

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146941.18 + phase: 0 /pseudo/partial
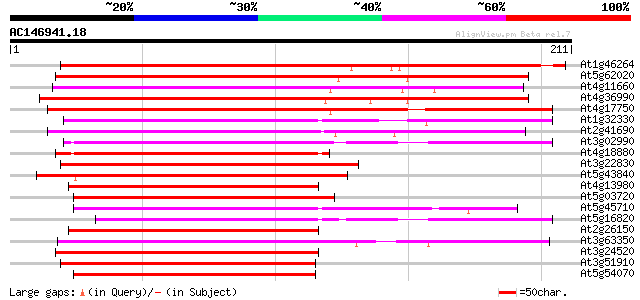
(211 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g46264 heat shock transcription factor, putative 264 2e-71
At5g62020 heat shock factor 6 171 3e-43
At4g11660 heat shock transcription factor - like protein 171 3e-43
At4g36990 heat shock transcription factor HSF4 169 1e-42
At4g17750 heat shock transcription factor HSF1 161 2e-40
At1g32330 heat shock transcription factor HSF8 like protein 152 1e-37
At2g41690 putative heat shock transcription factor 149 1e-36
At3g02990 putative heat shock transcription factor 145 2e-35
At4g18880 heat shock transcription factor 21 (AtHSF21) 144 3e-35
At3g22830 putative heat shock protein 144 4e-35
At5g43840 heat shock transcription factor-like protein 137 4e-33
At4g13980 unknown protein 137 5e-33
At5g03720 heat shock transcription factor -like protein 136 7e-33
At5g45710 heat shock transcription factor 133 8e-32
At5g16820 Heat Shock Factor 3 132 2e-31
At2g26150 heat shock transcription factor like protein 131 3e-31
At3g63350 heat shock transcription factor-like protein 129 1e-30
At3g24520 heat shock transcription factor HSF1, putative 127 4e-30
At3g51910 putative heat shock transcription factor 126 7e-30
At5g54070 putative protein 125 2e-29
>At1g46264 heat shock transcription factor, putative
Length = 348
Score = 264 bits (675), Expect = 2e-71
Identities = 133/218 (61%), Positives = 155/218 (71%), Gaps = 32/218 (14%)
Query: 20 KSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSFVR 79
K+VPAPFLTKTYQLVDDP TDH+VSW DD+TTFVVWRPPEFARDLLPN+FKHNNFSSFVR
Sbjct: 29 KAVPAPFLTKTYQLVDDPATDHVVSWGDDDTTFVVWRPPEFARDLLPNYFKHNNFSSFVR 88
Query: 80 QLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQHYQQQYYE------------ 127
QLNTYGF+K+V DRWEFAN++FK+G KHLLCEIHRRKT Q QQ+
Sbjct: 89 QLNTYGFRKIVPDRWEFANEFFKRGEKHLLCEIHRRKTSQMIPQQHSPFMSHHHAPPQIP 148
Query: 128 ---------QSPQIFQPDESICWI-DSP------LPSPKSNTDILTALSEDNQRLRRKNF 171
P++ P+E W DSP +P +TALSEDN+RLRR N
Sbjct: 149 FSGGSFFPLPPPRVTTPEEDHYWCDDSPPSRPRVIPQQIDTAAQVTALSEDNERLRRSNT 208
Query: 172 MLLSELSHMKNLYNDIIYFIQNHVSPASPFEQRSNNSA 209
+L+SEL+HMK LYNDIIYF+QNHV P +P SNNS+
Sbjct: 209 VLMSELAHMKKLYNDIIYFVQNHVKPVAP----SNNSS 242
>At5g62020 heat shock factor 6
Length = 299
Score = 171 bits (433), Expect = 3e-43
Identities = 90/196 (45%), Positives = 121/196 (60%), Gaps = 18/196 (9%)
Query: 18 SQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSF 77
SQ+S+P PFLTKT+ LV+D D ++SW++D ++F+VW P +FA+DLLP FKHNNFSSF
Sbjct: 16 SQRSIPTPFLTKTFNLVEDSSIDDVISWNEDGSSFIVWNPTDFAKDLLPKHFKHNNFSSF 75
Query: 78 VRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQHYQ----------QQYYE 127
VRQLNTYGFKKVV DRWEF+ND+FK+G K LL EI RRK +Q Q
Sbjct: 76 VRQLNTYGFKKVVPDRWEFSNDFFKRGEKRLLREIQRRKITTTHQTVVAPSSEQRNQTMV 135
Query: 128 QSPQIFQPDESICWIDSPLPS--------PKSNTDILTALSEDNQRLRRKNFMLLSELSH 179
SP D + + S PS N + L E+N++LR +N L EL+
Sbjct: 136 VSPSNSGEDNNNNQVMSSSPSSWYCHQTKTTGNGGLSVELLEENEKLRSQNIQLNRELTQ 195
Query: 180 MKNLYNDIIYFIQNHV 195
MK++ ++I + N+V
Sbjct: 196 MKSICDNIYSLMSNYV 211
>At4g11660 heat shock transcription factor - like protein
Length = 377
Score = 171 bits (432), Expect = 3e-43
Identities = 95/213 (44%), Positives = 123/213 (57%), Gaps = 36/213 (16%)
Query: 17 ESQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSS 76
+SQ+S+P PFLTKTYQLV+DP+ D ++SW++D TTF+VWRP EFARDLLP +FKHNNFSS
Sbjct: 51 DSQRSIPTPFLTKTYQLVEDPVYDELISWNEDGTTFIVWRPAEFARDLLPKYFKHNNFSS 110
Query: 77 FVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQ----------------- 119
FVRQLNTYGF+KVV DRWEF+ND FK+G K LL +I RRK Q
Sbjct: 111 FVRQLNTYGFRKVVPDRWEFSNDCFKRGEKILLRDIQRRKISQPAMAAAAAAAAAAVAAS 170
Query: 120 -----------HYQQQYYEQSPQIFQPDESICWIDSPL------PSPKSNTDILTA--LS 160
H Q+ + S + + S + T TA L
Sbjct: 171 AVTVAAVPVVAHIVSPSNSGEEQVISSNSSPAAAAAAIGGVVGGGSLQRTTSCTTAPELV 230
Query: 161 EDNQRLRRKNFMLLSELSHMKNLYNDIIYFIQN 193
E+N+RLR+ N L E++ +K LY +I + N
Sbjct: 231 EENERLRKDNERLRKEMTKLKGLYANIYTLMAN 263
>At4g36990 heat shock transcription factor HSF4
Length = 284
Score = 169 bits (427), Expect = 1e-42
Identities = 90/192 (46%), Positives = 124/192 (63%), Gaps = 8/192 (4%)
Query: 12 MVFTMESQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKH 71
M +Q+SVPAPFL+KTYQLVDD TD +VSW+++ T FVVW+ EFA+DLLP +FKH
Sbjct: 1 MTAVTAAQRSVPAPFLSKTYQLVDDHSTDDVVSWNEEGTAFVVWKTAEFAKDLLPQYFKH 60
Query: 72 NNFSSFVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKT--PQHYQQQYYEQS 129
NNFSSF+RQLNTYGF+K V D+WEFANDYF++G + LL +I RRK+ + S
Sbjct: 61 NNFSSFIRQLNTYGFRKTVPDKWEFANDYFRRGGEDLLTDIRRRKSVIASTAGKCVVVGS 120
Query: 130 PQIFQ----PDESICWIDSPLPS--PKSNTDILTALSEDNQRLRRKNFMLLSELSHMKNL 183
P D SP S P S +++ LS +N++L+R+N L SEL+ K
Sbjct: 121 PSESNSGGGDDHGSSSTSSPGSSKNPGSVENMVADLSGENEKLKRENNNLSSELAAAKKQ 180
Query: 184 YNDIIYFIQNHV 195
++++ F+ H+
Sbjct: 181 RDELVTFLTGHL 192
>At4g17750 heat shock transcription factor HSF1
Length = 495
Score = 161 bits (408), Expect = 2e-40
Identities = 89/191 (46%), Positives = 117/191 (60%), Gaps = 7/191 (3%)
Query: 15 TMESQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNF 74
T+ + S+P PFL+KTY +V+DP TD IVSWS +F+VW PPEF+RDLLP +FKHNNF
Sbjct: 42 TLLNANSLPPPFLSKTYDMVEDPATDAIVSWSPTNNSFIVWDPPEFSRDLLPKYFKHNNF 101
Query: 75 SSFVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQ-HYQQQYYEQSPQIF 133
SSFVRQLNTYGF+KV DRWEFAN+ F +G KHLL +I RRK+ Q H QS Q+
Sbjct: 102 SSFVRQLNTYGFRKVDPDRWEFANEGFLRGQKHLLKKISRRKSVQGHGSSSSNPQSQQLS 161
Query: 134 QPDESICWIDSPLPSPKSNTDILTALSEDNQRLRRKNFMLLSELSHMKNLYNDIIYFIQN 193
Q S+ + S + K L E+ ++L+R +L+ EL ++ +Q
Sbjct: 162 QGQGSMAALSSCVEVGK------FGLEEEVEQLKRDKNVLMQELVKLRQQQQTTDNKLQV 215
Query: 194 HVSPASPFEQR 204
V EQR
Sbjct: 216 LVKHLQVMEQR 226
>At1g32330 heat shock transcription factor HSF8 like protein
Length = 482
Score = 152 bits (384), Expect = 1e-37
Identities = 85/185 (45%), Positives = 111/185 (59%), Gaps = 12/185 (6%)
Query: 21 SVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSFVRQ 80
+ P PFL+KTY +VDD TD IVSWS + +F+VW+PPEFARDLLP FKHNNFSSFVRQ
Sbjct: 33 NAPPPFLSKTYDMVDDHNTDSIVSWSANNNSFIVWKPPEFARDLLPKNFKHNNFSSFVRQ 92
Query: 81 LNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQHYQQQYYEQSPQIFQPDESIC 140
LNTYGF+KV DRWEFAN+ F +G KHLL I RRK P H Q Q +++S + S+
Sbjct: 93 LNTYGFRKVDPDRWEFANEGFLRGQKHLLQSITRRK-PAHGQGQGHQRSQHSNGQNSSV- 150
Query: 141 WIDSPLPSPKSNTDI-LTALSEDNQRLRRKNFMLLSELSHMKNLYNDIIYFIQNHVSPAS 199
+ ++ L E+ +RL+R +L+ EL ++ +Q V
Sbjct: 151 ---------SACVEVGKFGLEEEVERLKRDKNVLMQELVRLRQQQQSTDNQLQTMVQRLQ 201
Query: 200 PFEQR 204
E R
Sbjct: 202 GMENR 206
>At2g41690 putative heat shock transcription factor
Length = 244
Score = 149 bits (375), Expect = 1e-36
Identities = 79/189 (41%), Positives = 113/189 (58%), Gaps = 10/189 (5%)
Query: 15 TMESQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNF 74
T ++ P PFL KTY++V+DP TD ++SW++ T FVVW+P EFARDLLP FKH NF
Sbjct: 30 TSTAELQPPPPFLVKTYKVVEDPTTDGVISWNEYGTGFVVWQPAEFARDLLPTLFKHCNF 89
Query: 75 SSFVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQHY-----QQQYYEQS 129
SSFVRQLNTYGF+KV RWEF+N+ F+KG + L+ I RRK+ QH+ Q +
Sbjct: 90 SSFVRQLNTYGFRKVTTIRWEFSNEMFRKGQRELMSNIRRRKS-QHWSHNKSNHQVVPTT 148
Query: 130 PQIFQPDESICWID----SPLPSPKSNTDILTALSEDNQRLRRKNFMLLSELSHMKNLYN 185
+ Q ID S S++ + TAL ++N+ L+ +N +L EL K
Sbjct: 149 TMVNQEGHQRIGIDHHHEDQQSSATSSSFVYTALLDENKCLKNENELLSCELGKTKKKCK 208
Query: 186 DIIYFIQNH 194
++ ++ +
Sbjct: 209 QLMELVERY 217
>At3g02990 putative heat shock transcription factor
Length = 468
Score = 145 bits (366), Expect = 2e-35
Identities = 82/184 (44%), Positives = 103/184 (55%), Gaps = 16/184 (8%)
Query: 21 SVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSFVRQ 80
S+P PFL+KTY +VDDPLTD +VSWS +FVVW PEFA+ LP +FKHNNFSSFVRQ
Sbjct: 20 SIP-PFLSKTYDMVDDPLTDDVVSWSSGNNSFVVWNVPEFAKQFLPKYFKHNNFSSFVRQ 78
Query: 81 LNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQHYQQQYYEQSPQIFQPDESIC 140
LNTYGF+KV DRWEFAN+ F +G K +L I RRK Q Q PQ+ C
Sbjct: 79 LNTYGFRKVDPDRWEFANEGFLRGQKQILKSIVRRKPAQVQP----PQQPQVQHSSVGAC 134
Query: 141 WIDSPLPSPKSNTDILTALSEDNQRLRRKNFMLLSELSHMKNLYNDIIYFIQNHVSPASP 200
L E+ +RL+R +L+ EL ++ + +QN
Sbjct: 135 VEVGKF-----------GLEEEVERLQRDKNVLMQELVRLRQQQQVTEHHLQNVGQKVHV 183
Query: 201 FEQR 204
EQR
Sbjct: 184 MEQR 187
>At4g18880 heat shock transcription factor 21 (AtHSF21)
Length = 401
Score = 144 bits (363), Expect = 3e-35
Identities = 67/103 (65%), Positives = 80/103 (77%), Gaps = 2/103 (1%)
Query: 18 SQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSF 77
S S+P PFLTKTY++VDD +D IVSWS +F+VW PPEF+RDLLP FFKHNNFSSF
Sbjct: 9 SSSSLP-PFLTKTYEMVDDSSSDSIVSWSQSNKSFIVWNPPEFSRDLLPRFFKHNNFSSF 67
Query: 78 VRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQH 120
+RQLNTYGF+K ++WEFAND F +G HL+ IHRRK P H
Sbjct: 68 IRQLNTYGFRKADPEQWEFANDDFVRGQPHLMKNIHRRK-PVH 109
>At3g22830 putative heat shock protein
Length = 406
Score = 144 bits (362), Expect = 4e-35
Identities = 68/112 (60%), Positives = 80/112 (70%)
Query: 20 KSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSFVR 79
+S P PFLTKTY LV+D T+H+VSWS +F+VW P F+ LLP FFKHNNFSSFVR
Sbjct: 56 ESGPPPFLTKTYDLVEDSRTNHVVSWSKSNNSFIVWDPQAFSVTLLPRFFKHNNFSSFVR 115
Query: 80 QLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQHYQQQYYEQSPQ 131
QLNTYGF+KV DRWEFAN+ F +G KHLL I RRKT + Q QS +
Sbjct: 116 QLNTYGFRKVNPDRWEFANEGFLRGQKHLLKNIRRRKTSNNSNQMQQPQSSE 167
>At5g43840 heat shock transcription factor-like protein
Length = 251
Score = 137 bits (345), Expect = 4e-33
Identities = 67/118 (56%), Positives = 80/118 (67%), Gaps = 1/118 (0%)
Query: 11 NMVFTMESQKSVP-APFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFF 69
N+ +E K P FLTKTY +V+D T++IVSWS D +F+VW P FA LP F
Sbjct: 4 NLPIPLEGLKETPPTAFLTKTYNIVEDSSTNNIVSWSRDNNSFIVWEPETFALICLPRCF 63
Query: 70 KHNNFSSFVRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQHYQQQYYE 127
KHNNFSSFVRQLNTYGFKK+ +RWEFAN++F KG +HLL I RRKT Q Q E
Sbjct: 64 KHNNFSSFVRQLNTYGFKKIDTERWEFANEHFLKGERHLLKNIKRRKTSSQTQTQSLE 121
>At4g13980 unknown protein
Length = 466
Score = 137 bits (344), Expect = 5e-33
Identities = 62/94 (65%), Positives = 73/94 (76%)
Query: 23 PAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSFVRQLN 82
PAPFL KTY++VDD TD IVSWS + +F+VW EF+R LLP +FKHNNFSSF+RQLN
Sbjct: 21 PAPFLVKTYEMVDDSSTDQIVSWSANNNSFIVWNHAEFSRLLLPTYFKHNNFSSFIRQLN 80
Query: 83 TYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRK 116
TYGF+K+ +RWEF ND F K KHLL IHRRK
Sbjct: 81 TYGFRKIDPERWEFLNDDFIKDQKHLLKNIHRRK 114
>At5g03720 heat shock transcription factor -like protein
Length = 476
Score = 136 bits (343), Expect = 7e-33
Identities = 62/98 (63%), Positives = 75/98 (76%)
Query: 25 PFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSFVRQLNTY 84
PFL+KT+ LVDDP D ++SW +FVVW P EFAR +LP FKHNNFSSFVRQLNTY
Sbjct: 119 PFLSKTFDLVDDPTLDPVISWGLTGASFVVWDPLEFARIILPRNFKHNNFSSFVRQLNTY 178
Query: 85 GFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQHYQ 122
GF+K+ D+WEFAN+ F +G KHLL IHRR++PQ Q
Sbjct: 179 GFRKIDTDKWEFANEAFLRGKKHLLKNIHRRRSPQSNQ 216
>At5g45710 heat shock transcription factor
Length = 345
Score = 133 bits (334), Expect = 8e-32
Identities = 73/172 (42%), Positives = 96/172 (55%), Gaps = 8/172 (4%)
Query: 25 PFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSFVRQLNTY 84
PFLTKTY++VDD +D +V+WS++ +F+V P EF+RDLLP FFKH NFSSF+RQLNTY
Sbjct: 13 PFLTKTYEMVDDSSSDSVVAWSENNKSFIVKNPAEFSRDLLPRFFKHKNFSSFIRQLNTY 72
Query: 85 GFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQHYQQQYYEQSPQIFQPDESICWIDS 144
GF+KV ++WEF ND F +G +L+ IHRRK P H Q+ E D
Sbjct: 73 GFRKVDPEKWEFLNDDFVRGRPYLMKNIHRRK-PVHSHSLVNLQAQNPLTESERRSMEDQ 131
Query: 145 PLPSPKSNTDILTALSEDNQRLRRKNF-----MLLSELSHMKNLYNDIIYFI 191
+L L NQ RK F L L HM+ I+ ++
Sbjct: 132 IERLKNEKEGLLAEL--QNQEQERKEFELQVTTLKDRLQHMEQHQKSIVAYV 181
>At5g16820 Heat Shock Factor 3
Length = 447
Score = 132 bits (331), Expect = 2e-31
Identities = 75/172 (43%), Positives = 96/172 (55%), Gaps = 14/172 (8%)
Query: 33 LVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSFVRQLNTYGFKKVVAD 92
+VDDPLT+ +VSWS +FVVW PEF++ LLP +FKHNNFSSFVRQLNTYGF+KV D
Sbjct: 1 MVDDPLTNEVVSWSSGNNSFVVWSAPEFSKVLLPKYFKHNNFSSFVRQLNTYGFRKVDPD 60
Query: 93 RWEFANDYFKKGAKHLLCEIHRRKTPQHYQQQYYEQSPQIFQPDESICWIDSPLPSPKSN 152
RWEFAN+ F +G K LL I RRK P H QQ +Q Q+ C
Sbjct: 61 RWEFANEGFLRGRKQLLKSIVRRK-PSHVQQN--QQQTQVQSSSVGACVEVGKF------ 111
Query: 153 TDILTALSEDNQRLRRKNFMLLSELSHMKNLYNDIIYFIQNHVSPASPFEQR 204
+ E+ +RL+R +L+ EL ++ +QN EQR
Sbjct: 112 -----GIEEEVERLKRDKNVLMQELVRLRQQQQATENQLQNVGQKVQVMEQR 158
>At2g26150 heat shock transcription factor like protein
Length = 345
Score = 131 bits (329), Expect = 3e-31
Identities = 58/94 (61%), Positives = 73/94 (76%)
Query: 23 PAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSFVRQLN 82
P PFLTKTY++V+DP TD +VSWS+ +FVVW +F+ LLP +FKH+NFSSF+RQLN
Sbjct: 42 PPPFLTKTYEMVEDPATDTVVSWSNGRNSFVVWDSHKFSTTLLPRYFKHSNFSSFIRQLN 101
Query: 83 TYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRK 116
TYGF+K+ DRWEFAN+ F G KHLL I RR+
Sbjct: 102 TYGFRKIDPDRWEFANEGFLAGQKHLLKNIKRRR 135
>At3g63350 heat shock transcription factor-like protein
Length = 282
Score = 129 bits (324), Expect = 1e-30
Identities = 73/190 (38%), Positives = 106/190 (55%), Gaps = 12/190 (6%)
Query: 19 QKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSFV 78
Q++ P+PFLTKT+++V DP T+HIVSW+ +FVVW P F+ +LP +FKHNNFSSFV
Sbjct: 22 QEAGPSPFLTKTFEMVGDPNTNHIVSWNRGGISFVVWDPHSFSATILPLYFKHNNFSSFV 81
Query: 79 RQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRKTPQHYQQQYYEQS-PQIFQPDE 137
RQLNTYGF+K+ A+RWEF N+ F G + LL I RR + Y QS P+ P
Sbjct: 82 RQLNTYGFRKIEAERWEFMNEGFLMGQRDLLKSIKRRTSSSSPPSLNYSQSQPEAHDPGV 141
Query: 138 SICWIDSPLPSPKSNTDIL----TALSEDNQRLRRKNFMLLSELSHMKNLYNDIIYFIQN 193
LP + +L + L ++ QR R + ++ + ++ F++
Sbjct: 142 E-------LPQLREERHVLMMEISTLRQEEQRARGYVQAMEQRINGAEKKQRHMMSFLRR 194
Query: 194 HVSPASPFEQ 203
V S +Q
Sbjct: 195 AVENPSLLQQ 204
>At3g24520 heat shock transcription factor HSF1, putative
Length = 330
Score = 127 bits (319), Expect = 4e-30
Identities = 57/99 (57%), Positives = 76/99 (76%)
Query: 18 SQKSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSF 77
+ +V APF+ KTYQ+V+DP TD +++W +F+V P +F++ +LP +FKHNNFSSF
Sbjct: 10 NNNNVIAPFIVKTYQMVNDPSTDWLITWGPAHNSFIVVDPLDFSQRILPAYFKHNNFSSF 69
Query: 78 VRQLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRRK 116
VRQLNTYGF+KV DRWEFAN++F +G KHLL I RRK
Sbjct: 70 VRQLNTYGFRKVDPDRWEFANEHFLRGQKHLLNNIARRK 108
>At3g51910 putative heat shock transcription factor
Length = 272
Score = 126 bits (317), Expect = 7e-30
Identities = 58/96 (60%), Positives = 73/96 (75%)
Query: 20 KSVPAPFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSFVR 79
++ P PFLTKT+++VDDP TDHIVSW+ T+FVVW F+ LLP FKH+NFSSF+R
Sbjct: 24 ENAPPPFLTKTFEMVDDPNTDHIVSWNRGGTSFVVWDLHSFSTILLPRHFKHSNFSSFIR 83
Query: 80 QLNTYGFKKVVADRWEFANDYFKKGAKHLLCEIHRR 115
QLNTYGF+K+ A+RWEFAN+ F G + LL I RR
Sbjct: 84 QLNTYGFRKIEAERWEFANEEFLLGQRQLLKNIKRR 119
>At5g54070 putative protein
Length = 331
Score = 125 bits (313), Expect = 2e-29
Identities = 55/91 (60%), Positives = 71/91 (77%)
Query: 25 PFLTKTYQLVDDPLTDHIVSWSDDETTFVVWRPPEFARDLLPNFFKHNNFSSFVRQLNTY 84
PFL KT+++VDD +TD +VSWS +F++W EF+ +LLP +FKH NFSSF+RQLN+Y
Sbjct: 71 PFLRKTFEIVDDKVTDPVVSWSPTRKSFIIWDSYEFSENLLPKYFKHKNFSSFIRQLNSY 130
Query: 85 GFKKVVADRWEFANDYFKKGAKHLLCEIHRR 115
GFKKV +DRWEFAN+ F+ G KHLL I RR
Sbjct: 131 GFKKVDSDRWEFANEGFQGGKKHLLKNIKRR 161
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.134 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,283,082
Number of Sequences: 26719
Number of extensions: 237454
Number of successful extensions: 600
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 561
Number of HSP's gapped (non-prelim): 42
length of query: 211
length of database: 11,318,596
effective HSP length: 95
effective length of query: 116
effective length of database: 8,780,291
effective search space: 1018513756
effective search space used: 1018513756
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146941.18


