

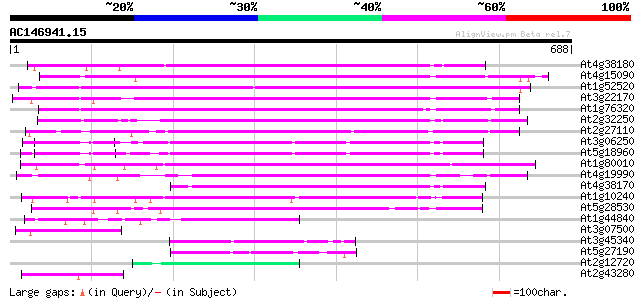
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146941.15 - phase: 0
(688 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g38180 unknown protein (At4g38180) 374 e-104
At4g15090 unknown protein 368 e-102
At1g52520 F6D8.26 361 e-100
At3g22170 far-red impaired response protein, putative 346 3e-95
At1g76320 putative phytochrome A signaling protein 332 6e-91
At2g32250 Mutator-like transposase 312 5e-85
At2g27110 Mutator-like transposase 311 1e-84
At3g06250 unknown protein 301 8e-82
At5g18960 FAR1 - like protein 292 4e-79
At1g80010 hypothetical protein 291 1e-78
At4g19990 putative protein 271 1e-72
At4g38170 hypothetical protein 260 2e-69
At1g10240 unknown protein 200 2e-51
At5g28530 far-red impaired response protein (FAR1) - like 195 6e-50
At1g44840 hypothetical protein 59 7e-09
At3g07500 hypothetical protein 59 9e-09
At3g45340 putative protein 57 3e-08
At5g27190 putative protein 56 6e-08
At2g12720 putative protein 56 8e-08
At2g43280 unknown protein 54 3e-07
>At4g38180 unknown protein (At4g38180)
Length = 788
Score = 374 bits (961), Expect = e-104
Identities = 207/582 (35%), Positives = 312/582 (53%), Gaps = 28/582 (4%)
Query: 23 SDDDEAC----WKPATGMRFSCIDDVKTLYREYALKKGFRWKTRTSKK-RDDGETCYAIL 77
+ ++EAC +P G+ F + K Y YA + GF + +S++ R DG
Sbjct: 60 NQEEEACDLLDLEPYDGLEFESEEAAKAFYNSYARRIGFSTRVSSSRRSRRDGAIIQRQF 119
Query: 78 VCTREGSQGSEIPCT----LKTPPSKTK-NCPAKICIKLEKDGLWYISKFESRHSHKTSP 132
VC +EG + T +K P + T+ C A + +K++ G W +S F H+H+ P
Sbjct: 120 VCAKEGFRNMNEKRTKDREIKRPRTITRVGCKASLSVKMQDSGKWLVSGFVKDHNHELVP 179
Query: 133 T---------KRMDLHVKRTIEINEDAGVRTIKTFRSIVNNAEGHENIPFCEKDMINYV- 182
+++ K I+ + AG+ + +++ G + F E D NY+
Sbjct: 180 PDQVHCLRSHRQISGPAKTLIDTLQAAGMGPRRIMSALIKEYGGISKVGFTEVDCRNYMR 239
Query: 183 NNEQHLIGKEGDGKALMSYFSKMREQNSNFFYDIDLDDDFHVRNVFWADARSRATYEYFG 242
NN Q I EG+ + L+ Y +M N NFFY + +D V NVFWAD ++ + +FG
Sbjct: 240 NNRQKSI--EGEIQLLLDYLRQMNADNPNFFYSVQGSEDQSVGNVFWADPKAIMDFTHFG 297
Query: 243 DVVTFDTTYLTNKFDMPFATFVGVNHHGQSTLLGCGLLSGEDTESFVWLFKSWLRCMLGK 302
D VTFDTTY +N++ +PFA F GVNHHGQ L GC + E SFVWLF +WL M
Sbjct: 298 DTVTFDTTYRSNRYRLPFAPFTGVNHHGQPILFGCAFIINETEASFVWLFNTWLAAMSAH 357
Query: 303 APVGIVTDQCKAVQNAIELVFPTTRHRWCLWHIMKKIPEKLNE-YSEYKRIKSAMEGAVY 361
PV I TD ++ AI VFP RHR+C WHI+KK EKL+ + ++ +S V
Sbjct: 358 PPVSITTDHDAVIRAAIMHVFPGARHRFCKWHILKKCQEKLSHVFLKHPSFESDFHKCVN 417
Query: 362 DTHTTTGFEEKWCSFIDKFMLQQNDWLSGLYEERHRWAPTFVRKYFWAGMSTTQRSESMH 421
T + FE W S +DK+ L+ ++WL +Y +R +W P ++R F+A MS T RS+S++
Sbjct: 418 LTESVEDFERCWFSLLDKYELRDHEWLQAIYSDRRQWVPVYLRDTFFADMSLTHRSDSIN 477
Query: 422 AFFDGYINSTTSLNQFVKQYDNALSSRAEKEFEADFNSLDTTIPCVSNSSIEKQLQGEYT 481
++FDGYIN++T+L+QF K Y+ AL SR EKE +AD++++++ + S +EKQ YT
Sbjct: 478 SYFDGYINASTNLSQFFKLYEKALESRLEKEVKADYDTMNSPPVLKTPSPMEKQASELYT 537
Query: 482 HAKFKEVQAEFISKMNCAPSLNVVEGCFATYHVLEEVVVGGRHKENVFKVVFNQENQDFS 541
F Q E + + S +G TY V + G HK + K FN +
Sbjct: 538 RKLFMRFQEELVGTLTFMASKADDDGDLVTYQVAK---YGEAHKAHFVK--FNVLEMRAN 592
Query: 542 CECSLFEFRGILCCHVLSVCTRERVKNVPEKYVLTRWKKNIK 583
C C +FEF GI+C H+L+V + +P Y+L RW +N K
Sbjct: 593 CSCQMFEFSGIICRHILAVFRVTNLLTLPPYYILKRWTRNAK 634
>At4g15090 unknown protein
Length = 768
Score = 368 bits (945), Expect = e-102
Identities = 212/641 (33%), Positives = 342/641 (53%), Gaps = 37/641 (5%)
Query: 37 RFSCIDDVKTLYREYALKKGFRWKTRTSKKRDDGETCYAILVCTREGSQGSEIPCTLKTP 96
R S I +++ L+ K R +T D C V S GS + +
Sbjct: 4 RISFIRNMQNLWVFTTSIKNSRRSKKTKDFIDAKFACSRYGVTPESESSGS----SSRRS 59
Query: 97 PSKTKNCPAKICIKLEKDGLWYISKFESRHSHKTSPTKRMDLHVKRTIEINEDAGV---- 152
K +C A + +K DG W I +F H+H+ P ++R +++ E +
Sbjct: 60 TVKKTDCKASMHVKRRPDGKWIIHEFVKDHNHELLPALAYHFRIQRNVKLAEKNNIDILH 119
Query: 153 ----RTIKTFRSIVNNAEGHENI-PFCEKDMINYVNNEQHLIGKEGDGKALMSYFSKMRE 207
RT K + + + G++NI + D+ + V+ ++L +EGD + L+ YF ++++
Sbjct: 120 AVSERTKKMYVEMSRQSGGYKNIGSLLQTDVSSQVDKGRYLALEEGDSQVLLEYFKRIKK 179
Query: 208 QNSNFFYDIDLDDDFHVRNVFWADARSRATYEYFGDVVTFDTTYLTNKFDMPFATFVGVN 267
+N FFY IDL++D +RN+FWADA+SR Y F DVV+FDTTY+ +P A F+GVN
Sbjct: 180 ENPKFFYAIDLNEDQRLRNLFWADAKSRDDYLSFNDVVSFDTTYVKFNDKLPLALFIGVN 239
Query: 268 HHGQSTLLGCGLLSGEDTESFVWLFKSWLRCMLGKAPVGIVTDQCKAVQNAIELVFPTTR 327
HH Q LLGC L++ E E+FVWL K+WLR M G+AP I+TDQ K + +A+ + P TR
Sbjct: 240 HHSQPMLLGCALVADESMETFVWLIKTWLRAMGGRAPKVILTDQDKFLMSAVSELLPNTR 299
Query: 328 HRWCLWHIMKKIPEKLNE-YSEYKRIKSAMEGAVYDTHTTTGFEEKWCSFIDKFMLQQND 386
H + LWH+++KIPE + ++ ++ + T F+ +W + +F L+ ++
Sbjct: 300 HCFALWHVLEKIPEYFSHVMKRHENFLLKFNKCIFRSWTDDEFDMRWWKMVSQFGLENDE 359
Query: 387 WLSGLYEERHRWAPTFVRKYFWAGMSTTQRSESMHAFFDGYINSTTSLNQFVKQYDNALS 446
WL L+E R +W PTF+ F AGMST+QRSES+++FFD YI+ +L +F++QY L
Sbjct: 360 WLLWLHEHRQKWVPTFMSDVFLAGMSTSQRSESVNSFFDKYIHKKITLKEFLRQYGVILQ 419
Query: 447 SRAEKEFEADFNSLDTTIPCVSNSSIEKQLQGEYTHAKFKEVQAEFISKMNCAPSLNVVE 506
+R E+E ADF++ S S EKQ+ YTH FK+ Q E + + C P +
Sbjct: 420 NRYEEESVADFDTCHKQPALKSPSPWEKQMATTYTHTIFKKFQVEVLGVVACHPRKEKED 479
Query: 507 GCFATYHVLEEVVVGGRHKENVFKVVFNQENQDFSCECSLFEFRGILCCHVLSVCTRERV 566
AT+ V + K++ F V +++ + C C +FE++G LC H L +
Sbjct: 480 ENMATFRVQD------CEKDDDFLVTWSKTKSELCCFCRMFEYKGFLCRHALMILQMCGF 533
Query: 567 KNVPEKYVLTRWKKNIKRKHSYIKSSYCARELKPKMDRFDKLCKHFYEIAEVAAESED-- 624
++P +Y+L RW K+ K S + + A +++ ++ R++ LC E++E SE+
Sbjct: 534 ASIPPQYILKRWTKDAK---SGVLAGEGADQIQTRVQRYNDLCSRATELSEEGCVSEENY 590
Query: 625 --ATKALHETLH---QFNSNKDGITDIVNRSFIDDSNPNNG 660
A + L ETL N+ ++ IT+ +S NNG
Sbjct: 591 NIALRTLVETLKNCVDMNNARNNITE-------SNSQLNNG 624
>At1g52520 F6D8.26
Length = 703
Score = 361 bits (927), Expect = e-100
Identities = 219/637 (34%), Positives = 329/637 (51%), Gaps = 15/637 (2%)
Query: 11 GSIHDEGMEVDNSDDDEACWKPATGMRFSCIDDVKTLYREYALKKGFRWKTRTSK-KRDD 69
G G+ VD + +A PA GM F DD Y YA + GFR + + S KR
Sbjct: 67 GFDEQSGLLVDERKEFDA---PAVGMEFESYDDAYNYYNCYASEVGFRVRVKNSWFKRRS 123
Query: 70 GETCYAILVCTREGSQGSEIPCTLKTPPSKTKNCPAKICIKLEKDGLWYISKFESRHSHK 129
E A+L C+ +G + I + CPA I ++ W + + H+H
Sbjct: 124 KEKYGAVLCCSSQGFK--RINDVNRVRKETRTGCPAMIRMRQVDSKRWRVVEVTLDHNHL 181
Query: 130 TSPTKRMDLHVKRTIEINEDAGVRTIKTFRS-IVNNAEGHENIPFCEKDMINYVNNEQHL 188
+ KR + + +TIK +R+ +V+N K N + L
Sbjct: 182 LGCKLYKSVKRKRKCVSSPVSDAKTIKLYRACVVDNGSNVNPNSTLNKKFQNSTGSPDLL 241
Query: 189 IGKEGDGKALMSYFSKMREQNSNFFYDIDLDDDFHVRNVFWADARSRATYEYFGDVVTFD 248
K GD A+ +YF +M+ N NFFY +D++D+ +RNVFWADA S+ + YFGDV+ D
Sbjct: 242 NLKRGDSAAIYNYFCRMQLTNPNFFYLMDVNDEGQLRNVFWADAFSKVSCSYFGDVIFID 301
Query: 249 TTYLTNKFDMPFATFVGVNHHGQSTLLGCGLLSGEDTESFVWLFKSWLRCMLGKAPVGIV 308
++Y++ KF++P TF GVNHHG++TLL CG L+GE ES+ WL K WL M ++P IV
Sbjct: 302 SSYISGKFEIPLVTFTGVNHHGKTTLLSCGFLAGETMESYHWLLKVWLSVM-KRSPQTIV 360
Query: 309 TDQCKAVQNAIELVFPTTRHRWCLWHIMKKIPEKLNEYSEYKRIKSAMEGAVYDTHTTTG 368
TD+CK ++ AI VFP + R+ L HIM+KIPEKL Y ++ A AVY+T
Sbjct: 361 TDRCKPLEAAISQVFPRSHQRFSLTHIMRKIPEKLGGLHNYDAVRKAFTKAVYETLKVVE 420
Query: 369 FEEKWCSFIDKFMLQQNDWLSGLYEERHRWAPTFVRKYFWAGMSTTQRSESMHAFFDGYI 428
FE W + F + +N+WL LYEER +WAP +++ F+AG++ E++ FF+ Y+
Sbjct: 421 FEAAWGFMVHNFGVIENEWLRSLYEERAKWAPVYLKDTFFAGIAAAHPGETLKPFFERYV 480
Query: 429 NSTTSLNQFVKQYDNALSSRAEKEFEADFNSLD-TTIPCVSNSSIEKQLQGEYTHAKFKE 487
+ T L +F+ +Y+ AL + +E +D S T + S E QL YT FK+
Sbjct: 481 HKQTPLKEFLDKYELALQKKHREETLSDIESQTLNTAELKTKCSFETQLSRIYTRDMFKK 540
Query: 488 VQAEFISKMNCAPSLNV-VEGCFATYHVLEEV-VVGGRHKENVFKVVFNQENQDFSCECS 545
Q E +C + V V+G F + V E V R + F+V++N+ + C CS
Sbjct: 541 FQIEVEEMYSCFSTTQVHVDGPFVIFLVKERVRGESSRREIRDFEVLYNRSVGEVRCICS 600
Query: 546 LFEFRGILCCHVLSVCTRERVKNVPEKYVLTRWKKNIKRKHSYIKSSYCARELKPKMDRF 605
F F G LC H L V V+ +P +Y+L RW+K+ KR H + ++ F
Sbjct: 601 CFNFYGYLCRHALCVLNFNGVEEIPLRYILPRWRKDYKRLHFADNGLTGFVDGTDRVQWF 660
Query: 606 DKLCKHFYEIAEVAAESED----ATKALHETLHQFNS 638
D+L K+ ++ E A S D A + L E+L + +S
Sbjct: 661 DQLYKNSLQVVEEGAVSLDHYKVAMQVLQESLDKVHS 697
>At3g22170 far-red impaired response protein, putative
Length = 814
Score = 346 bits (887), Expect = 3e-95
Identities = 201/654 (30%), Positives = 328/654 (49%), Gaps = 57/654 (8%)
Query: 4 SENLVDCGSIHDEGMEVDNSDD--------------DEACWKPATGMRFSCIDDVKTLYR 49
+E +D G I D +EV+ D + +P GM F + + Y+
Sbjct: 28 NEEDMDIGKIEDVSVEVNTDDSVGMGVPTGELVEYTEGMNLEPLNGMEFESHGEAYSFYQ 87
Query: 50 EYALKKGFRWKTRTSKK-RDDGETCYAILVCTREGSQGSEIPCTLKTPPSKTK------- 101
EY+ GF + S++ + E A C+R G++ E + P ++
Sbjct: 88 EYSRAMGFNTAIQNSRRSKTTREFIDAKFACSRYGTK-REYDKSFNRPRARQSKQDPENM 146
Query: 102 ---------NCPAKICIKLEKDGLWYISKFESRHSHKTSPTKRMDLHVKRTIEINEDAGV 152
+C A + +K DG W I F H+H+ P + +
Sbjct: 147 AGRRTCAKTDCKASMHVKRRPDGKWVIHSFVREHNHELLPAQAVS--------------E 192
Query: 153 RTIKTFRSIVNNAEGHENIPFCEKDMINYVNNEQHLIGKEGDGKALMSYFSKMREQNSNF 212
+T K + ++ ++ + + D + + L + GD K L+ + S+M+ NSNF
Sbjct: 193 QTRKIYAAMAKQFAEYKTVISLKSDSKSSFEKGRTLSVETGDFKILLDFLSRMQSLNSNF 252
Query: 213 FYDIDLDDDFHVRNVFWADARSRATYEYFGDVVTFDTTYLTNKFDMPFATFVGVNHHGQS 272
FY +DL DD V+NVFW DA+SR Y F DVV+ DTTY+ NK+ MP A FVGVN H Q
Sbjct: 253 FYAVDLGDDQRVKNVFWVDAKSRHNYGSFCDVVSLDTTYVRNKYKMPLAIFVGVNQHYQY 312
Query: 273 TLLGCGLLSGEDTESFVWLFKSWLRCMLGKAPVGIVTDQCKAVQNAIELVFPTTRHRWCL 332
+LGC L+S E ++ WL ++WLR + G+AP ++T+ + + + +FP TRH L
Sbjct: 313 MVLGCALISDESAATYSWLMETWLRAIGGQAPKVLITELDVVMNSIVPEIFPNTRHCLFL 372
Query: 333 WHIMKKIPEKLNE-YSEYKRIKSAMEGAVYDTHTTTGFEEKWCSFIDKFMLQQNDWLSGL 391
WH++ K+ E L + ++ E +Y + F KW + +F L+ + W+ L
Sbjct: 373 WHVLMKVSENLGQVVKQHDNFMPKFEKCIYKSGKDEDFARKWYKNLARFGLKDDQWMISL 432
Query: 392 YEERHRWAPTFVRKYFWAGMSTTQRSESMHAFFDGYINSTTSLNQFVKQYDNALSSRAEK 451
YE+R +WAPT++ AGMST+QR++S++AFFD Y++ TS+ +FVK YD L R E+
Sbjct: 433 YEDRKKWAPTYMTDVLLAGMSTSQRADSINAFFDKYMHKKTSVQEFVKVYDTVLQDRCEE 492
Query: 452 EFEADFNSLDTTIPCVSNSSIEKQLQGEYTHAKFKEVQAEFISKMNCAPSLNVVEGCFAT 511
E +AD + S S EK + YT A FK+ Q E + + C+P + +T
Sbjct: 493 EAKADSEMWNKQPAMKSPSPFEKSVSEVYTPAVFKKFQIEVLGAIACSPREENRDATCST 552
Query: 512 YHVLEEVVVGGRHKENVFKVVFNQENQDFSCECSLFEFRGILCCHVLSVCTRERVKNVPE 571
+ V + F V +NQ + SC C LFE++G LC H L+V + ++P
Sbjct: 553 FRVQD------FENNQDFMVTWNQTKAEVSCICRLFEYKGYLCRHTLNVLQCCHLSSIPS 606
Query: 572 KYVLTRWKKNIKRKHSYIKSSYCARELKPKMDRFDKLCKHFYEIAEVAAESEDA 625
+Y+L RW K+ K +H S ++L+ ++ R++ LC+ ++ E A+ S+++
Sbjct: 607 QYILKRWTKDAKSRH----FSGEPQQLQTRLLRYNDLCERALKLNEEASLSQES 656
>At1g76320 putative phytochrome A signaling protein
Length = 670
Score = 332 bits (850), Expect = 6e-91
Identities = 191/595 (32%), Positives = 306/595 (51%), Gaps = 17/595 (2%)
Query: 36 MRFSCIDDVKTLYREYALKKGFRWKTRTSKK-RDDGETCYAILVCTREGSQGSEIPCTLK 94
M F +D Y++YA GF +S++ R E A C R GS+ + +
Sbjct: 1 MEFETHEDAYLFYKDYAKSVGFGTAKLSSRRSRASKEFIDAKFSCIRYGSK-QQSDDAIN 59
Query: 95 TPPSKTKNCPAKICIKLEKDGLWYISKFESRHSHKTSPTKRMDLHVKRTIEI--NEDAGV 152
S C A + +K DG WY+ F H+H P + R E+ + D+ +
Sbjct: 60 PRASPKIGCKASMHVKRRPDGKWYVYSFVKEHNHDLLPEQAHYFRSHRNTELVKSNDSRL 119
Query: 153 RTIK-TFRSIVNNAEGHENIPFCEKDMINYVNNEQHLIGKEGDGKALMSYFSKMREQNSN 211
R K T + + + ++ F + M N + + L+ GD + L+ + +M+E+N
Sbjct: 120 RRKKNTPLTDCKHLSAYHDLDFIDGYMRNQHDKGRRLVLDTGDAEILLEFLMRMQEENPK 179
Query: 212 FFYDIDLDDDFHVRNVFWADARSRATYEYFGDVVTFDTTYLTNKFDMPFATFVGVNHHGQ 271
FF+ +D +D +RNVFW DA+ Y+ F DVV+F+T+Y +K+ +P FVGVNHH Q
Sbjct: 180 FFFAVDFSEDHLLRNVFWVDAKGIEDYKSFSDVVSFETSYFVSKYKVPLVLFVGVNHHVQ 239
Query: 272 STLLGCGLLSGEDTESFVWLFKSWLRCMLGKAPVGIVTDQCKAVQNAIELVFPTTRHRWC 331
LLGCGLL+ + ++VWL +SWL M G+ P ++TDQ A++ AI V P TRH +C
Sbjct: 240 PVLLGCGLLADDTVYTYVWLMQSWLVAMGGQKPKVMLTDQNNAIKAAIAAVLPETRHCYC 299
Query: 332 LWHIMKKIPEKLNEYSEYK-RIKSAMEGAVYDTHTTTGFEEKWCSFIDKFMLQQNDWLSG 390
LWH++ ++P L+ +S ++ + +Y + + F+ +W IDKF L+ W+
Sbjct: 300 LWHVLDQLPRNLDYWSMWQDTFMKKLFKCIYRSWSEEEFDRRWLKLIDKFHLRDVPWMRS 359
Query: 391 LYEERHRWAPTFVRKYFWAGMSTTQRSESMHAFFDGYINSTTSLNQFVKQYDNALSSRAE 450
LYEER WAPTF+R +AG+S RSES+++ FD Y++ TSL +F++ Y L R E
Sbjct: 360 LYEERKFWAPTFMRGITFAGLSMRCRSESVNSLFDRYVHPETSLKEFLEGYGLMLEDRYE 419
Query: 451 KEFEADFNSLDTTIPCVSNSSIEKQLQGEYTHAKFKEVQAEFISKMNCAPSLNVVEGCFA 510
+E +ADF++ S S EKQ+ Y+H F+ Q E + C + EG
Sbjct: 420 EEAKADFDAWHEAPELKSPSPFEKQMLLVYSHEIFRRFQLEVLGAAACHLTKESEEG--T 477
Query: 511 TYHVLEEVVVGGRHKENVFKVVFNQENQDFSCECSLFEFRGILCCHVLSVCTRERVKNVP 570
TY V + E + V +++ D C C FE++G LC H + V V +P
Sbjct: 478 TYSVKD------FDDEQKYLVDWDEFKSDIYCSCRSFEYKGYLCRHAIVVLQMSGVFTIP 531
Query: 571 EKYVLTRWKKNIKRKHSYIKSSYCARELKPKMDRFDKLCKHFYEIAEVAAESEDA 625
YVL RW + +H ++ ++ + RF+ LC+ + E + S+++
Sbjct: 532 INYVLQRWTNAARNRHQISRN---LELVQSNIRRFNDLCRRAIILGEEGSLSQES 583
>At2g32250 Mutator-like transposase
Length = 684
Score = 312 bits (799), Expect = 5e-85
Identities = 196/604 (32%), Positives = 297/604 (48%), Gaps = 45/604 (7%)
Query: 35 GMRFSCIDDVKTLYREYALKKGFRWKTRTSKK-RDDGETCYAILVCTREGSQGSEIPCTL 93
GM F + YREYA GF + S++ + G+ + C+R G++ + T
Sbjct: 41 GMDFESKEAAYYFYREYARSVGFGITIKASRRSKRSGKFIDVKIACSRFGTKREK--ATA 98
Query: 94 KTPPSKTKN-CPAKICIKLEKDGLWYISKFESRHSHKTSPTKRMDLHVKRTIEINEDAGV 152
P S K C A + +K ++D W I F H+H+ P D +V + N+ AG
Sbjct: 99 INPRSCPKTGCKAGLHMKRKEDEKWVIYNFVKEHNHEICPD---DFYVSVRGK-NKPAGA 154
Query: 153 RTIKTFRSIVNNAEGHENIPFCEKDMINYVNNEQHLIGKEGDGKALMSYFSKMREQNSNF 212
IK L +E D K L+ +F +M+++ F
Sbjct: 155 LAIK---------------------------KGLQLALEEEDLKLLLEHFMEMQDKQPGF 187
Query: 213 FYDIDLDDDFHVRNVFWADARSRATYEYFGDVVTFDTTYLTNKFDMPFATFVGVNHHGQS 272
FY +D D D VRNVFW DA+++ Y F DVV FDT Y+ N + +PFA F+GV+HH Q
Sbjct: 188 FYAVDFDSDKRVRNVFWLDAKAKHDYCSFSDVVLFDTFYVRNGYRIPFAPFIGVSHHRQY 247
Query: 273 TLLGCGLLSGEDTESFVWLFKSWLRCMLGKAPVGIVTDQCKAVQNAIELVFPTTRHRWCL 332
LLGC L+ ++ WLF++WL+ + G+AP ++TDQ K + + + VFP RH +CL
Sbjct: 248 VLLGCALIGEVSESTYSWLFRTWLKAVGGQAPGVMITDQDKLLSDIVVEVFPDVRHIFCL 307
Query: 333 WHIMKKIPEKLNEY-SEYKRIKSAMEGAVYDTHTTTGFEEKWCSFIDKFMLQQNDWLSGL 391
W ++ KI E LN + S+ + V + T FE +W + I KF L +N+W+ L
Sbjct: 308 WSVLSKISEMLNPFVSQDDGFMESFGNCVASSWTDEHFERRWSNMIGKFELNENEWVQLL 367
Query: 392 YEERHRWAPTFVRKYFWAGMSTTQRSESMHAFFDGYINSTTSLNQFVKQYDNALSSRAEK 451
+ +R +W P + AG+S +RS S+ + FD Y+NS + F + Y L R +
Sbjct: 368 FRDRKKWVPHYFHGICLAGLSGPERSGSIASHFDKYMNSEATFKDFFELYMKFLQYRCDV 427
Query: 452 EFEADFNSLDTTIPCVSNSSIEKQLQGEYTHAKFKEVQAEFISKMNCAPSLNVVEGCFAT 511
E + D S+ + EKQL YT A FK+ QAE ++C +G A
Sbjct: 428 EAKDDLEYQSKQPTLRSSLAFEKQLSLIYTDAAFKKFQAEVPGVVSCQLQKEREDGTTAI 487
Query: 512 YHVLEEVVVGGRHKENVFKVVFNQENQDFSCECSLFEFRGILCCHVLSVCTRERVKNVPE 571
+ + + ++N F V N E D C C LFE++G LC H + V V VP
Sbjct: 488 FRIED-----FEERQNFF-VALNNELLDACCSCHLFEYQGFLCKHAILVLQSADVSRVPS 541
Query: 572 KYVLTRWKKNIKRKHSYIKSSYCARELKPKMDRFDKLCKHFYEIAEVAAESEDATKALHE 631
+Y+L RW K K K+ CA + +M RFD LC+ F ++ VA+ S++A K +
Sbjct: 542 QYILKRWSKKGNNKED--KNDKCA-TIDNRMARFDDLCRRFVKLGVVASLSDEACKTALK 598
Query: 632 TLHQ 635
L +
Sbjct: 599 LLEE 602
>At2g27110 Mutator-like transposase
Length = 851
Score = 311 bits (796), Expect = 1e-84
Identities = 195/619 (31%), Positives = 297/619 (47%), Gaps = 45/619 (7%)
Query: 20 VDNS---DDDEACWKPATGMRFSCIDDVKTLYREYALKKGFRWKTRTSK--KRDDGETCY 74
+DNS D+ +P GM F+ + K+ Y EY+ + GF TSK R DG
Sbjct: 34 MDNSLGVQDEIGIAEPCVGMEFNSEKEAKSFYDEYSRQLGF-----TSKLLPRTDGSVSV 88
Query: 75 AILVCTREGSQGSEIPCTLKTPPSKTKNCPAKICIKLEKDGLWYISKFESRHSHKTSPTK 134
VC+ + +++C A + I+L+ W ++KF H+H + +
Sbjct: 89 REFVCSSSSKRSKR---------RLSESCDAMVRIELQGHEKWVVTKFVKEHTHGLASSN 139
Query: 135 RMDLHVKRTIEIN-------EDAGVRTIKTFRSIVNNAEGHENIPFCEKDMINYVNNEQH 187
+ R N E V + + S+ N+ G N + N +
Sbjct: 140 MLHCLRPRRHFANSEKSSYQEGVNVPSGMMYVSMDANSRGARNA--------SMATNTKR 191
Query: 188 LIGKEGDGKALMSYFSKMREQNSNFFYDIDLDDDFHVRNVFWADARSRATYEYFGDVVTF 247
IG+ D L+ YF +M+ +N FFY + LD+D + NVFWAD+RSR Y +FGD VT
Sbjct: 192 TIGR--DAHNLLEYFKRMQAENPGFFYAVQLDEDNQMSNVFWADSRSRVAYTHFGDTVTL 249
Query: 248 DTTYLTNKFDMPFATFVGVNHHGQSTLLGCGLLSGEDTESFVWLFKSWLRCMLGKAPVGI 307
DT Y N+F +PFA F GVNHHGQ+ L GC L+ E SF+WLFK++L M + PV +
Sbjct: 250 DTRYRCNQFRVPFAPFTGVNHHGQAILFGCALILDESDTSFIWLFKTFLTAMRDQPPVSL 309
Query: 308 VTDQCKAVQNAIELVFPTTRHRWCLWHIMKKIPEKLNEYS-EYKRIKSAMEGAVYDTHTT 366
VTDQ +A+Q A VFP RH W ++++ EKL Y + + + T T
Sbjct: 310 VTDQDRAIQIAAGQVFPGARHCINKWDVLREGQEKLAHVCLAYPSFQVELYNCINFTETI 369
Query: 367 TGFEEKWCSFIDKFMLQQNDWLSGLYEERHRWAPTFVRKYFWAGMSTTQRSESMHAFFDG 426
FE W S IDK+ L +++WL+ LY R +W P + R F+A + +Q +FFDG
Sbjct: 370 EEFESSWSSVIDKYDLGRHEWLNSLYNARAQWVPVYFRDSFFAAVFPSQGYSG--SFFDG 427
Query: 427 YINSTTSLNQFVKQYDNALSSRAEKEFEADFNSLDTTIPCVSNSSIEKQLQGEYTHAKFK 486
Y+N T+L F + Y+ A+ S E E EAD ++++T + S +E Q +T F
Sbjct: 428 YVNQQTTLPMFFRLYERAMESWFEMEIEADLDTVNTPPVLKTPSPMENQAANLFTRKIFG 487
Query: 487 EVQAEFISKMNCAPSLNVVEGCFATYHVLEEVVVGGRHKENVFKVVFNQENQDFSCECSL 546
+ Q E + + +G +T+ V + + V F +C C +
Sbjct: 488 KFQEELVETFAHTANRIEDDGTTSTFR-----VANFENDNKAYIVTFCYPEMRANCSCQM 542
Query: 547 FEFRGILCCHVLSVCTRERVKNVPEKYVLTRWKKNIKRKHSYIKSSYCARELKPKMDRFD 606
FE GILC HVL+V T + +P Y+L RW +N K + + R++
Sbjct: 543 FEHSGILCRHVLTVFTVTNILTLPPHYILRRWTRNAKSMVE-LDEHVSENGHDSSIHRYN 601
Query: 607 KLCKHFYEIAEVAAESEDA 625
LC+ + AE A + +A
Sbjct: 602 HLCREAIKYAEEGAITAEA 620
>At3g06250 unknown protein
Length = 764
Score = 301 bits (771), Expect = 8e-82
Identities = 177/569 (31%), Positives = 284/569 (49%), Gaps = 43/569 (7%)
Query: 16 EGMEVDNSDDDEACWKPATGMRFSCIDDVKTLYREYALKKGFRWKT-RTSKKRDDGETCY 74
+G E + +P G+ F+ ++ Y+ YA GFR + + + + DG
Sbjct: 174 KGAEDSDGQTQPKATEPYAGLEFNSANEACQFYQAYAEVVGFRVRIGQLFRSKVDGSITS 233
Query: 75 AILVCTREGSQGSEIPCTLKTPPSKTKNCPAKICIKLEKDGLWYISKFESRHSHKTSPTK 134
VC++EG Q PS+ C A + IK + G W + + H+H P K
Sbjct: 234 RRFVCSKEGFQH----------PSRM-GCGAYMRIKRQDSGGWIVDRLNKDHNHDLEPGK 282
Query: 135 RMDLHVKRTIEINEDAGVRTIKTFRSIVNNAEGHENIPFCE-KDMINYVNN-EQHLIGKE 192
+ +AG++ I + G +++ E D+ N++++ ++ IGKE
Sbjct: 283 K-------------NAGMKKITD-----DVTGGLDSVDLIELNDLSNHISSTRENTIGKE 324
Query: 193 GDGKALMSYFSKMREQNSNFFYDIDLDDDFHVRNVFWADARSRATYEYFGDVVTFDTTYL 252
L+ YF + ++ FFY I+LD + ++FWAD+RSR FGD V FDT+Y
Sbjct: 325 WY-PVLLDYFQSKQAEDMGFFYAIELDSNGSCMSIFWADSRSRFACSQFGDAVVFDTSYR 383
Query: 253 TNKFDMPFATFVGVNHHGQSTLLGCGLLSGEDTESFVWLFKSWLRCMLGKAPVGIVTDQC 312
+ +PFATF+G NHH Q LLG L++ E E+F WLF++WLR M G+ P +V DQ
Sbjct: 384 KGDYSVPFATFIGFNHHRQPVLLGGALVADESKEAFSWLFQTWLRAMSGRRPRSMVADQD 443
Query: 313 KAVQNAIELVFPTTRHRWCLWHIMKKIPEKLNEYSEYKRIKSAMEGAVYDTHTTTGFEEK 372
+Q A+ VFP T HR+ W I K E L + K E +Y + TT F+
Sbjct: 444 LPIQQAVAQVFPGTHHRFSAWQIRSKERENLRSFP--NEFKYEYEKCLYQSQTTVEFDTM 501
Query: 373 WCSFIDKFMLQQNDWLSGLYEERHRWAPTFVRKYFWAGMSTTQRSESMHAFFDGYINSTT 432
W S ++K+ L+ N WL +YE+R +W P ++R F+ G+ + F+ +NS T
Sbjct: 502 WSSLVNKYGLRDNMWLREIYEKREKWVPAYLRASFFGGIHV---DGTFDPFYGTSLNSLT 558
Query: 433 SLNQFVKQYDNALSSRAEKEFEADFNSLDTTIPCVSNSSIEKQLQGEYTHAKFKEVQAEF 492
SL +F+ +Y+ L R E+E + DFNS + + +E+Q + YT F+ Q+E
Sbjct: 559 SLREFISRYEQGLEQRREEERKEDFNSYNLQPFLQTKEPVEEQCRRLYTLTIFRIFQSEL 618
Query: 493 ISKMNCAPSLNVVEGCFATYHVLEEVVVGGRHKENVFKVVFNQENQDFSCECSLFEFRGI 552
N EG + + V + G ++++ V F+ N + SC C +FE+ G+
Sbjct: 619 AQSYNYLGLKTYEEGAISRFLVRK---CGNENEKHA--VTFSASNLNASCSCQMFEYEGL 673
Query: 553 LCCHVLSVCTRERVKNVPEKYVLTRWKKN 581
LC H+L V ++ +P +Y+L RW KN
Sbjct: 674 LCRHILKVFNLLDIRELPSRYILHRWTKN 702
Score = 48.1 bits (113), Expect = 2e-05
Identities = 27/99 (27%), Positives = 47/99 (47%), Gaps = 12/99 (12%)
Query: 31 KPATGMRFSCIDDVKTLYREYALKKGFRWKT-RTSKKRDDGETCYAILVCTREGSQGSEI 89
+P G+ F ++ + Y YA + GF+ +T + + R DG VC++EG Q +
Sbjct: 27 EPYVGLEFDTAEEARDYYNSYATRTGFKVRTGQLYRSRTDGTVSSRRFVCSKEGFQLN-- 84
Query: 90 PCTLKTPPSKTKNCPAKICIKLEKDGLWYISKFESRHSH 128
S+T CPA I ++ G W + + + H+H
Sbjct: 85 --------SRT-GCPAFIRVQRRDTGKWVLDQIQKEHNH 114
>At5g18960 FAR1 - like protein
Length = 788
Score = 292 bits (748), Expect = 4e-79
Identities = 173/553 (31%), Positives = 275/553 (49%), Gaps = 38/553 (6%)
Query: 31 KPATGMRFSCIDDVKTLYREYALKKGFRWKT-RTSKKRDDGETCYAILVCTREGSQGSEI 89
+P G+ F ++ Y+ YA GFR + + + + DG VC+REG Q
Sbjct: 210 EPYAGLEFGSANEACQFYQAYAEVVGFRVRIGQLFRSKVDGSITSRRFVCSREGFQH--- 266
Query: 90 PCTLKTPPSKTKNCPAKICIKLEKDGLWYISKFESRHSHKTSPTKRMDLHVKRTIEINED 149
PS+ C A + IK + G W + + H+H P K+ D +K+ D
Sbjct: 267 -------PSRM-GCGAYMRIKRQDSGGWIVDRLNKDHNHDLEPGKKNDAGMKKI----PD 314
Query: 150 AGVRTIKTFRSIVNNAEGHENIPFCEKDMINYVNNEQHLIGKEGDGKALMSYFSKMREQN 209
G + + I N G+ +I ++ IGKE L+ YF + ++
Sbjct: 315 DGTGGLDSVDLIELNDFGNNHIK----------KTRENRIGKEWY-PLLLDYFQSRQTED 363
Query: 210 SNFFYDIDLD-DDFHVRNVFWADARSRATYEYFGDVVTFDTTYLTNKFDMPFATFVGVNH 268
FFY ++LD ++ ++FWAD+R+R FGD V FDT+Y + +PFAT +G NH
Sbjct: 364 MGFFYAVELDVNNGSCMSIFWADSRARFACSQFGDSVVFDTSYRKGSYSVPFATIIGFNH 423
Query: 269 HGQSTLLGCGLLSGEDTESFVWLFKSWLRCMLGKAPVGIVTDQCKAVQNAIELVFPTTRH 328
H Q LLGC +++ E E+F+WLF++WLR M G+ P IV DQ +Q A+ VFP H
Sbjct: 424 HRQPVLLGCAMVADESKEAFLWLFQTWLRAMSGRRPRSIVADQDLPIQQALVQVFPGAHH 483
Query: 329 RWCLWHIMKKIPEKLNEYSEYKRIKSAMEGAVYDTHTTTGFEEKWCSFIDKFMLQQNDWL 388
R+ W I +K E L + K E +Y T T F+ W + I+K+ L+ + WL
Sbjct: 484 RYSAWQIREKERENLIPFP--SEFKYEYEKCIYQTQTIVEFDSVWSALINKYGLRDDVWL 541
Query: 389 SGLYEERHRWAPTFVRKYFWAGMSTTQRSESMHAFFDGYINSTTSLNQFVKQYDNALSSR 448
+YE+R W P ++R F+AG+ + ++ FF +++ T L +F+ +Y+ AL R
Sbjct: 542 REIYEQRENWVPAYLRASFFAGIPI---NGTIEPFFGASLDALTPLREFISRYEQALEQR 598
Query: 449 AEKEFEADFNSLDTTIPCVSNSSIEKQLQGEYTHAKFKEVQAEFISKMNCAPSLNVVEGC 508
E+E + DFNS + + +E+Q + YT F+ Q E + N EG
Sbjct: 599 REEERKEDFNSYNLQPFLQTKEPVEEQCRRLYTLTVFRIFQNELVQSYNYLCLKTYEEGA 658
Query: 509 FATYHVLEEVVVGGRHKENVFKVVFNQENQDFSCECSLFEFRGILCCHVLSVCTRERVKN 568
+ + V + G +++ V F+ N + SC C +FE G+LC H+L V ++
Sbjct: 659 ISRFLVRK---CGNESEKHA--VTFSASNLNSSCSCQMFEHEGLLCRHILKVFNLLDIRE 713
Query: 569 VPEKYVLTRWKKN 581
+P +Y+L RW KN
Sbjct: 714 LPSRYILHRWTKN 726
Score = 46.6 bits (109), Expect = 5e-05
Identities = 29/117 (24%), Positives = 52/117 (43%), Gaps = 14/117 (11%)
Query: 14 HDEGMEVDNSDDDEACWKPATGMRFSCIDDVKTLYREYALKKGFRWKT-RTSKKRDDGET 72
H+ G+ D +P G+ F ++ + Y YA + GF+ +T + + R DG
Sbjct: 27 HNNGISEDEEGGSGV--EPYVGLEFDTAEEAREFYNAYAARTGFKVRTGQLYRSRTDGTV 84
Query: 73 CYAILVCTREGSQGSEIPCTLKTPPSKTKNCPAKICIKLEKDGLWYISKFESRHSHK 129
VC++EG Q + S+T C A I ++ G W + + + H+H+
Sbjct: 85 SSRRFVCSKEGFQLN----------SRT-GCTAFIRVQRRDTGKWVLDQIQKEHNHE 130
>At1g80010 hypothetical protein
Length = 696
Score = 291 bits (744), Expect = 1e-78
Identities = 191/659 (28%), Positives = 314/659 (46%), Gaps = 36/659 (5%)
Query: 14 HDEGMEVD-NSDDDEACWK---PATGMRFSCIDDVKTLYREYALKKGFRWKTRTS-KKRD 68
H+E D N+ +E C P GM F DD + Y YA + GF + ++S KR+
Sbjct: 44 HEEETGCDENAFANEKCLMAPPPTPGMEFESYDDAYSFYNSYARELGFAIRVKSSWTKRN 103
Query: 69 DGETCYAILVCTREGSQGSEIPCTLKTPPSKTKN----CPAKICIKLEKDGLWYISKFES 124
E A+L C +G + LK S+ K C A I ++L W + + +
Sbjct: 104 SKEKRGAVLCCNCQGFK------LLKDAHSRRKETRTGCQAMIRLRLIHFDRWKVDQVKL 157
Query: 125 RHSHKTSPTKRMDLH------------VKRTIEINEDAGVRTIKTFRSIVNNAEGHENIP 172
H+H P + + K E VRTIK +R++ +
Sbjct: 158 DHNHSFDPQRAHNSKSHKKSSSSASPATKTNPEPPPHVQVRTIKLYRTLALDTPPALGTS 217
Query: 173 FCEKDM----INYVNNEQHLIGKEGDGKALMSYFSKMREQNSNFFYDIDLDDDFHVRNVF 228
+ +++ + + L G +AL +F +++ + NF Y +DL DD +RNVF
Sbjct: 218 LSSGETSDLSLDHFQSSRRL-ELRGGFRALQDFFFQIQLSSPNFLYLMDLADDGSLRNVF 276
Query: 229 WADARSRATYEYFGDVVTFDTTYLTNKFDMPFATFVGVNHHGQSTLLGCGLLSGEDTESF 288
W DAR+RA Y +FGDV+ FDTT L+N +++P FVG+NHHG + LLGCGLL+ + E++
Sbjct: 277 WIDARARAAYSHFGDVLLFDTTCLSNAYELPLVAFVGINHHGDTILLGCGLLADQSFETY 336
Query: 289 VWLFKSWLRCMLGKAPVGIVTDQCKAVQNAIELVFPTTRHRWCLWHIMKKIPEKLNEYSE 348
VWLF++WL CMLG+ P +T+QCKA++ A+ VFP HR L H++ I + + + +
Sbjct: 337 VWLFRAWLTCMLGRPPQIFITEQCKAMRTAVSEVFPRAHHRLSLTHVLHNICQSVVQLQD 396
Query: 349 YKRIKSAMEGAVYDTHTTTGFEEKWCSFIDKFMLQQNDWLSGLYEERHRWAPTFVRKYFW 408
A+ VY FE W I +F + N+ + ++++R WAP +++ F
Sbjct: 397 SDLFPMALNRVVYGCLKVEEFETAWEEMIIRFGMTNNETIRDMFQDRELWAPVYLKDTFL 456
Query: 409 AGMSTTQRSESMHAF-FDGYINSTTSLNQFVKQYDNALSSRAEKEFEADFNSLDTTIPCV 467
AG T F F GY++ TSL +F++ Y++ L + +E D SL
Sbjct: 457 AGALTFPLGNVAAPFIFSGYVHENTSLREFLEGYESFLDKKYTREALCDSESLKLIPKLK 516
Query: 468 SNSSIEKQLQGEYTHAKFKEVQAEFISKMNCAPSLNV-VEGCFATYHVLEEVVVGGRHKE 526
+ E Q+ +T F+ Q E + +C V G ++Y V E R E
Sbjct: 517 TTHPYESQMAKVFTMEIFRRFQDEVSAMSSCFGVTQVHSNGSASSYVVKEREGDKVRDFE 576
Query: 527 NVFKVVFNQENQDFSCECSLFEFRGILCCHVLSVCTRERVKNVPEKYVLTRWKKNIKRKH 586
+++ + + F C C F F G C HVL + + ++ VP +Y+L RW+K++KR +
Sbjct: 577 VIYETSAAAQVRCF-CVCGGFSFNGYQCRHVLLLLSHNGLQEVPPQYILQRWRKDVKRLY 635
Query: 587 SYIKSSYCARELKPKMDRFDKLCKHFYEIAEVAAESEDATKALHETLHQFNSNKDGITD 645
S + P ++ L + ++ E S++ +A E + + +T+
Sbjct: 636 VAEFGSGRVDIMNPD-QWYEHLHRRAMQVVEQGMRSKEHCRAAWEAFRECANKVQFVTE 693
>At4g19990 putative protein
Length = 672
Score = 271 bits (692), Expect = 1e-72
Identities = 182/650 (28%), Positives = 302/650 (46%), Gaps = 98/650 (15%)
Query: 9 DCGSIHDEGMEVDNSDDDEACWKPATGMRFSCIDDVKTLYREYALKKGFRWKTRTSKK-R 67
+C ++ + +N + DE G F ++ Y+EYA GF + S++ R
Sbjct: 5 ECSNVQLDDHRKNNLEIDE-------GREFESKEEAFEFYKEYANSVGFTTIIKASRRSR 57
Query: 68 DDGETCYAILVCTREGSQGSEIPCTLKTP---------------PSKTKNCPAKICIKLE 112
G+ A VCTR GS+ +I L T S +C A + +K
Sbjct: 58 MTGKFIDAKFVCTRYGSKKEDIDTGLGTDGFNIPQARKRGRINRSSSKTDCKAFLHVKRR 117
Query: 113 KDGLWYISKFESRHSHKT---SPTKRMDLHVKRTIEINEDAGVRTIKTFRSIVNNAEGHE 169
+DG W + H+H+ +L +R +E A V+ +K+ +
Sbjct: 118 QDGRWVVRSLVKEHNHEIFTGQADSLRELSGRRKLEKLNGAIVKEVKSRKL--------- 168
Query: 170 NIPFCEKDMINYVNNEQHLIGKEGDGKALMSYFSKMREQNSNFFYDIDLDDDFHVRNVFW 229
++GD + L+++F+ M+ +RN+FW
Sbjct: 169 ---------------------EDGDVERLLNFFTDMQS----------------LRNIFW 191
Query: 230 ADARSRATYEYFGDVVTFDTTYLTNKFDMPFATFVGVNHHGQSTLLGCGLLSGEDTES-F 288
DA+ R Y F DVV+ DTT++ N++ +P F GVNHHGQ LLG GLL ++++S F
Sbjct: 192 VDAKGRFDYTCFSDVVSIDTTFIKNEYKLPLVAFTGVNHHGQFLLLGFGLLLTDESKSGF 251
Query: 289 VWLFKSWLRCMLGKAPVGIVTDQCKAVQNAIELVFPTTRHRWCLWHIMKKIPEKLNEYSE 348
VWLF++WL+ M G P I+T + ++ A+ VFP++RH + +W + ++PEKL
Sbjct: 252 VWLFRAWLKAMHGCRPRVILTKHDQMLKEAVLEVFPSSRHCFYMWDTLGQMPEKLGHVIR 311
Query: 349 Y-KRIKSAMEGAVYDTHTTTGFEEKWCSFIDKFMLQQNDWLSGLYEERHRWAPTFVRKYF 407
K++ + A+Y + + FE+ W +D+F ++ N WL LYE+R W P +++
Sbjct: 312 LEKKLVDEINDAIYGSCQSEDFEKNWWEVVDRFHMRDNVWLQSLYEDREYWVPVYMKDVS 371
Query: 408 WAGMSTTQRSESMHAFFDGYINSTTSLNQFVKQYDNALSSRAEKEFEADFNSLDTTIPCV 467
AGM T QRS+S+++ D YI T+ F++QY + R E+E +++ +L
Sbjct: 372 LAGMCTAQRSDSVNSGLDKYIQRKTTFKAFLEQYKKMIQERYEEEEKSEIETLYKQPGLK 431
Query: 468 SNSSIEKQLQGEYTHAKFKEVQAEFISKMNCAPSLNVVEGCF--ATYHVLEEVVVGGRHK 525
S S KQ+ YT FK+ Q E + + C P E T+ V + +
Sbjct: 432 SPSPFGKQMAEVYTREMFKKFQVEVLGGVACHPKKESEEDGVNKRTFRVQD------YEQ 485
Query: 526 ENVFKVVFNQENQDFSCECSLFEFRGILCCHVLSVCTRERVKNVPEKYVLTRWKKNIKRK 585
F VV+N E+ + C C LFE +G L ++P +YVL RW K+ K +
Sbjct: 486 NRSFVVVWNSESSEVVCSCRLFELKGEL--------------SIPSQYVLKRWTKDAKSR 531
Query: 586 HSYIKSSYCARELKPKMDRFDKLCKHFYEIAEVAAESEDATKALHETLHQ 635
+ S K R+ LC +++E A+ SE++ A+ L++
Sbjct: 532 E--VMESDQTDVESTKAQRYKDLCLRSLKLSEEASLSEESYNAVVNVLNE 579
>At4g38170 hypothetical protein
Length = 531
Score = 260 bits (665), Expect = 2e-69
Identities = 134/388 (34%), Positives = 210/388 (53%), Gaps = 11/388 (2%)
Query: 198 LMSYFSKMREQNSNFFYDIDLDDDFHVRNVFWADARSRATYEYFGDVVTFDTTYLTNK-F 256
+++Y + + +N F Y I+ D NVFWAD R Y YFGD + FDTTY K +
Sbjct: 7 VLNYLKRRQLENPGFLYAIEDD----CGNVFWADPTCRLNYTYFGDTLVFDTTYRRGKRY 62
Query: 257 DMPFATFVGVNHHGQSTLLGCGLLSGEDTESFVWLFKSWLRCMLGKAPVGIVTDQCKAVQ 316
+PFA F G NHHGQ L GC L+ E SF WLF++WL+ M P I + + +Q
Sbjct: 63 QVPFAAFTGFNHHGQPVLFGCALILNESESSFAWLFQTWLQAMSAPPPPSITVEPDRLIQ 122
Query: 317 NAIELVFPTTRHRWCLWHIMKKIPEKL-NEYSEYKRIKSAMEGAVYDTHTTTGFEEKWCS 375
A+ VF TR R+ I ++ EKL + + + +S V +T T FE W S
Sbjct: 123 VAVSRVFSQTRLRFSQPLIFEETEEKLAHVFQAHPTFESEFINCVTETETAAEFEASWDS 182
Query: 376 FIDKFMLQQNDWLSGLYEERHRWAPTFVRKYFWAGMSTTQRSESMHAFFDGYINSTTSLN 435
+ ++ ++ NDWL +Y R +W F+R F+ +ST + S +++FF G+++++T++
Sbjct: 183 IVRRYYMEDNDWLQSIYNARQQWVRVFIRDTFYGELSTNEGSSILNSFFQGFVDASTTMQ 242
Query: 436 QFVKQYDNALSSRAEKEFEADFNSLDTTIPCVSNSSIEKQLQGEYTHAKFKEVQAEFISK 495
+KQY+ A+ S EKE +AD+ + ++T + S +EKQ YT A F + Q EF+
Sbjct: 243 MLIKQYEKAIDSWREKELKADYEATNSTPVMKTPSPMEKQAASLYTRAAFIKFQEEFVET 302
Query: 496 MNCAPSLNVVEGCFATYHVLEEVVVGGRHKENVFKVVFNQENQDFSCECSLFEFRGILCC 555
+ ++ G TY V + G HK + V F+ +C C +FE+ GI+C
Sbjct: 303 LAIPANIISDSGTHTTYRVAK---FGEVHKGHT--VSFDSLEVKANCSCQMFEYSGIICR 357
Query: 556 HVLSVCTRERVKNVPEKYVLTRWKKNIK 583
H+L+V + + V +P +Y+L RW K K
Sbjct: 358 HILAVFSAKNVLALPSRYLLRRWTKEAK 385
>At1g10240 unknown protein
Length = 680
Score = 200 bits (509), Expect = 2e-51
Identities = 160/599 (26%), Positives = 273/599 (44%), Gaps = 51/599 (8%)
Query: 15 DEGMEVDNSDDD-----EACWK--PATGMRFSCIDDVKTLYREYALKKGFRWKTRTSKKR 67
D+ + S DD EA P G F D Y +A + GF + ++ +
Sbjct: 24 DDASSTEESPDDNNLSLEAVHNAIPYLGQIFLTHDTAYEFYSTFAKRCGFSIRRHRTEGK 83
Query: 68 DD---GETCYAILVCTREGSQGSEIPCTLKTPPSKTKN-----CPAKICI-KLEKDGL-- 116
D G T VC R G+ + I + P + + C A + I KL + G
Sbjct: 84 DGVGKGLT-RRYFVCHRAGN--TPIKTLSEGKPQRNRRSSRCGCQAYLRISKLTELGSTE 140
Query: 117 WYISKFESRHSHKT-SPTKRMDLHVKRTIEINEDAGV----RTIKTFRSIVNNAEGHENI 171
W ++ F + H+H+ P + L R+I + + + +T + + ++ E + +
Sbjct: 141 WRVTGFANHHNHELLEPNQVRFLPAYRSISDADKSRILMFSKTGISVQQMMRLLELEKCV 200
Query: 172 -----PFCEKDMINYVNNEQHLIGKEGDGKALMSYFSKMREQNSNFFYDIDLDDDFHVRN 226
PF EKD+ N + + + L E + + ++E++ NF ++ LD + + N
Sbjct: 201 EPGFLPFTEKDVRNLLQSFKKL-DPEDENIDFLRMCQSIKEKDPNFKFEFTLDANDKLEN 259
Query: 227 VFWADARSRATYEYFGDVVTFDTTYLTNKFDMPFATFVGVNHHGQSTLLGCGLLSGEDTE 286
+ W+ A S +YE FGD V FDTT+ + +MP +VGVN++G GC LL E+
Sbjct: 260 IAWSYASSIQSYELFGDAVVFDTTHRLSAVEMPLGIWVGVNNYGVPCFFGCVLLRDENLR 319
Query: 287 SFVWLFKSWLRCMLGKAPVGIVTDQCKAVQNAIELVFPTTRHRWCLWHIMKKIPEKLN-- 344
S+ W +++ M GKAP I+TD ++ AI P T+H C+W ++ K P N
Sbjct: 320 SWSWALQAFTGFMNGKAPQTILTDHNMCLKEAIAGEMPATKHALCIWMVVGKFPSWFNAG 379
Query: 345 ---EYSEYKRIKSAMEGAVYDTHTTTGFEEKWCSFIDKFMLQQNDWLSGLYEERHRWAPT 401
Y+++K A +Y + FE W ++ F L N ++ LY R W+
Sbjct: 380 LGERYNDWK----AEFYRLYHLESVEEFELGWRDMVNSFGLHTNRHINNLYASRSLWSLP 435
Query: 402 FVRKYFWAGMSTTQRSESMHAFFDGYINSTTSLNQFVKQYDNALSSRAEKEFEADFNSLD 461
++R +F AGM+ T RS++++AF ++++ T L FV+Q + + + +
Sbjct: 436 YLRSHFLAGMTLTGRSKAINAFIQRFLSAQTRLAHFVEQVAVVVDFKDQATEQQTMQQNL 495
Query: 462 TTIPCVSNSSIEKQLQGEYTHAKFKEVQAEFISKMNCAPSLNVVEGCFATYHVLEEVVVG 521
I + + +E T F ++Q + + + A S + EG +H + G
Sbjct: 496 QNISLKTGAPMESHAASVLTPFAFSKLQEQLVLAAHYA-SFQMDEGYLVRHHTKLD---G 551
Query: 522 GRHKENVFKVVFNQENQDFSCECSLFEFRGILCCHVLSVCTRERVKNVPEKYVLTRWKK 580
GR KV + + SC C LFEF G LC H L V + VP++Y+ RW++
Sbjct: 552 GR------KVYWVPQEGIISCSCQLFEFSGFLCRHALRVLSTGNCFQVPDRYLPLRWRR 604
>At5g28530 far-red impaired response protein (FAR1) - like
Length = 700
Score = 195 bits (496), Expect = 6e-50
Identities = 153/592 (25%), Positives = 266/592 (44%), Gaps = 61/592 (10%)
Query: 27 EACWKPATGMRFSCIDDVKTLYREYALKKGFRWKTRTSKKRDDGETCYAILVCTREGSQG 86
+ + P G F+ D+ Y +A K GF + S + + VC R G
Sbjct: 65 DTVFTPYVGQIFTTDDEAFEYYSTFARKSGFSIRKARSTESQNLGVYRRDFVCYRSGFNQ 124
Query: 87 SEIPCTLKTPPSKTK---NCPAKICIKLEK-DGL--WYISKFESRHSHKT---------- 130
++ P + C K+ + E DG+ WY+S+F + H+H+
Sbjct: 125 PRKKANVEHPRERKSVRCGCDGKLYLTKEVVDGVSHWYVSQFSNVHNHELLEDDQVRLLP 184
Query: 131 -------SPTKRMDLHVKRTIEINEDAGVRTIKTFRSIVNNAEGHENIPFCEKDMINYVN 183
S +R+ L K +N V+ ++ + +V+ +PF EKD+ N+V
Sbjct: 185 AYRKIQQSDQERILLLSKAGFPVNRI--VKLLELEKGVVSG-----QLPFIEKDVRNFVR 237
Query: 184 --------NEQHLIGK-EGDGKALMSYFSKMREQNSNFFYDIDLDDDFHVRNVFWADARS 234
N+ + K E D L+ + E++ +F YD D++ V N+ WA S
Sbjct: 238 ACKKSVQENDAFMTEKRESDTLELLECCKGLAERDMDFVYDCTSDENQKVENIAWAYGDS 297
Query: 235 RATYEYFGDVVTFDTTYLTNKFDMPFATFVGVNHHGQSTLLGCGLLSGEDTESFVWLFKS 294
Y FGDVV FDT+Y + + + F G++++G++ LLGC LL E SF W ++
Sbjct: 298 VRGYSLFGDVVVFDTSYRSVPYGLLLGVFFGIDNNGKAMLLGCVLLQDESCRSFTWALQT 357
Query: 295 WLRCMLGKAPVGIVTDQCKAVQNAIELVFPTTRHRWCLWHIMKKIPEKLNEY--SEYKRI 352
++R M G+ P I+TD +++AI P T H + HI+ K+ ++ S Y+
Sbjct: 358 FVRFMRGRHPQTILTDIDTGLKDAIGREMPNTNHVVFMSHIVSKLASWFSQTLGSHYEEF 417
Query: 353 KSAMEGAVYDTHTTTGFEEKWCSFIDKFMLQQNDWLSGLYEERHRWAPTFVRKYFWAGMS 412
++ + + FE++W + +F L + + LY R W P +R++F A
Sbjct: 418 RAGFD-MLCRAGNVDEFEQQWDLLVTRFGLVPDRHAALLYSCRASWLPCCIREHFVAQTM 476
Query: 413 TTQRSESMHAFFDGYINSTTSLNQFVKQYDNALSSRAE--KEFEADFN--SLDTTIPCVS 468
T++ + S+ +F ++ T + +++ +S+ A K+ F SL T +P
Sbjct: 477 TSEFNLSIDSFLKRVVDGATCMQLLLEESALQVSAAASLAKQILPRFTYPSLKTCMP--- 533
Query: 469 NSSIEKQLQGEYTHAKFKEVQAEFISKMNCAPSLNVVEGCFATYHVLEEVVVGGRHKENV 528
+E +G T F +Q E + + A + + G F +H + E
Sbjct: 534 ---MEDHARGILTPYAFSVLQNEMVLSVQYAVA-EMANGPFIVHHY--------KKMEGE 581
Query: 529 FKVVFNQENQDFSCECSLFEFRGILCCHVLSVCTRERVKNVPEKYVLTRWKK 580
V++N EN++ C C FE GILC H L V T + ++PE+Y L RW++
Sbjct: 582 CCVIWNPENEEIQCSCKEFEHSGILCRHTLRVLTVKNCFHIPEQYFLLRWRQ 633
>At1g44840 hypothetical protein
Length = 926
Score = 59.3 bits (142), Expect = 7e-09
Identities = 75/355 (21%), Positives = 144/355 (40%), Gaps = 51/355 (14%)
Query: 19 EVDNSDDDEACWKPATGMRFSCIDDVKTLYREYALKKGFRWKTRTSKKR------DDGET 72
++D DD + +K G ++ +D + YA+K F +K +K D +
Sbjct: 364 DLDIEDDGKGIYK---GKVYASKEDCQIGLAIYAIKNMFHFKQTRTKWNYFVLSCSDEKC 420
Query: 73 CYAILVCTREGSQGSEIP-------CTLKTPPSKTKNCPAKICIKLEKDGLWYISKFESR 125
+ IL +G+ EI C+L T + +K+ S F+++
Sbjct: 421 DWRILATLMKGTGYYEIKKASLDHTCSLDTRGQFMQKATSKVIA----------SVFKAK 470
Query: 126 HSHKTSPTKRMDLHVKRTIEINEDAGVRTIKTFR---SIVNNAEGHENIPFCEKDMINYV 182
+S TS MDL ++ + + V K +R S + + G + +C Y+
Sbjct: 471 YSDPTSGPVPMDL--QQLVLEDLRVSVSYSKCWRARESALTSVAGSDEESYC------YL 522
Query: 183 NNEQHLIGKEGDGKALMSYFSKMREQNSNFFYDIDLDDDFHVR--NVFWADARSRATYEY 240
HL+ G ++ + D++D+ R +F A S A + +
Sbjct: 523 AEYLHLLKLTNPGTI------------THIETERDVEDESKERFLYMFLAFGASIAGFRH 570
Query: 241 FGDVVTFDTTYLTNKFDMPFATFVGVNHHGQSTLLGCGLLSGEDTESFVWLFKSWLRCML 300
++ D T+L K+ T G + + Q LG ++ E+ ES+ W F R +
Sbjct: 571 LRRILVVDGTHLKGKYKGVLLTSSGQDANFQVYPLGFAVVDSENDESWTWFFTKLERIIA 630
Query: 301 GKAPVGIVTDQCKAVQNAIELVFPTTRHRWCLWHIMKKIPEKLNEYSEYKRIKSA 355
+ I++D+ ++ A++ VFP H C+ H+ + I K + + +K+A
Sbjct: 631 DSKTLTILSDRHSSILVAVKRVFPQANHGACIIHLCRNIQTKYKNKALTQLVKNA 685
>At3g07500 hypothetical protein
Length = 214
Score = 58.9 bits (141), Expect = 9e-09
Identities = 44/141 (31%), Positives = 65/141 (45%), Gaps = 11/141 (7%)
Query: 8 VDCGSIHDEGMEV-DNSD-----DDEAC--WKPATGMRFSCIDDVKTLYREYALKKGFRW 59
V+ S+ +E +E+ +NSD DEA +P GM F + K+ Y YA GF
Sbjct: 9 VEGNSLPEEDVEMMENSDVMKTVTDEASPMVEPFIGMEFESEEAAKSFYDNYATCMGFVM 68
Query: 60 KTRTSKKR-DDGETCYAILVCTREGSQGSEIPCTLKTPPSKT--KNCPAKICIKLEKDGL 116
+ ++ DG + LVC +EG + S + P + C A I +K EK G
Sbjct: 69 RVDAFRRSMRDGTVVWRRLVCNKEGFRRSRPRRSESRKPRAITREGCKALIVVKREKSGT 128
Query: 117 WYISKFESRHSHKTSPTKRMD 137
W ++KFE H+H P D
Sbjct: 129 WLVTKFEKEHNHPLLPLSPND 149
>At3g45340 putative protein
Length = 693
Score = 57.0 bits (136), Expect = 3e-08
Identities = 55/235 (23%), Positives = 96/235 (40%), Gaps = 18/235 (7%)
Query: 196 KALMSYFSKMREQNSNFFYDIDLDDDFHVRNVFWADARSRATYEYFGDVVTFDTTYLTNK 255
K L +Y +R N +++LD D + +F A S ++ V+ D T+L
Sbjct: 338 KLLFAYLHILRSVNHGTITEVELDADDRFKFLFIALGASIEGFKAMRKVIVIDATFLKTI 397
Query: 256 FD--MPFATFVGVNHHGQSTLLGCGLLSGEDTESFVWLFKSWLRCMLGKAPVGIVTDQCK 313
+ + AT NHH + G++ E+ S+ W F+ + + + D
Sbjct: 398 YGGMLVIATAQDPNHHHYP--IAFGVIDSENHASWNWFFRKLNSIIPDDPELVFIRDIHG 455
Query: 314 AVQNAIELVFPTTRHRWCLWHIMKKIPEKLNEYSEYKRIKSAMEGAVYDTHTTTGFEEKW 373
++ + VFP H C+WH+ + I + L E K +Y T T F ++
Sbjct: 456 SIIKGVADVFPKASHGHCVWHLSQNIKKMLAGDKEAGMAKFMELAHIY---TATEFNIRY 512
Query: 374 CSFIDKF--MLQQNDWLSGLYEERHRWAPTFVR--KYFWAGMSTTQRSESMHAFF 424
F K + D G+ +WA + + KY + T+ +ESM+A F
Sbjct: 513 AEFRRKHPKVATYLDRAIGI----EKWARCYFQGDKY---NIDTSNSAESMNAVF 560
>At5g27190 putative protein
Length = 779
Score = 56.2 bits (134), Expect = 6e-08
Identities = 54/238 (22%), Positives = 100/238 (41%), Gaps = 26/238 (10%)
Query: 198 LMSYFSKMREQNSNFFYDIDLDDDFHVRNVFWADARSRATYEYFGDVVTFDTTYLTNKFD 257
L SY ++E N +++D + +F A ++ VV D T+L +
Sbjct: 347 LYSYLHVLKEVNPGTISYVEVDAQQKFKYLFVALGACIEGFKVMRKVVIVDATFLKTVYG 406
Query: 258 --MPFATFVGVNHHGQSTLLGCGLLSGEDTESFVWLFKSWLRCMLGKAP-VGIVTDQCKA 314
+ FAT NHH ++ ++ E+ S+ W F L+ ++ P + V+D+ ++
Sbjct: 407 GMLVFATAQDPNHHNY--IIASAVIDRENDASWSWFFNK-LKTVIPDVPGLVFVSDRHQS 463
Query: 315 VQNAIELVFPTTRHRWCLWHIMKKIPEKLNEYSEYKRIKSAMEGAVYDTHTTTGFE--EK 372
+ +I VFP RH C+WH+ + + ++ E A + H T FE +
Sbjct: 464 IIKSIMQVFPNARHGHCVWHLSQNVKVRVKTEKE-----EAAANFIACAHVYTQFEFTRE 518
Query: 373 WCSFIDKFMLQQNDWLSGLYEERHRWAP--TFVRKYFWA---GMSTTQRSESMHAFFD 425
+ F +F + RWAP + YF + T+ ES+++ F+
Sbjct: 519 YARFRRRF--------PNVGPYLDRWAPVENWAMCYFEGDRYNIDTSNACESLNSTFE 568
>At2g12720 putative protein
Length = 819
Score = 55.8 bits (133), Expect = 8e-08
Identities = 45/205 (21%), Positives = 83/205 (39%), Gaps = 9/205 (4%)
Query: 151 GVRTIKTFRSIVNNAEGHENIPFCEKDMINYVNNEQHLIGKEGDGKALMSYFSKMREQNS 210
G+ K FR ++ ++ ++ + F E L + + L SY +R N
Sbjct: 365 GIIITKHFRVKMSYSKSYKTLRFAR---------ELTLGTPDSSFEELPSYLYMIRRANP 415
Query: 211 NFFYDIDLDDDFHVRNVFWADARSRATYEYFGDVVTFDTTYLTNKFDMPFATFVGVNHHG 270
+ +D+ +F S A + Y VV D T+L + T + + +
Sbjct: 416 GTVARLQIDESGRFNYMFIVFGASIAGFHYMRRVVVVDGTFLHGSYKGTLLTALAQDGNF 475
Query: 271 QSTLLGCGLLSGEDTESFVWLFKSWLRCMLGKAPVGIVTDQCKAVQNAIELVFPTTRHRW 330
Q L G++ E+ +S+ WLF + + I++D+ K++ AI V+P
Sbjct: 476 QIFPLAFGVVDTENDDSWRWLFTQLKVVIPDATDLAIISDRHKSIGKAIGEVYPLAARGI 535
Query: 331 CLWHIMKKIPEKLNEYSEYKRIKSA 355
C +H+ K I K + +K A
Sbjct: 536 CTYHLYKNILVKFKRKDLFPLVKKA 560
>At2g43280 unknown protein
Length = 206
Score = 53.9 bits (128), Expect = 3e-07
Identities = 33/129 (25%), Positives = 58/129 (44%), Gaps = 4/129 (3%)
Query: 15 DEGMEVDNSDDDEACWKPATGMRFSCIDDVKTLYREYALKKGFRWKTRTSKKRD-DGETC 73
D+ ++++ D+ +P G+ F D K Y +Y+ + GF + + ++ + DG
Sbjct: 5 DDPTTKNSNNTDDFAIEPREGIIFESEDAAKMFYDDYSRRLGFVMRVMSCRRSEKDGRIL 64
Query: 74 YAILVCTREG---SQGSEIPCTLKTPPSKTKNCPAKICIKLEKDGLWYISKFESRHSHKT 130
C +EG S + K PS + C A I +K ++ G W I+KF H+H
Sbjct: 65 ARRFGCNKEGHCVSIRGKFGSVRKPRPSTREGCKAMIHVKYDRSGKWVITKFVKEHNHPL 124
Query: 131 SPTKRMDLH 139
+ R H
Sbjct: 125 VVSPREARH 133
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,561,397
Number of Sequences: 26719
Number of extensions: 755873
Number of successful extensions: 2012
Number of sequences better than 10.0: 85
Number of HSP's better than 10.0 without gapping: 57
Number of HSP's successfully gapped in prelim test: 28
Number of HSP's that attempted gapping in prelim test: 1842
Number of HSP's gapped (non-prelim): 125
length of query: 688
length of database: 11,318,596
effective HSP length: 106
effective length of query: 582
effective length of database: 8,486,382
effective search space: 4939074324
effective search space used: 4939074324
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146941.15


