

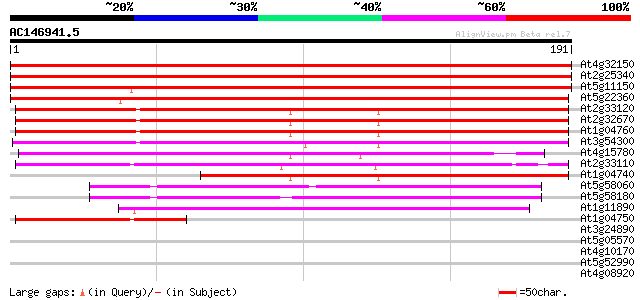
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146941.5 - phase: 0
(191 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g32150 vesicle-associated membrane protein 7C (At VAMP7C) 328 1e-90
At2g25340 putative synaptobrevin 323 3e-89
At5g11150 unknown protein (At5g11150) 316 5e-87
At5g22360 synaptobrevin-like protein 250 4e-67
At2g33120 putative synaptobrevin 147 4e-36
At2g32670 synaptobrevin like protein 144 3e-35
At1g04760 synaptobrevin 7B, putative 142 8e-35
At3g54300 synaptobrevin -like protein 138 2e-33
At4g15780 SYBL1 like protein 120 6e-28
At2g33110 synaptobrevin like protein 110 3e-25
At1g04740 putative vesicle-associated membrane protein, synaptob... 101 3e-22
At5g58060 ATGP1 59 1e-09
At5g58180 ATGP1-like protein 55 2e-08
At1g11890 putative vesicle transport protein 51 3e-07
At1g04750 synaptobrevin 7B, putative, 3' partial 47 6e-06
At3g24890 synaptobrevin, putative 39 0.002
At5g05570 putative protein 30 1.0
At4g10170 unknown protein 29 1.3
At5g52990 unknown protein 29 1.7
At4g08920 flavin-type blue-light photoreceptor (HY4) 28 2.3
>At4g32150 vesicle-associated membrane protein 7C (At VAMP7C)
Length = 219
Score = 328 bits (841), Expect = 1e-90
Identities = 158/191 (82%), Positives = 181/191 (94%)
Query: 1 MGILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDG 60
M ILYALVARGTVVL+EFT T+TNA+ IA+QILEK+PG+ND++VSYSQDRY+FHVKRTDG
Sbjct: 1 MAILYALVARGTVVLSEFTATSTNASTIAKQILEKVPGDNDSNVSYSQDRYVFHVKRTDG 60
Query: 61 LTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFSS 120
LTVLCMA+++ GRRIPFAFLE+IHQRFVR+YGRAV TA AYAMN+EFSRVL+QQ++Y+S+
Sbjct: 61 LTVLCMAEETAGRRIPFAFLEDIHQRFVRTYGRAVHTALAYAMNEEFSRVLSQQIDYYSN 120
Query: 121 DPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFR 180
DPNADRINR+KGEM+QVR VMIENIDKVLDRG+RLELLVDK ANMQGNTFRFRKQARRFR
Sbjct: 121 DPNADRINRIKGEMNQVRGVMIENIDKVLDRGERLELLVDKTANMQGNTFRFRKQARRFR 180
Query: 181 STVWWRNVKLT 191
S VWWRN KLT
Sbjct: 181 SNVWWRNCKLT 191
>At2g25340 putative synaptobrevin
Length = 219
Score = 323 bits (828), Expect = 3e-89
Identities = 154/191 (80%), Positives = 177/191 (92%)
Query: 1 MGILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDG 60
M ILYALVARGTVVLAE + T+TNA+ IA+QILEKIPGN D+HVSYSQDRY+FHVKRTDG
Sbjct: 1 MSILYALVARGTVVLAELSTTSTNASTIAKQILEKIPGNGDSHVSYSQDRYVFHVKRTDG 60
Query: 61 LTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFSS 120
LTVLCMAD+ GRRIPF+FLE+IHQRFVR+YGRA+ +A+AYAMNDEFSRVLNQQ+EY+S+
Sbjct: 61 LTVLCMADEDAGRRIPFSFLEDIHQRFVRTYGRAIHSAQAYAMNDEFSRVLNQQIEYYSN 120
Query: 121 DPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFR 180
DPNAD I+R+KGEM+QVR+VMIENID +LDRG+RLELLVDK ANMQGNTFRFRKQ RRF
Sbjct: 121 DPNADTISRIKGEMNQVRDVMIENIDNILDRGERLELLVDKTANMQGNTFRFRKQTRRFN 180
Query: 181 STVWWRNVKLT 191
+TVWWRN KLT
Sbjct: 181 NTVWWRNCKLT 191
>At5g11150 unknown protein (At5g11150)
Length = 221
Score = 316 bits (809), Expect = 5e-87
Identities = 149/192 (77%), Positives = 183/192 (94%), Gaps = 1/192 (0%)
Query: 1 MGILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNN-DTHVSYSQDRYIFHVKRTD 59
M I++ALVARGTVVL+EF+ T+TNA++I++QILEK+PGN+ D+H+SYSQDRYIFHVKRTD
Sbjct: 1 MAIIFALVARGTVVLSEFSATSTNASSISKQILEKLPGNDSDSHMSYSQDRYIFHVKRTD 60
Query: 60 GLTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFS 119
GLTVLCMAD++ GR IPFAFL++IHQRFV++YGRA+ +A+AY+MNDEFSRVL+QQME++S
Sbjct: 61 GLTVLCMADETAGRNIPFAFLDDIHQRFVKTYGRAIHSAQAYSMNDEFSRVLSQQMEFYS 120
Query: 120 SDPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRF 179
+DPNADR++R+KGEMSQVRNVMIENIDKVLDRG+RLELLVDK NMQGNTFRFRKQARR+
Sbjct: 121 NDPNADRMSRIKGEMSQVRNVMIENIDKVLDRGERLELLVDKTENMQGNTFRFRKQARRY 180
Query: 180 RSTVWWRNVKLT 191
R+ +WWRNVKLT
Sbjct: 181 RTIMWWRNVKLT 192
>At5g22360 synaptobrevin-like protein
Length = 221
Score = 250 bits (638), Expect = 4e-67
Identities = 115/191 (60%), Positives = 158/191 (82%), Gaps = 1/191 (0%)
Query: 1 MGILYALVARGTVVLAEFTGTTTNAAAIARQILEKI-PGNNDTHVSYSQDRYIFHVKRTD 59
M I+YA+VARGTVVLAEF+ T N A+ R+ILEK+ P +D + +SQDRYIFH+ R+D
Sbjct: 1 MAIVYAVVARGTVVLAEFSAVTGNTGAVVRRILEKLSPEISDERLCFSQDRYIFHILRSD 60
Query: 60 GLTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFS 119
GLT LCMA+D+ GRR+PF++LEEIH RF+++YG+ A AYAMNDEFSRVL+QQME+FS
Sbjct: 61 GLTFLCMANDTFGRRVPFSYLEEIHMRFMKNYGKVAHNAPAYAMNDEFSRVLHQQMEFFS 120
Query: 120 SDPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRF 179
S+P+ D +NR++GE+S++R+VM+ENI+K+++RGDR+ELLVDK A MQ ++F FRKQ++R
Sbjct: 121 SNPSVDTLNRVRGEVSEIRSVMVENIEKIMERGDRIELLVDKTATMQDSSFHFRKQSKRL 180
Query: 180 RSTVWWRNVKL 190
R +W +N KL
Sbjct: 181 RRALWMKNAKL 191
>At2g33120 putative synaptobrevin
Length = 221
Score = 147 bits (370), Expect = 4e-36
Identities = 76/190 (40%), Positives = 122/190 (64%), Gaps = 3/190 (1%)
Query: 3 ILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDGLT 62
++Y+ VARGTV+L EFT N +IA Q L+K+P +N+ +Y+ D + F+ +G T
Sbjct: 6 LIYSFVARGTVILVEFTDFKGNFTSIAAQCLQKLPSSNNKF-TYNCDGHTFNYLVENGFT 64
Query: 63 VLCMADDSVGRRIPFAFLEEIHQRFVRSYGRA-VLTAEAYAMNDEFSRVLNQQMEYFSSD 121
+A DS GR+IP AFLE + + F + YG TA+A ++N EF L + M+Y
Sbjct: 65 YCVVAVDSAGRQIPMAFLERVKEDFNKRYGGGKAATAQANSLNKEFGSKLKEHMQYCMDH 124
Query: 122 PNA-DRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFR 180
P+ ++ ++K ++S+V+ VM+ENI+KVLDRG+++ELLVDK N++ FR Q + R
Sbjct: 125 PDEISKLAKVKAQVSEVKGVMMENIEKVLDRGEKIELLVDKTENLRSQAQDFRTQGTQMR 184
Query: 181 STVWWRNVKL 190
+W++N+K+
Sbjct: 185 RKMWFQNMKI 194
>At2g32670 synaptobrevin like protein
Length = 285
Score = 144 bits (363), Expect = 3e-35
Identities = 74/190 (38%), Positives = 122/190 (63%), Gaps = 3/190 (1%)
Query: 3 ILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDGLT 62
++Y+ VARGTV+L E+T N A+A Q L+K+P +N+ +Y+ D + F+ +G T
Sbjct: 71 LIYSFVARGTVILVEYTEFKGNFTAVAAQCLQKLPSSNNKF-TYNCDGHTFNYLVENGFT 129
Query: 63 VLCMADDSVGRRIPFAFLEEIHQRFVRSYGRA-VLTAEAYAMNDEFSRVLNQQMEYFSSD 121
+A +SVGR+IP AFLE + + F + YG TA+A ++N EF L + M+Y
Sbjct: 130 YCVVAVESVGRQIPMAFLERVKEDFNKRYGGGKATTAQANSLNREFGSKLKEHMQYCVDH 189
Query: 122 PNA-DRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFR 180
P+ ++ ++K ++++V+ VM+ENI+KVLDRG+++ELLVDK N++ FR Q + R
Sbjct: 190 PDEISKLAKVKAQVTEVKGVMMENIEKVLDRGEKIELLVDKTENLRSQAQDFRTQGTKIR 249
Query: 181 STVWWRNVKL 190
+W+ N+K+
Sbjct: 250 RKMWFENMKI 259
>At1g04760 synaptobrevin 7B, putative
Length = 220
Score = 142 bits (359), Expect = 8e-35
Identities = 71/190 (37%), Positives = 122/190 (63%), Gaps = 3/190 (1%)
Query: 3 ILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDGLT 62
++Y+ VARGTV+LAE+T N ++A Q L+K+P +N+ +Y+ D + F+ +G T
Sbjct: 6 LIYSFVARGTVILAEYTEFKGNFTSVAAQCLQKLPSSNNKF-TYNCDGHTFNYLADNGFT 64
Query: 63 VLCMADDSVGRRIPFAFLEEIHQRFVRSYGRA-VLTAEAYAMNDEFSRVLNQQMEYFSSD 121
+ +S GR+IP AFLE + + F + YG TA+A ++N EF L + M+Y +
Sbjct: 65 YCVVVIESAGRQIPMAFLERVKEDFNKRYGGGKASTAKANSLNKEFGSKLKEHMQYCADH 124
Query: 122 PNA-DRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFR 180
P +++++K ++++V+ VM+ENI+KVLDRG+++ELLVDK N++ FR Q + +
Sbjct: 125 PEEISKLSKVKAQVTEVKGVMMENIEKVLDRGEKIELLVDKTENLRSQAQDFRTQGTKMK 184
Query: 181 STVWWRNVKL 190
+W+ N+K+
Sbjct: 185 RKLWFENMKI 194
>At3g54300 synaptobrevin -like protein
Length = 240
Score = 138 bits (348), Expect = 2e-33
Identities = 73/210 (34%), Positives = 126/210 (59%), Gaps = 22/210 (10%)
Query: 2 GILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDGL 61
G++Y+ VA+GTVVLAE T + N + IA Q L+K+P N+ + +YS D + F+ +G
Sbjct: 5 GLIYSFVAKGTVVLAEHTPYSGNFSTIAVQCLQKLPTNSSKY-TYSCDGHTFNFLVDNGF 63
Query: 62 TVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVLTAE--------------------AY 101
L +AD+S GR +PF FLE + + F + Y ++ E AY
Sbjct: 64 VFLVVADESTGRSVPFVFLERVKEDFKKRYEASIKNDERHPLADEDEDDDLFGDRFSVAY 123
Query: 102 AMNDEFSRVLNQQMEYFSSDPNA-DRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVD 160
++ EF +L + M+Y S P ++++LK ++++V+ +M++NI+KVLDRG+++ELLVD
Sbjct: 124 NLDREFGPILKEHMQYCMSHPEEMSKLSKLKAQITEVKGIMMDNIEKVLDRGEKIELLVD 183
Query: 161 KAANMQGNTFRFRKQARRFRSTVWWRNVKL 190
K N+Q F++Q R+ R +W +++++
Sbjct: 184 KTENLQFQADSFQRQGRQLRRKMWLQSLQM 213
>At4g15780 SYBL1 like protein
Length = 194
Score = 120 bits (300), Expect = 6e-28
Identities = 64/181 (35%), Positives = 108/181 (59%), Gaps = 9/181 (4%)
Query: 4 LYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDGLTV 63
+Y+ VARGT++LAE+T T N +IA Q L+K+P ++++ +Y+ D + F+ D
Sbjct: 7 IYSFVARGTMILAEYTEFTGNFPSIAAQCLQKLPSSSNSKFTYNCDHHTFNFLVEDAYAY 66
Query: 64 LCMADDSVGRRIPFAFLEEIHQRFVRSYGRA-VLTAEAYAMNDEFSRVLNQQMEYF-SSD 121
+A DS+ ++I AFLE + F + YG TA A ++N EF V+ + M Y
Sbjct: 67 CVVAKDSLSKQISIAFLERVKADFKKRYGGGKASTAIAKSLNKEFGPVMKEHMNYIVDHA 126
Query: 122 PNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFRS 181
+++ ++K ++S+V+++M+ENIDK +DRG+ L +L DK N+ R QAR ++
Sbjct: 127 EEIEKLIKVKAQVSEVKSIMLENIDKAIDRGENLTVLTDKTENL-------RSQAREYKK 179
Query: 182 T 182
T
Sbjct: 180 T 180
>At2g33110 synaptobrevin like protein
Length = 217
Score = 110 bits (276), Expect = 3e-25
Identities = 66/190 (34%), Positives = 109/190 (56%), Gaps = 7/190 (3%)
Query: 3 ILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDGLT 62
+ Y+ +ARGTV+L EFT N ++A Q LE +P +N+ +Y+ D + F+ +G T
Sbjct: 6 LFYSFIARGTVILVEFTDFKGNFTSVAAQYLENLPSSNN-KFTYNCDGHTFNDLVENGFT 64
Query: 63 VLCMADDSVGRRIPFAFLEEIHQRFVRSY-GRAVLTAEAYAMNDEFSRVLNQQMEYFSSD 121
+A DS GR IP AFLE + + F + Y G T +A ++N EF L + M+Y
Sbjct: 65 YCVVAVDSAGREIPMAFLERVKEDFYKRYGGEKAATDQANSLNKEFGSNLKEHMQYCMDH 124
Query: 122 PN-ADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFR 180
P+ + + K ++S+V+++M+ENI+KVL RG E+L + Q + R Q +R +
Sbjct: 125 PDEISNLAKAKAQVSEVKSLMMENIEKVLARGVICEMLGSSESQPQAFYIK-RTQMKRKK 183
Query: 181 STVWWRNVKL 190
W++N+K+
Sbjct: 184 ---WFQNMKI 190
>At1g04740 putative vesicle-associated membrane protein,
synaptobrevin 7B, 5' partial
Length = 166
Score = 101 bits (251), Expect = 3e-22
Identities = 50/127 (39%), Positives = 82/127 (64%), Gaps = 2/127 (1%)
Query: 66 MADDSVGRRIPFAFLEEIHQRFVRSYGRA-VLTAEAYAMNDEFSRVLNQQMEYFSSDPNA 124
+A DS GR+IP +FLE + + F + YG TA+A ++N EF L + M+Y P+
Sbjct: 15 VAVDSAGRQIPMSFLERVKEDFNKRYGGGKAATAQANSLNKEFGSKLKEHMQYCMDHPDE 74
Query: 125 -DRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRFRKQARRFRSTV 183
++ ++K ++S+V+ VM+ENI+KVLDRG+++ELLVDK N++ FR + R +
Sbjct: 75 ISKLAKVKAQVSEVKGVMMENIEKVLDRGEKIELLVDKTENLRSQAQDFRTTGTQMRRKM 134
Query: 184 WWRNVKL 190
W +N+K+
Sbjct: 135 WLQNMKI 141
>At5g58060 ATGP1
Length = 199
Score = 59.3 bits (142), Expect = 1e-09
Identities = 35/154 (22%), Positives = 79/154 (50%), Gaps = 4/154 (2%)
Query: 28 IARQILEKIPGNNDTHVSYSQDRYIFHVKRTDGLTVLCMADDSVGRRIPFAFLEEIHQRF 87
+ R + + P + V + + Y H +GL + DD R F+ L ++ +
Sbjct: 45 VGRTVASRTPPSQRQSVQHEE--YKVHAYNRNGLCAVGFMDDHYPVRSAFSLLNQVLDEY 102
Query: 88 VRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFSSDPNADRINRLKGEMSQVRNVMIENIDK 147
+S+G + +A+ + ++ L + + F AD++ +++ E+ + + ++ + ID
Sbjct: 103 QKSFGESWRSAKEDS--NQPWPYLTEALNKFQDPAEADKLLKIQRELDETKIILHKTIDS 160
Query: 148 VLDRGDRLELLVDKAANMQGNTFRFRKQARRFRS 181
VL RG++L+ LV+K++++ + F KQA++ S
Sbjct: 161 VLARGEKLDSLVEKSSDLSMASQMFYKQAKKTNS 194
>At5g58180 ATGP1-like protein
Length = 199
Score = 55.1 bits (131), Expect = 2e-08
Identities = 36/154 (23%), Positives = 73/154 (47%), Gaps = 6/154 (3%)
Query: 28 IARQILEKIPGNNDTHVSYSQDRYIFHVKRTDGLTVLCMADDSVGRRIPFAFLEEIHQRF 87
IAR + + P V + + Y H +GL + DD R F+ L ++ +
Sbjct: 47 IARTVARRTPPGQRQSVKHEE--YKVHAYNINGLCAVGFMDDHYPVRSAFSLLNQVLDVY 104
Query: 88 VRSYGRAVLTAEAYAMNDEFSRVLNQQMEYFSSDPNADRINRLKGEMSQVRNVMIENIDK 147
+ YG + + + L + + F AD++ +++ E+ + + ++ + ID
Sbjct: 105 QKDYG----DTWRFENSSQPWPYLKEASDKFRDPAEADKLLKIQRELDETKIILHKTIDG 160
Query: 148 VLDRGDRLELLVDKAANMQGNTFRFRKQARRFRS 181
VL RG++L+ LV+K++ + + F KQA++ S
Sbjct: 161 VLARGEKLDSLVEKSSELSLASKMFYKQAKKTNS 194
>At1g11890 putative vesicle transport protein
Length = 218
Score = 51.2 bits (121), Expect = 3e-07
Identities = 31/141 (21%), Positives = 65/141 (45%), Gaps = 1/141 (0%)
Query: 38 GNND-THVSYSQDRYIFHVKRTDGLTVLCMADDSVGRRIPFAFLEEIHQRFVRSYGRAVL 96
G ND + +S Y+FH + L M D S +++ F +LE++ F R G +
Sbjct: 46 GQNDASRMSVETGPYVFHYIIEGRVCYLTMCDRSYPKKLAFQYLEDLKNEFERVNGPNIE 105
Query: 97 TAEAYAMNDEFSRVLNQQMEYFSSDPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLE 156
TA +F + + + + I +L E+ +V +M N+ +VL G++L+
Sbjct: 106 TAARPYAFIKFDTFIQKTKKLYQDTRTQRNIAKLNDELYEVHQIMTRNVQEVLGVGEKLD 165
Query: 157 LLVDKAANMQGNTFRFRKQAR 177
+ + ++ + + + +A+
Sbjct: 166 QVSEMSSRLTSESRIYADKAK 186
>At1g04750 synaptobrevin 7B, putative, 3' partial
Length = 63
Score = 47.0 bits (110), Expect = 6e-06
Identities = 24/58 (41%), Positives = 37/58 (63%), Gaps = 1/58 (1%)
Query: 3 ILYALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHVSYSQDRYIFHVKRTDG 60
++Y+ VARGTV+L EFT N +IA Q L+K+P +N+ +Y+ D + F+ DG
Sbjct: 6 LIYSFVARGTVILVEFTDFKGNFTSIAAQCLQKLPSSNN-KFTYNCDGHTFNYLVEDG 62
>At3g24890 synaptobrevin, putative
Length = 114
Score = 38.9 bits (89), Expect = 0.002
Identities = 22/84 (26%), Positives = 47/84 (55%), Gaps = 9/84 (10%)
Query: 111 LNQQMEY-----FSSDPNADRINRLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANM 165
L QQ+E + ++ ++K +++++ VM+ENI+K LDR +++++LVD +
Sbjct: 11 LTQQLEQRVLVLLAHPEEISKLAKVKALVTKMKGVMMENIEKALDRSEKIKILVDLRSKY 70
Query: 166 QGNTFRFRKQARRFRSTVWWRNVK 189
+ K R+ +W++N+K
Sbjct: 71 KDIITPGTKITRK----MWFQNMK 90
>At5g05570 putative protein
Length = 1124
Score = 29.6 bits (65), Expect = 1.0
Identities = 17/61 (27%), Positives = 30/61 (48%), Gaps = 7/61 (11%)
Query: 129 RLKGEMSQVRNVMIENIDKVLDRGDRLELLVDKAANMQGNTFRF----RKQARRFRSTVW 184
R GE S + + + DK+ +RG++LE + + A +Q N F + A++ W
Sbjct: 1065 RKAGETSAIAS---QAKDKLHERGEKLERISQRTAELQDNAENFASMAHELAKQMEKRKW 1121
Query: 185 W 185
W
Sbjct: 1122 W 1122
>At4g10170 unknown protein
Length = 254
Score = 29.3 bits (64), Expect = 1.3
Identities = 34/145 (23%), Positives = 57/145 (38%), Gaps = 12/145 (8%)
Query: 5 YALVARGTVVLAEFTGTTTNAAAIARQILEKIPGNNDTHV-SYSQDRYIFHVKRTDGLTV 63
Y V+R +L + G ++A LEK P + + + + R+ F + DG
Sbjct: 10 YCCVSRDNQILYSYNGGDQTNESLAALCLEKSPPFHTWYFETIGKRRFGFLIG--DGFVY 67
Query: 64 LCMADDSVGRRIPFAFLEEIHQRF---VRSYGRAVLTAEAYAMN--DEFSRVLNQQMEYF 118
+ D+ + R FLE + F R R TA ++N D+ V+ + +
Sbjct: 68 FAIVDEVLKRSSVLKFLEHLRDEFKKAARENSRGSFTAMIGSINVEDQLVPVVTRLIASL 127
Query: 119 SSDPNADRINRLK----GEMSQVRN 139
+ N LK GE S+ N
Sbjct: 128 ERVAESSSNNELKSSNLGEQSEGSN 152
>At5g52990 unknown protein
Length = 272
Score = 28.9 bits (63), Expect = 1.7
Identities = 23/82 (28%), Positives = 39/82 (47%), Gaps = 5/82 (6%)
Query: 5 YALVARGTVVLAEFTG-TTTNAAAIARQILEKIPGNND--THVSYSQDRYIFHVKRTDGL 61
Y +A+GTVVLAEF AIA + +E P ++ +H + + Y F + D
Sbjct: 10 YTCIAKGTVVLAEFVSRQEPGIEAIALRCIENTPPHHSMFSHTVHKK-TYTFAID-DDSF 67
Query: 62 TVLCMADDSVGRRIPFAFLEEI 83
++D+S+ + F L +
Sbjct: 68 VYFSISDESMEKPESFWVLNRL 89
>At4g08920 flavin-type blue-light photoreceptor (HY4)
Length = 681
Score = 28.5 bits (62), Expect = 2.3
Identities = 12/32 (37%), Positives = 19/32 (58%)
Query: 106 EFSRVLNQQMEYFSSDPNADRINRLKGEMSQV 137
EF R+ N Q E + DPN + + R E+S++
Sbjct: 412 EFDRIDNPQFEGYKFDPNGEYVRRWLPELSRL 443
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,785,806
Number of Sequences: 26719
Number of extensions: 143950
Number of successful extensions: 539
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 496
Number of HSP's gapped (non-prelim): 29
length of query: 191
length of database: 11,318,596
effective HSP length: 94
effective length of query: 97
effective length of database: 8,807,010
effective search space: 854279970
effective search space used: 854279970
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146941.5


