIAEAVKNESFVHIIDFQISQGGQWV 349
Query: 256 TLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFKIIFSD 315
+L+ AL R P++R+T + S + Q ++ +G R+ K A + GVPF+F
Sbjct: 350 SLIRALGARPGGPPNVRITGIDDPRSSFARQGGLELVGQRLGKLAEMCGVPFEFHGAALC 409
Query: 316 LRELNLCDLDIKEDEALAINCVNSLHSISGAG----NHRDLFISLLRGLEPRVLTIVEEE 371
E+ + L ++ EALA+N LH + NHRD + L++ L P V+T+VE+E
Sbjct: 410 CTEVEIEKLGVRNGEALAVNFPLVLHHMPDESVTVENHRDRLLRLVKHLSPNVVTLVEQE 469
Query: 372 ADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVDLVACDP 430
A+ + F+ F E + + FE++D +R ER+ +E+ R +V+L+AC+
Sbjct: 470 ANTNT---APFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLAREVVNLIACEG 526
Query: 431 YESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDTGIFLS 490
E ER E +WR R H GF S V ++ LL Y E +++ DG ++L
Sbjct: 527 VEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKGLLESYSEKYTLEERDG--ALYLG 584
Query: 491 WKDKPVVWASVWR 503
WK++P++ + WR
Sbjct: 585 WKNQPLITSCAWR 597
>At5g48150 SCARECROW gene regulator-like
Length = 490
Score = 188 bits (478), Expect = 5e-48
Identities = 108/375 (28%), Positives = 196/375 (51%), Gaps = 12/375 (3%)
Query: 134 LLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTL 193
L+ A+A S+N+ +M L ++ + G+ Q+L +Y L+ L +++ +G YK L
Sbjct: 123 LVSCAKAMSENDLMMAHSMMEKLRQMVSVSGEPIQRLGAYLLEGLVAQLASSGSSIYKAL 182
Query: 194 TTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCTQ 253
E + + +L EV P+ FG+++ANGAI EA++ ++HIID +Q
Sbjct: 183 NRCPEPASTELLSYMHIL--YEVCPYFKFGYMSANGAIAEAMKEENRVHIIDFQIGQGSQ 240
Query: 254 WPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFKIIF 313
W TL++A A R P +R+T + S + + +G+R+ K A+ VPF+F +
Sbjct: 241 WVTLIQAFAARPGGPPRIRITGIDDMTSAYARGGGLSIVGNRLAKLAKQFNVPFEFNSVS 300
Query: 314 SDLRELNLCDLDIKEDEALAINCVNSLHSIS----GAGNHRDLFISLLRGLEPRVLTIVE 369
+ E+ +L ++ EALA+N LH + NHRD + +++ L P+V+T+VE
Sbjct: 301 VSVSEVKPKNLGVRPGEALAVNFAFVLHHMPDESVSTENHRDRLLRMVKSLSPKVVTLVE 360
Query: 370 EEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVDLVAC 428
+E++ + F F E + ++ FE++D + R +R+ +E+ R +V+++AC
Sbjct: 361 QESNTNT---AAFFPRFMETMNYYAAMFESIDVTLPRDHKQRINVEQHCLARDVVNIIAC 417
Query: 429 DPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDTGIF 488
+ + VER E +WR R GF S V +++LLR Y + + + DG ++
Sbjct: 418 EGADRVERHELLGKWRSRFGMAGFTPYPLSPLVNSTIKSLLRNYSDKYRLEERDG--ALY 475
Query: 489 LSWKDKPVVWASVWR 503
L W + +V + W+
Sbjct: 476 LGWMHRDLVASCAWK 490
>At1g21450 scarecrow-like 1 (SCL1)
Length = 593
Score = 178 bits (452), Expect = 5e-45
Identities = 116/379 (30%), Positives = 196/379 (51%), Gaps = 14/379 (3%)
Query: 131 QNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTY 190
+ IL+ ARA S+ ++ L ++ + GD Q++++Y ++ L +RM +G Y
Sbjct: 223 KQILISCARALSEGKLEEALSMVNELRQIVSIQGDPSQRIAAYMVEGLAARMAASGKFIY 282
Query: 191 KTLTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTY 250
+ L + + ++L EV P FG +AANGAILEA++G ++HIID
Sbjct: 283 RALKCKEPPSDERLAAMQVLF---EVCPCFKFGFLAANGAILEAIKGEEEVHIIDFDINQ 339
Query: 251 CTQWPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFK 310
Q+ TL+ ++A P LRLT + S ++ IG R+E+ A GV FKFK
Sbjct: 340 GNQYMTLIRSIAELPGKRPRLRLTGIDDPESVQRSIGGLRIIGLRLEQLAEDNGVSFKFK 399
Query: 311 IIFSDLRELNLCDLDIKEDEALAINCVNSLHSISGAG----NHRDLFISLLRGLEPRVLT 366
+ S ++ L K E L +N LH + N RD + +++ L P+++T
Sbjct: 400 AMPSKTSIVSPSTLGCKPGETLIVNFAFQLHHMPDESVTTVNQRDELLHMVKSLNPKLVT 459
Query: 367 IVEEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVDL 425
+VE++ + S F F E ++ FE+LD + R S ER+ +ER+ R IV++
Sbjct: 460 VVEQDVNTNT---SPFFPRFIEAYEYYSAVFESLDMTLPRESQERMNVERQCLARDIVNI 516
Query: 426 VACDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLR-RYKEGWSMTSSDGD 484
VAC+ E +ER E A +WR R+ GFN S +V ++++ L++ +Y + + G+
Sbjct: 517 VACEGEERIERYEAAGKWRARMMMAGFNPKPMSAKVTNNIQNLIKQQYCNKYKLKEEMGE 576
Query: 485 TGIFLSWKDKPVVWASVWR 503
+ W++K ++ AS WR
Sbjct: 577 --LHFCWEEKSLIVASAWR 593
>At3g03450 RGA1-like protein
Length = 547
Score = 173 bits (438), Expect = 2e-43
Identities = 113/376 (30%), Positives = 192/376 (51%), Gaps = 20/376 (5%)
Query: 134 LLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTL 193
L+ A A N N L+ + L+ K+++YF QAL R+ D T +T
Sbjct: 184 LVACAEAIHQENLNLADALVKRVGTLAGSQAGAMGKVATYFAQALARRIYR--DYTAETD 241
Query: 194 TTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCTQ 253
A+ SF+ +M F E P+ F H AN AILEA+ ++H+ID+ Q
Sbjct: 242 VCAAVNP-SFEEVLEM--HFYESCPYLKFAHFTANQAILEAVTTARRVHVIDLGLNQGMQ 298
Query: 254 WPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFKIIF 313
WP L++ALA R P RLT + + S ++++G ++ +FA+ MGV F+FK +
Sbjct: 299 WPALMQALALRPGGPPSFRLTGIGPPQTENSDS--LQQLGWKLAQFAQNMGVEFEFKGLA 356
Query: 314 SDLRELNLCDLDIK------EDEALAINCVNSLHSISGAGNHRDLFISLLRGLEPRVLTI 367
++ +L DL+ + E E L +N V LH + + ++ ++ ++P ++T+
Sbjct: 357 AE----SLSDLEPEMFETRPESETLVVNSVFELHRLLARSGSIEKLLNTVKAIKPSIVTV 412
Query: 368 VEEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREAGRGIVDLVA 427
VE+EA+ G F++ F E L ++ F++L++S+S S +R+M E GR I+++VA
Sbjct: 413 VEQEANHN---GIVFLDRFNEALHYYSSLFDSLEDSYSLPSQDRVMSEVYLGRQILNVVA 469
Query: 428 CDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDTGI 487
+ + VER ETAA+WR R+ GF+ + LL Y G + D +
Sbjct: 470 AEGSDRVERHETAAQWRIRMKSAGFDPIHLGSSAFKQASMLLSLYATGDGYRVEENDGCL 529
Query: 488 FLSWKDKPVVWASVWR 503
+ W+ +P++ S W+
Sbjct: 530 MIGWQTRPLITTSAWK 545
>At1g66350 gibberellin regulatory protein like
Length = 511
Score = 170 bits (431), Expect = 1e-42
Identities = 115/374 (30%), Positives = 191/374 (50%), Gaps = 27/374 (7%)
Query: 134 LLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTL 193
LL A A NN L+ + L++ +K+++YF + L R+ Y+
Sbjct: 156 LLACAEAVQQNNLKLADALVKHVGLLASSQAGAMRKVATYFAEGLARRI-------YRIY 208
Query: 194 TTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCTQ 253
SF T + + F E P+ F H AN AILE K+H+ID+ + Q
Sbjct: 209 PRDDVALSSFSDT--LQIHFYESCPYLKFAHFTANQAILEVFATAEKVHVIDLGLNHGLQ 266
Query: 254 WPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFK-II 312
WP L++ALA R + P RLT + +++ ++E+G ++ + A +GV F+FK I
Sbjct: 267 WPALIQALALRPNGPPDFRLTGIGYSLTD------IQEVGWKLGQLASTIGVNFEFKSIA 320
Query: 313 FSDLRELNLCDLDIKED-EALAINCVNSLHSISGAGNHRDLFISLLRGLEPRVLTIVEEE 371
++L +L LDI+ E++A+N V LH + D F+S ++ + P ++T+VE+E
Sbjct: 321 LNNLSDLKPEMLDIRPGLESVAVNSVFELHRLLAHPGSIDKFLSTIKSIRPDIMTVVEQE 380
Query: 372 ADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREAGRGIVDLVACDPY 431
A+ G+ F++ F E L ++ F++L+ S +R+M E GR I++LVAC+
Sbjct: 381 ANHN---GTVFLDRFTESLHYYSSLFDSLE---GPPSQDRVMSELFLGRQILNLVACEGE 434
Query: 432 ESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRY--KEGWSMTSSDGDTGIFL 489
+ VER ET +WR R GGF VS LL Y +G+++ ++G + L
Sbjct: 435 DRVERHETLNQWRNRFGLGGFKPVSIGSNAYKQASMLLALYAGADGYNVEENEG--CLLL 492
Query: 490 SWKDKPVVWASVWR 503
W+ +P++ S WR
Sbjct: 493 GWQTRPLIATSAWR 506
>At5g17490 RGA-like protein
Length = 523
Score = 162 bits (409), Expect = 5e-40
Identities = 107/376 (28%), Positives = 188/376 (49%), Gaps = 26/376 (6%)
Query: 134 LLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTL 193
L+ A A N + L+ + L+ K+++YF +AL R+ Y+
Sbjct: 161 LVACAEAVQLENLSLADALVKRVGLLAASQAGAMGKVATYFAEALARRI-------YRIH 213
Query: 194 TTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCTQ 253
+A+ SF+ +M F + P+ F H AN AILEA+ + +H+ID+ Q
Sbjct: 214 PSAAAIDPSFEEILQM--NFYDSCPYLKFAHFTANQAILEAVTTSRVVHVIDLGLNQGMQ 271
Query: 254 WPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFKIIF 313
WP L++ALA R P RLT + S ++ ++E+G ++ + A+ +GV FKF +
Sbjct: 272 WPALMQALALRPGGPPSFRLT----GVGNPSNREGIQELGWKLAQLAQAIGVEFKFNGLT 327
Query: 314 SDLRELNLCDLDIK------EDEALAINCVNSLHSISGAGNHRDLFISLLRGLEPRVLTI 367
++ L DL+ E E L +N V LH + + ++ ++ ++P ++T+
Sbjct: 328 TE----RLSDLEPDMFETRTESETLVVNSVFELHPVLSQPGSIEKLLATVKAVKPGLVTV 383
Query: 368 VEEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREAGRGIVDLVA 427
VE+EA+ G F++ F E L ++ F++L++ S +R+M E GR I++LVA
Sbjct: 384 VEQEANHN---GDVFLDRFNEALHYYSSLFDSLEDGVVIPSQDRVMSEVYLGRQILNLVA 440
Query: 428 CDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDTGI 487
+ + +ER ET A+WR+R+ GF+ V+ + LL G + D +
Sbjct: 441 TEGSDRIERHETLAQWRKRMGSAGFDPVNLGSDAFKQASLLLALSGGGDGYRVEENDGSL 500
Query: 488 FLSWKDKPVVWASVWR 503
L+W+ KP++ AS W+
Sbjct: 501 MLAWQTKPLIAASAWK 516
>At3g13840 hypothetical protein
Length = 510
Score = 159 bits (403), Expect = 3e-39
Identities = 117/386 (30%), Positives = 196/386 (50%), Gaps = 30/386 (7%)
Query: 129 WSQNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDR 188
W++ +L A A + +N++R+Q + +L+EL++ GD +++L+++ L+AL ++ +
Sbjct: 144 WAEKLLNPCALAITASNSSRVQHYLCVLSELASSSGDANRRLAAFGLRALQHHLSSSSVS 203
Query: 189 T--YKTLTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPK----LH 242
+ + T AS + F T LLKF EVSPW + AN AIL+ L +PK LH
Sbjct: 204 SSFWPVFTFASAEVKMFQKT---LLKFYEVSPWFALPNNMANSAILQILAQDPKDKKDLH 260
Query: 243 IIDISNTYCTQWPTLLEALATRSDDTP-HLRLTTV--VTAISGGSVQKVMKEIGSRMEKF 299
IIDI ++ QWPTLLEAL+ R + P +R+T + +TA SV GS++ F
Sbjct: 261 IIDIGVSHGMQWPTLLEALSCRLEGPPPRVRITVISDLTADIPFSVGPPGYNYGSQLLGF 320
Query: 300 ARLMGVPFKFKIIFSDLRELNLCDLDIKEDEALAINCVNSLHSISGAGN-HRDLFISLLR 358
AR + K + S L +L L +D E L + LH + + N R + +R
Sbjct: 321 ARSL----KINLQISVLDKLQL--IDTSPHENLIVCAQFRLHHLKHSINDERGETLKAVR 374
Query: 359 GLEPRVLTIVEEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSE-RLMLERE 417
L P+ + + E E +DF GF + L + + ++ F +SE R ++E E
Sbjct: 375 SLRPKGVVLCENNG--ECSSSADFAAGFSKKLEYVWKFLDSTSSGFKEENSEERKLMEGE 432
Query: 418 AGRGIVDLVACDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWS 477
A + +++ + + E +W R+ GF +F ++ D ++LLR+Y W
Sbjct: 433 ATKVLMN--------AGDMNEGKEKWYERMREAGFFVEAFEEDAVDGAKSLLRKYDNNWE 484
Query: 478 MTSSDGDTGIFLSWKDKPVVWASVWR 503
+ DGDT L WK + V + S+W+
Sbjct: 485 IRMEDGDTFAGLMWKGEAVSFCSLWK 510
>At2g01570 putative RGA1, giberellin repsonse modulation protein
Length = 587
Score = 159 bits (402), Expect = 3e-39
Identities = 118/444 (26%), Positives = 204/444 (45%), Gaps = 25/444 (5%)
Query: 65 NNNNDEDLSSSSSKQHYYTYPYASTTTITTPNTTYNTINTPTTTDNYSFSPSHDYFNFEF 124
+NN ++ L S SS T ST+T T T T TTT + S
Sbjct: 158 SNNQNKRLKSCSSPDSMVT----STSTGTQIGGVIGTTVTTTTTTTTAAGESTRSVILVD 213
Query: 125 SGHSWSQNI--LLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRM 182
S + + + L+ A A NN + L+ + L+ +K+++YF +AL R+
Sbjct: 214 SQENGVRLVHALMACAEAIQQNNLTLAEALVKQIGCLAVSQAGAMRKVATYFAEALARRI 273
Query: 183 NDAGDRTYKTLTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLH 242
Y+ ++ T +M F E P+ F H AN AILEA EG ++H
Sbjct: 274 -------YRLSPPQNQIDHCLSDTLQM--HFYETCPYLKFAHFTANQAILEAFEGKKRVH 324
Query: 243 IIDISNTYCTQWPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARL 302
+ID S QWP L++ALA R P RLT + S + E+G ++ + A
Sbjct: 325 VIDFSMNQGLQWPALMQALALREGGPPTFRLTGIGPPAPDNSDH--LHEVGCKLAQLAEA 382
Query: 303 MGVPFKFK-IIFSDLRELNLCDLDIK--EDEALAINCVNSLHSISGAGNHRDLFISLLRG 359
+ V F+++ + + L +L+ L+++ + EA+A+N V LH + G + + +++
Sbjct: 383 IHVEFEYRGFVANSLADLDASMLELRPSDTEAVAVNSVFELHKLLGRPGGIEKVLGVVKQ 442
Query: 360 LEPRVLTIVEEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREAG 419
++P + T+VE+E++ G F++ F E L ++ F++L+ S +++M E G
Sbjct: 443 IKPVIFTVVEQESNHN---GPVFLDRFTESLHYYSTLFDSLEG--VPNSQDKVMSEVYLG 497
Query: 420 RGIVDLVACDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMT 479
+ I +LVAC+ + VER ET ++W R G LL + G
Sbjct: 498 KQICNLVACEGPDRVERHETLSQWGNRFGSSGLAPAHLGSNAFKQASMLLSVFNSGQGYR 557
Query: 480 SSDGDTGIFLSWKDKPVVWASVWR 503
+ + + L W +P++ S W+
Sbjct: 558 VEESNGCLMLGWHTRPLITTSAWK 581
>At3g54220 SCARECROW1
Length = 653
Score = 157 bits (398), Expect = 1e-38
Identities = 129/507 (25%), Positives = 233/507 (45%), Gaps = 45/507 (8%)
Query: 5 FRLVNFQQQQQQYQPDPSLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDH 64
+RL + DPS + L S++ S Q + QQ++ + +
Sbjct: 183 YRLRSLMLLDPSSSSDPSPQTFEPLYQISNNPSPPQQQQQHQQQQQQHKPPPPPIQQQER 242
Query: 65 NNNNDEDLSSSSSKQHYYTYPYASTTTITTPNTTYNTINTPTTTDNYSFSPSHDYFNFEF 124
N++ + P T T T P NT D
Sbjct: 243 ENSSTD------------APPQPETVTATVPAVQTNTAEALRERKEEIKRQKQDEEGLHL 290
Query: 125 SGHSWSQNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMND 184
+LL+ A A S +N +L+ +++LSTPYG + Q++++YF +A+ +R+ +
Sbjct: 291 L------TLLLQCAEAVSADNLEEANKLLLEISQLSTPYGTSAQRVAAYFSEAMSARLLN 344
Query: 185 AGDRTYKTLTTASEKTCSFDSTRKMLLKFQE---VSPWTTFGHVAANGAILEALEGNPKL 241
+ Y L + + + KM+ FQ +SP F H AN AI EA E +
Sbjct: 345 SCLGIYAALPS---RWMPQTHSLKMVSAFQVFNGISPLVKFSHFTANQAIQEAFEKEDSV 401
Query: 242 HIIDISNTYCTQWPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFAR 301
HIID+ QWP L LA+R PH+RLT + G+ + ++ G R+ FA
Sbjct: 402 HIIDLDIMQGLQWPGLFHILASRPGGPPHVRLTGL------GTSMEALQATGKRLSDFAD 455
Query: 302 LMGVPFKFKIIFSDLRELNLCDLDIKEDEALAINCV-NSLHSISGAGNHRDLFISLLRGL 360
+G+PF+F + + L+ L++++ EA+A++ + +SL+ ++G+ H + LL+ L
Sbjct: 456 KLGLPFEFCPLAEKVGNLDTERLNVRKREAVAVHWLQHSLYDVTGSDAHT---LWLLQRL 512
Query: 361 EPRVLTIVEEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLERE-AG 419
P+V+T+VE+ ++ F+ F E + ++ F++L S+ S ER ++E++
Sbjct: 513 APKVVTVVEQ----DLSHAGSFLGRFVEAIHYYSALFDSLGASYGEESEERHVVEQQLLS 568
Query: 420 RGIVDLVAC-DPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRY-KEGWS 477
+ I +++A P S E + + WR ++ GF +S + LL + +G++
Sbjct: 569 KEIRNVLAVGGPSRSGEVKFES--WREKMQQCGFKGISLAGNAATQATLLLGMFPSDGYT 626
Query: 478 MTSSDGDTGIFLSWKDKPVVWASVWRP 504
+ +G + L WKD ++ AS W P
Sbjct: 627 LVDDNGT--LKLGWKDLSLLTASAWTP 651
>At5g66770 SCARECROW gene regulator
Length = 584
Score = 153 bits (386), Expect = 2e-37
Identities = 108/370 (29%), Positives = 180/370 (48%), Gaps = 21/370 (5%)
Query: 142 SDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTLTTASEKTC 201
SD++ N + + + E + GD ++++ YF +AL +R++ T +
Sbjct: 228 SDSDPNEASKTLLQIRESVSELGDPTERVAFYFTEALSNRLSPNSPAT----------SS 277
Query: 202 SFDSTRKMLLKFQEVS---PWTTFGHVAANGAILEALEGNPKLHIIDISNTYCTQWPTLL 258
S ST ++L ++ ++ P++ F H+ AN AILEA E + K+HI+D QWP LL
Sbjct: 278 SSSSTEDLILSYKTLNDACPYSKFAHLTANQAILEATEKSNKIHIVDFGIVQGIQWPALL 337
Query: 259 EALATRSDDTP-HLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFKIIFSDLR 317
+ALATR+ P +R++ + G S + + G+R+ FA+++ + F F I + +
Sbjct: 338 QALATRTSGKPTQIRVSGIPAPSLGESPEPSLIATGNRLRDFAKVLDLNFDFIPILTPIH 397
Query: 318 ELNLCDLDIKEDEALAINCVNSLHS-ISGAGNHRDLFISLLRGLEPRVLTIVEEEADLEV 376
LN + DE LA+N + L+ + D + L + L PRV+T+ E E L
Sbjct: 398 LLNGSSFRVDPDEVLAVNFMLQLYKLLDETPTIVDTALRLAKSLNPRVVTLGEYEVSLN- 456
Query: 377 CFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVDLVACDPYE-SV 434
F K L+++ FE+L+ + R S ER+ +ERE GR I L+ +
Sbjct: 457 --RVGFANRVKNALQFYSAVFESLEPNLGRDSEERVRVERELFGRRISGLIGPEKTGIHR 514
Query: 435 ERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDTG-IFLSWKD 493
ER E +WR + GF +V S+ + LL Y + + G I L+W D
Sbjct: 515 ERMEEKEQWRVLMENAGFESVKLSNYAVSQAKILLWNYNYSNLYSIVESKPGFISLAWND 574
Query: 494 KPVVWASVWR 503
P++ S WR
Sbjct: 575 LPLLTLSSWR 584
>At1g14920 signal response protein (GAI)
Length = 533
Score = 152 bits (385), Expect = 3e-37
Identities = 103/373 (27%), Positives = 177/373 (46%), Gaps = 19/373 (5%)
Query: 134 LLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTL 193
LL A A N + L+ + L+ +K+++YF +AL R+ Y+
Sbjct: 173 LLACAEAVQKENLTVAEALVKQIGFLAVSQIGAMRKVATYFAEALARRI-------YRLS 225
Query: 194 TTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCTQ 253
+ S S T +M F E P+ F H AN AILEA +G ++H+ID S + Q
Sbjct: 226 PSQSPIDHSLSDTLQM--HFYETCPYLKFAHFTANQAILEAFQGKKRVHVIDFSMSQGLQ 283
Query: 254 WPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFK-II 312
WP L++ALA R P RLT + + E+G ++ A + V F+++ +
Sbjct: 284 WPALMQALALRPGGPPVFRLTGIGPPAPDNF--DYLHEVGCKLAHLAEAIHVEFEYRGFV 341
Query: 313 FSDLRELNLCDLDIK--EDEALAINCVNSLHSISGAGNHRDLFISLLRGLEPRVLTIVEE 370
+ L +L+ L+++ E E++A+N V LH + G D + ++ ++P + T+VE+
Sbjct: 342 ANTLADLDASMLELRPSEIESVAVNSVFELHKLLGRPGAIDKVLGVVNQIKPEIFTVVEQ 401
Query: 371 EADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREAGRGIVDLVACDP 430
E++ F++ F E L ++ F++L+ S +++M E G+ I ++VACD
Sbjct: 402 ESNHN---SPIFLDRFTESLHYYSTLFDSLEGVPS--GQDKVMSEVYLGKQICNVVACDG 456
Query: 431 YESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDTGIFLS 490
+ VER ET ++WR R GF LL + G + D + L
Sbjct: 457 PDRVERHETLSQWRNRFGSAGFAAAHIGSNAFKQASMLLALFNGGEGYRVEESDGCLMLG 516
Query: 491 WKDKPVVWASVWR 503
W +P++ S W+
Sbjct: 517 WHTRPLIATSAWK 529
>At2g29060 putative SCARECROW gene regulator
Length = 1336
Score = 150 bits (380), Expect = 1e-36
Identities = 101/384 (26%), Positives = 184/384 (47%), Gaps = 14/384 (3%)
Query: 131 QNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGD--- 187
+ +L A+A S + + + + + S+P GD Q+L+ F AL +R+ +
Sbjct: 956 RTLLTHCAQAISTGDKTTALEFLLQIRQQSSPLGDAGQRLAHCFANALEARLQGSTGPMI 1015
Query: 188 RTYKTLTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDIS 247
+TY T+S K + D+ R + + SP+ T + + IL+ + P LHI+D
Sbjct: 1016 QTYYNALTSSLKDTAADTIRAYRV-YLSSSPFVTLMYFFSIWMILDVAKDAPVLHIVDFG 1074
Query: 248 NTYCTQWPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPF 307
Y QWP +++++ R D LR+T + G + ++E G R+ ++ + VPF
Sbjct: 1075 ILYGFQWPMFIQSISDRKDVPRKLRITGIELPQCGFRPAERIEETGRRLAEYCKRFNVPF 1134
Query: 308 KFKIIFSDLRE-LNLCDLDIKEDEALAINC---VNSLHSISGAGNH--RDLFISLLRGLE 361
++K I S E + + DLDI+ +E LA+N + +L +G+ + RD + L+R +
Sbjct: 1135 EYKAIASQNWETIRIEDLDIRPNEVLAVNAGLRLKNLQDETGSEENCPRDAVLKLIRNMN 1194
Query: 362 PRVLTIVEEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLERE-AGR 420
P V F F+ FKE + + F+ D + R + ER+ ERE GR
Sbjct: 1195 PDVFIHAIVNGSFNAPF---FISRFKEAVYHYSALFDMFDSTLPRDNKERIRFEREFYGR 1251
Query: 421 GIVDLVACDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTS 480
++++AC+ + VER ET +W+ R+ GF + E+ + R L++++
Sbjct: 1252 EAMNVIACEEADRVERPETYRQWQVRMVRAGFKQKTIKPELVELFRGKLKKWRYHKDFVV 1311
Query: 481 SDGDTGIFLSWKDKPVVWASVWRP 504
+ + WK + + +S W P
Sbjct: 1312 DENSKWLLQGWKGRTLYASSCWVP 1335
Score = 146 bits (368), Expect = 3e-35
Identities = 118/419 (28%), Positives = 198/419 (47%), Gaps = 33/419 (7%)
Query: 105 PTTTDNYSFSPSHDYFNFEFSGHSWSQN------ILLETARAFSDNNTNRIQQLMWMLNE 158
P +S SP + E SG+S+++ +L+ A+A S N+ +L+ + +
Sbjct: 289 PAKASTFSKSPKGE--KPEASGNSYTKETPDLRTMLVSCAQAVSINDRRTADELLSRIRQ 346
Query: 159 LSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTLTTASEKTCSFDSTRKMLLKFQ---E 215
S+ YGD ++L+ YF +L +R+ G + Y L+ S+KT ST ML +Q
Sbjct: 347 HSSSYGDGTERLAHYFANSLEARLAGIGTQVYTALS--SKKT----STSDMLKAYQTYIS 400
Query: 216 VSPWTTFGHVAANGAILE-ALEGNPK-LHIIDISNTYCTQWPTLLEALATRSDDTPHLRL 273
V P+ + AN +I+ A N K +HIID + QWP+L+ LA R + LR+
Sbjct: 401 VCPFKKIAIIFANHSIMRLASSANAKTIHIIDFGISDGFQWPSLIHRLAWRRGSSCKLRI 460
Query: 274 TTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFKIIFSDLRELNLCDLDIKEDEALA 333
T + G + + E G R+ K+ + +PF++ I + L DL +KE E +A
Sbjct: 461 TGIELPQRGFRPAEGVIETGRRLAKYCQKFNIPFEYNAIAQKWESIKLEDLKLKEGEFVA 520
Query: 334 INCV----NSLHSISGAGNHRDLFISLLRGLEPRVLTIVEEEADLEVCFGSDFVEGFKEC 389
+N + N L + RD + L+R ++P V F FV F+E
Sbjct: 521 VNSLFRFRNLLDETVAVHSPRDTVLKLIRKIKPDVFIPGILSGSYNAPF---FVTRFREV 577
Query: 390 LRWFRVYFEALDESFSRTSSERLMLERE-AGRGIVDLVACDPYESVERRETAARWRRRLH 448
L + F+ D + +R R+M E+E GR I+++VAC+ E VER E+ +W+ R
Sbjct: 578 LFHYSSLFDMCDTNLTREDPMRVMFEKEFYGREIMNVVACEGTERVERPESYKQWQARAM 637
Query: 449 GGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDTG---IFLSWKDKPVVWASVWRP 504
GF + E+ ++ ++ + G+ D D + WK + V +S+W P
Sbjct: 638 RAGFRQIPLEKELVQKLKLMV---ESGYKPKEFDVDQDCHWLLQGWKGRIVYGSSIWVP 693
>At1g55580 unknown protein
Length = 445
Score = 148 bits (373), Expect = 8e-36
Identities = 119/415 (28%), Positives = 198/415 (47%), Gaps = 53/415 (12%)
Query: 131 QNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMN-DAGDRT 189
+ +L A S +N Q L+ +L+ S+P+GD+ ++L F +AL R+N D+T
Sbjct: 42 RRLLFTAANFVSQSNFTAAQNLLSILSLNSSPHGDSTERLVHLFTKALSVRINRQQQDQT 101
Query: 190 YKTLT-------TASEKTCSFDSTRKMLLKFQ-----------------EVSPWTTFGHV 225
+T+ T S T S K F+ +++P+ FGH+
Sbjct: 102 AETVATWTTNEMTMSNSTVFTSSVCKEQFLFRTKNNNSDFESCYYLWLNQLTPFIRFGHL 161
Query: 226 AANGAILEALEGNPK--LHIIDISNTYCTQWPTLLEALATRSDD----TPHLRLTTVVTA 279
AN AIL+A E N LHI+D+ + QWP L++ALA RS + P LR+T
Sbjct: 162 TANQAILDATETNDNGALHILDLDISQGLQWPPLMQALAERSSNPSSPPPSLRITGCGRD 221
Query: 280 ISGGSVQKVMKEIGSRMEKFARLMGVPFKFK---IIFSDLR----ELNLCDLDIKEDEAL 332
++G + G R+ +FA +G+ F+F I+ DL ++ L L + E +
Sbjct: 222 VTG------LNRTGDRLTRFADSLGLQFQFHTLVIVEEDLAGLLLQIRLLALSAVQGETI 275
Query: 333 AINCVNSLHSI-SGAGNHRDLFISLLRGLEPRVLTIVEEEADLEVCFGSDFVEGFKECLR 391
A+NCV+ LH I + G+ F+S ++ L R++T+ E EA+ F+ F E +
Sbjct: 276 AVNCVHFLHKIFNDDGDMIGHFLSAIKSLNSRIVTMAEREANHG---DHSFLNRFSEAVD 332
Query: 392 WFRVYFEALDESFSRTSSERLMLE-REAGRGIVDLVACDPYESVERRETAARWRRRLHGG 450
+ F++L+ + S ERL LE R G+ I+D+VA + E +R W +
Sbjct: 333 HYMAIFDSLEATLPPNSRERLTLEQRWFGKEILDVVAAEETERKQRHRRFEIWEEMMKRF 392
Query: 451 GFNTVSFSDEVCDDVRALLRRY--KEGWSMTSSDGDTGIFLSWKDKPVVWASVWR 503
GF V + LLR + EG+++ + +FL W+++P+ S W+
Sbjct: 393 GFVNVPIGSFALSQAKLLLRLHYPSEGYNLQFL--NNSLFLGWQNRPLFSVSSWK 445
>At1g07530 transcription factor scarecrow-like 14, putative
Length = 769
Score = 142 bits (357), Expect = 6e-34
Identities = 102/382 (26%), Positives = 182/382 (46%), Gaps = 17/382 (4%)
Query: 131 QNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTY 190
+ +L+ A+A S ++ +++ + E S+P G+ ++L+ YF +L +R+ G + Y
Sbjct: 394 RTLLVLCAQAVSVDDRRTANEMLRQIREHSSPLGNGSERLAHYFANSLEARLAGTGTQIY 453
Query: 191 KTLTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTY 250
L+ S+KT + D K + V P+ + AN +++ +HIID +Y
Sbjct: 454 TALS--SKKTSAADML-KAYQTYMSVCPFKKAAIIFANHSMMRFTANANTIHIIDFGISY 510
Query: 251 CTQWPTLLEALA-TRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKF 309
QWP L+ L+ +R +P LR+T + G + ++E G R+ ++ + VPF++
Sbjct: 511 GFQWPALIHRLSLSRPGGSPKLRITGIELPQRGFRPAEGVQETGHRLARYCQRHNVPFEY 570
Query: 310 KIIFSDLRELNLCDLDIKEDEALAINCV----NSLHSISGAGNHRDLFISLLRGLEPRVL 365
I + + DL +++ E + +N + N L + RD + L+R + P V
Sbjct: 571 NAIAQKWETIQVEDLKLRQGEYVVVNSLFRFRNLLDETVLVNSPRDAVLKLIRKINPNVF 630
Query: 366 TIVEEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVD 424
+ F FV F+E L + F+ D +R RLM E+E GR IV+
Sbjct: 631 IPAILSGNYNAPF---FVTRFREALFHYSAVFDMCDSKLAREDEMRLMYEKEFYGREIVN 687
Query: 425 LVACDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGD 484
+VAC+ E VER ET +W+ RL GF + E+ +++ + + G+
Sbjct: 688 VVACEGTERVERPETYKQWQARLIRAGFRQLPLEKELMQNLKL---KIENGYDKNFDVDQ 744
Query: 485 TGIFL--SWKDKPVVWASVWRP 504
G +L WK + V +S+W P
Sbjct: 745 NGNWLLQGWKGRIVYASSLWVP 766
>At2g37650 scarecrow-like 9 (SCL9)
Length = 718
Score = 141 bits (356), Expect = 7e-34
Identities = 100/379 (26%), Positives = 174/379 (45%), Gaps = 14/379 (3%)
Query: 131 QNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTY 190
+++L+ A+A + ++ QL+ + STP+GD +Q+L+ F L +R+ G + Y
Sbjct: 345 RSLLIHCAQAVAADDRRCAGQLLKQIRLHSTPFGDGNQRLAHCFANGLEARLAGTGSQIY 404
Query: 191 KTLTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTY 250
K + + + ++ L P+ + N I + + + ++H+ID Y
Sbjct: 405 KGIVSKPRSAAAVLKAHQLFLA---CCPFRKLSYFITNKTIRDLVGNSQRVHVIDFGILY 461
Query: 251 CTQWPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFK 310
QWPTL+ + +P +R+T + G + ++E G R+ +A+L GVPF++K
Sbjct: 462 GFQWPTLIHRFSMYG--SPKVRITGIEFPQPGFRPAQRVEETGQRLAAYAKLFGVPFEYK 519
Query: 311 IIFSDLRELNLCDLDIKEDEALAINCV---NSLHSIS-GAGNHRDLFISLLRGLEPRVLT 366
I + L DLDI DE +NC+ +LH S + RD ++L+ + P +
Sbjct: 520 AIAKKWDAIQLEDLDIDRDEITVVNCLYRAENLHDESVKVESCRDTVLNLIGKINPDLFV 579
Query: 367 IVEEEADLEVCFGSDFVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVDL 425
F FV F+E L F F+ L+ R ER+ LE E GR +++
Sbjct: 580 FGIVNGAYNAPF---FVTRFREALFHFSSIFDMLETIVPREDEERMFLEMEVFGREALNV 636
Query: 426 VACDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDT 485
+AC+ +E VER ET +W R G V F + + + + D +
Sbjct: 637 IACEGWERVERPETYKQWHVRAMRSGLVQVPFDPSIMKTSLHKVHTFYHKDFVIDQD-NR 695
Query: 486 GIFLSWKDKPVVWASVWRP 504
+ WK + V+ SVW+P
Sbjct: 696 WLLQGWKGRTVMALSVWKP 714
>At1g63100 transcription factor SCARECROW, putative
Length = 658
Score = 141 bits (356), Expect = 7e-34
Identities = 108/389 (27%), Positives = 178/389 (44%), Gaps = 29/389 (7%)
Query: 132 NILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQ-KLSSYFLQALFSRMNDAGDRTY 190
N+L A N I + +L++P G T +L +Y+++AL R+ +
Sbjct: 276 NLLTGCLDAIRSRNIAAINHFIARTGDLASPRGRTPMTRLIAYYIEALALRVARMWPHIF 335
Query: 191 KTLTTASEKTCSFDSTRKMLLKFQEVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTY 250
D + L +V+P F H AN +L A EG ++HIID
Sbjct: 336 HIAPPREFDRTVEDESGNALRFLNQVTPIPKFIHFTANEMLLRAFEGKERVHIIDFDIKQ 395
Query: 251 CTQWPTLLEALATRSDDTPHLRLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFK 310
QWP+ ++LA+R + H+R+T + G + + E G R+ FA M + F+F
Sbjct: 396 GLQWPSFFQSLASRINPPHHVRITGI------GESKLELNETGDRLHGFAEAMNLQFEFH 449
Query: 311 IIFSDLRELNLCDLDIKEDEALAINCVNSLHS--ISGAGNHRDLFISLLRGLEPRVLTIV 368
+ L ++ L L +KE E++A+NCV +H G G F+ L+R P L +
Sbjct: 450 PVVDRLEDVRLWMLHVKEGESVAVNCVMQMHKTLYDGTGAAIRDFLGLIRSTNPIALVLA 509
Query: 369 EEEADLEVCFGSDFVEGFKEC--LRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVDL 425
E+EA+ S+ +E + C L+++ F+A+ + + S R+ +E GR I ++
Sbjct: 510 EQEAE----HNSEQLE-TRVCNSLKYYSAMFDAIHTNLATDSLMRVKVEEMLFGREIRNI 564
Query: 426 VACDPYESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRY---KEG-WSMTSS 481
VAC+ ER WRR L GF ++ S+ + LLR Y EG +++ S
Sbjct: 565 VACEGSHRQERHVGFRHWRRMLEQLGFRSLGVSEREVLQSKMLLRMYGSDNEGFFNVERS 624
Query: 482 DGDT--------GIFLSWKDKPVVWASVW 502
D D G+ L W ++P+ S W
Sbjct: 625 DEDNGGEGGRGGGVTLRWSEQPLYTISAW 653
>At5g52510 scarecrow-like 8 (SCL8)
Length = 525
Score = 136 bits (343), Expect = 2e-32
Identities = 116/482 (24%), Positives = 213/482 (44%), Gaps = 50/482 (10%)
Query: 47 NQQEEDEECFNNFYYMDHNNNNDEDLSSSSSKQHYYTYPYAS-TTTITTPNTTYNTINTP 105
N+ E E N+ ++ +D+D S T + + TPN N +
Sbjct: 69 NRVHESENMLNSLRELEKQLLDDDDESGGDDDVSVITNSNSDWIQNLVTPNPNPNPV--- 125
Query: 106 TTTDNYSFSPSHDYFNFEFSGHSWSQNI-----LLETARAFSDNNTNRIQQLMWMLNELS 160
SFSPS + S S + ++ ++E A A ++ T +++ +++
Sbjct: 126 -----LSFSPSSSSSSSSPSTASTTTSVCSRQTVMEIATAIAEGKTEIATEILARVSQTP 180
Query: 161 TPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTLTTASEKTCSFDSTRKMLLKFQEVSPWT 220
++++KL + + AL SR+ Y S + E+SP
Sbjct: 181 NLERNSEEKLVDFMVAALRSRIASPVTELYGKEHLISTQL------------LYELSPCF 228
Query: 221 TFGHVAANGAILEALEGNPK----LHIIDISNTYCTQWPTLLEALATR------SDDTPH 270
G AAN AIL+A + N H+ID Q+ LL L+TR S ++P
Sbjct: 229 KLGFEAANLAILDAADNNDGGMMIPHVIDFDIGEGGQYVNLLRTLSTRRNGKSQSQNSPV 288
Query: 271 LRLTTVVTAISGGSV----QKVMKEIGSRMEKFARLMGVPFKFKIIFS-DLRELNLCDLD 325
+++T V + G V ++ +K +G + + +G+ F ++ S L +LN L
Sbjct: 289 VKITAVANNVYGCLVDDGGEERLKAVGDLLSQLGDRLGISVSFNVVTSLRLGDLNRESLG 348
Query: 326 IKEDEALAINCVNSLHSISGAG----NHRDLFISLLRGLEPRVLTIVEEEADLEVCFGSD 381
DE LA+N L+ + N RD + ++GL+PRV+T+VE+E + +
Sbjct: 349 CDPDETLAVNLAFKLYRVPDESVCTENPRDELLRRVKGLKPRVVTLVEQEMNSNT---AP 405
Query: 382 FVEGFKECLRWFRVYFEALDESFSRTSSERLMLEREAGRGIVDLVACDPYESVERRETAA 441
F+ E + E+++ + T+S+R +E GR +V+ VAC+ + +ER E
Sbjct: 406 FLGRVSESCACYGALLESVESTVPSTNSDRAKVEEGIGRKLVNAVACEGIDRIERCEVFG 465
Query: 442 RWRRRLHGGGFNTVSFSDEVCDDVRALLRRYKEGWSMTSSDGDTGIFLSWKDKPVVWASV 501
+WR R+ GF + S+++ + +++ R G+++ +G G+ W + + AS
Sbjct: 466 KWRMRMSMAGFELMPLSEKIAESMKSRGNRVHPGFTVKEDNG--GVCFGWMGRALTVASA 523
Query: 502 WR 503
WR
Sbjct: 524 WR 525
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.131 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,959,553
Number of Sequences: 26719
Number of extensions: 522701
Number of successful extensions: 2253
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 34
Number of HSP's that attempted gapping in prelim test: 2062
Number of HSP's gapped (non-prelim): 94
length of query: 504
length of database: 11,318,596
effective HSP length: 104
effective length of query: 400
effective length of database: 8,539,820
effective search space: 3415928000
effective search space used: 3415928000
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147000.1


