

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146941.12 - phase: 0
(146 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
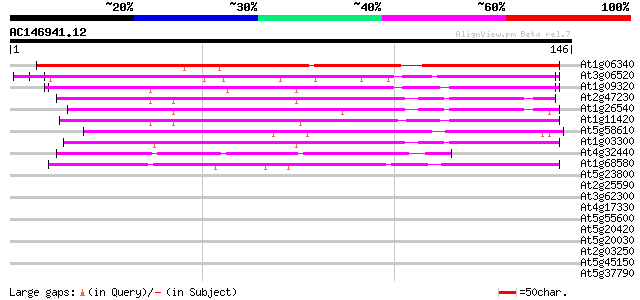
Score E
Sequences producing significant alignments: (bits) Value
At1g06340 unknown protein 116 5e-27
At3g06520 hypothetical protein 85 2e-17
At1g09320 unknown protein 83 5e-17
At2g47230 unknown protein 68 2e-12
At1g26540 unknown protein 66 7e-12
At1g11420 hypothetical protein 56 6e-09
At5g58610 putative protein 52 1e-07
At1g03300 unknown protein 41 3e-04
At4g32440 unknown protein 40 6e-04
At1g68580 unknown protein 39 7e-04
At5g23800 putative protein 37 0.004
At2g25590 hypothetical protein 36 0.006
At3g62300 putative protein 35 0.018
At4g17330 G2484-1 protein 33 0.041
At5g55600 unknown protein 32 0.12
At5g20420 putative protein 28 1.7
At5g20030 unknown protein 27 5.0
At2g03250 unknown protein 27 5.0
At5g45150 putative protein 26 6.6
At5g37790 protein kinase - like protein 26 6.6
>At1g06340 unknown protein
Length = 134
Score = 116 bits (290), Expect = 5e-27
Identities = 61/138 (44%), Positives = 85/138 (61%), Gaps = 8/138 (5%)
Query: 8 VDYKVGDKVEVCSKEEGFVGSYFEATIVSCLESGKYV-IRYKNLLKD-DESELLMETLFP 65
+++ GD+VEVCSKE+GF+GSYF AT+VS G Y I+YKNL+ D D+S+ L+E +
Sbjct: 1 MEFVKGDQVEVCSKEDGFLGSYFGATVVSKTPEGSYYKIKYKNLVSDTDQSKRLVEVISA 60
Query: 66 KDLRPLPPRVHNPWRFELNQKVDVFDNDGWWVGEIASEKILMEKSYYYSVYFDYCHQTIY 125
+LRP+PP+ + KVD FD DGWWVGE+ + + YSVYF + +
Sbjct: 61 DELRPMPPKSLHV-LIRCGDKVDAFDKDGWWVGEVTA-----VRRNIYSVYFSTTDEELE 114
Query: 126 YPCDQIRVHQELVWGDWI 143
YP +R H E V G W+
Sbjct: 115 YPLYSLRKHHEWVNGSWV 132
>At3g06520 hypothetical protein
Length = 466
Score = 84.7 bits (208), Expect = 2e-17
Identities = 53/150 (35%), Positives = 79/150 (52%), Gaps = 12/150 (8%)
Query: 2 RPPVKRVD--------YKVGDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLKD 53
+PP+K++ + G +VEV S E G+ S+F A IVS L +Y + Y+ L D
Sbjct: 317 KPPLKKLKSCERAEKVFNNGMEVEVRSDEPGYEASWFSAKIVSYLGENRYTVEYQTLKTD 376
Query: 54 DESELLMETLFPKDLR-PLPPRVHNPWRFELNQKVDVFDNDGWWVGEIASEKILMEKSYY 112
DE ELL E D+R P PP + +R+EL + VD + N+GWW G + KI K+
Sbjct: 377 DERELLKEEARGSDIRPPPPPLIPKGYRYELYELVDAWYNEGWWSGRV--YKINNNKT-R 433
Query: 113 YSVYFDYCHQTIYYPCDQIRVHQELVWGDW 142
Y VYF +++ + + +R Q G W
Sbjct: 434 YGVYFQTTDESLEFAYNDLRPCQVWRNGKW 463
Score = 65.9 bits (159), Expect = 7e-12
Identities = 43/140 (30%), Positives = 72/140 (50%), Gaps = 4/140 (2%)
Query: 6 KRVDYKVGDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLKDD-ESELLMETLF 64
+R Y+ G VEV S+E+ + GS++ A I+ L KY++ + +DD ES L + +
Sbjct: 165 RRDQYEKGALVEVRSEEKAYKGSWYCARILCLLGDDKYIVEHLKFSRDDGESIPLRDVVE 224
Query: 65 PKDLRPLPPRVHNP-WRFELNQKVDVFDNDGWWVGEIASEKILMEKSYYYSVYFDYCHQT 123
KD+RP+PP +P +E VD + N WW ++ K+L S YSV+ +
Sbjct: 225 AKDIRPVPPSELSPVVCYEPGVIVDAWFNKRWWTSRVS--KVLGGGSNKYSVFIISTGEE 282
Query: 124 IYYPCDQIRVHQELVWGDWI 143
+R H++ + G W+
Sbjct: 283 TTILNFNLRPHKDWINGQWV 302
Score = 47.8 bits (112), Expect = 2e-06
Identities = 42/139 (30%), Positives = 70/139 (50%), Gaps = 10/139 (7%)
Query: 10 YKVGDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKN---LLKDDESELLMETLFPK 66
++ GD+VEV G +YF A++VS K ++ ++ + S + E + P
Sbjct: 7 WRGGDRVEVERLVSGAT-AYFPASVVSAPSVRKKLVWVEHESLTVGGSVSVRMKEYVTPT 65
Query: 67 DLRPLPPRVHNPWRFELNQKVDVF-DNDGWWV-GEIASEKILMEKSYYYSVYFDYCHQTI 124
LRP PPR N RF+ + +VDVF D++G WV G + + ++E S Y + I
Sbjct: 66 RLRPSPPRELNR-RFKADDEVDVFRDSEGCWVRGNVTT---VLEDSRYIVEFKGENRPEI 121
Query: 125 YYPCDQIRVHQELVWGDWI 143
+R+H+E + G W+
Sbjct: 122 EVDQFNLRLHREWLDGGWV 140
>At1g09320 unknown protein
Length = 517
Score = 83.2 bits (204), Expect = 5e-17
Identities = 48/134 (35%), Positives = 70/134 (51%), Gaps = 4/134 (2%)
Query: 10 YKVGDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKDLR 69
+ +G +EV +EEGF S+F A ++ K ++ Y NL +D E L E + +R
Sbjct: 380 FSIGTPIEVSPEEEGFEDSWFLAKLIEYRGKDKCLVEYDNLKAEDGKEPLREEVNVSRIR 439
Query: 70 PLPPRVHNPWRFELNQKVDVFDNDGWWVGEIASEKILMEKSYYYSVYFDYCHQTIYYPCD 129
PLP FE + KV+ NDGWWVG I K+L + SY V F + + +
Sbjct: 440 PLPLESVMVSPFERHDKVNALYNDGWWVGVI--RKVLAKSSYL--VLFKNTQELLKFHHS 495
Query: 130 QIRVHQELVWGDWI 143
Q+R+HQE + G WI
Sbjct: 496 QLRLHQEWIDGKWI 509
Score = 73.9 bits (180), Expect = 3e-14
Identities = 43/133 (32%), Positives = 66/133 (49%), Gaps = 5/133 (3%)
Query: 10 YKVGDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKDLR 69
+ G VEV S EEGF G +F A +V + K+++ Y++L + D E L E +R
Sbjct: 221 FSSGTVVEVSSDEEGFQGCWFAAKVVEPVGEDKFLVEYRDLREKDGIEPLKEETDFLHIR 280
Query: 70 PLPPRVHNPWRFELNQKVDVFDNDGWWVGEIASEKILMEKSYYYSVYFDYCHQTIYYPCD 129
P PPR F + K++ F NDGWWVG + K +YF + + +
Sbjct: 281 PPPPR-DEDIDFAVGDKINAFYNDGWWVGVVIDGM----KHGTVGIYFRQSQEKMRFGRQ 335
Query: 130 QIRVHQELVWGDW 142
+R+H++ V G W
Sbjct: 336 GLRLHKDWVDGTW 348
Score = 63.9 bits (154), Expect = 3e-11
Identities = 44/141 (31%), Positives = 71/141 (50%), Gaps = 13/141 (9%)
Query: 11 KVGDKVEVCSKEEGFVGSYFEATIV-----SCLESGKYVIRYKNLLKDDE-SELLMETLF 64
K G VE+ S E GF GS++ ++ S +S K + Y L D E ++ L E +
Sbjct: 40 KPGSAVEISSDEIGFRGSWYMGKVITIPSSSDKDSVKCQVEYTTLFFDKEGTKPLKEVVD 99
Query: 65 PKDLRPLPP---RVHNPWRFELNQKVDVFDNDGWWVGEIASEKILMEKSYYYSVYFDYCH 121
LRP P + + + ++VD F NDGWW G++ ++L + +SV+F
Sbjct: 100 MSQLRPPAPPMSEIEKKKKIVVGEEVDAFYNDGWWEGDVT--EVLDDGK--FSVFFRSSK 155
Query: 122 QTIYYPCDQIRVHQELVWGDW 142
+ I + D++R H+E V G W
Sbjct: 156 EQIRFRKDELRFHREWVDGAW 176
>At2g47230 unknown protein
Length = 701
Score = 67.8 bits (164), Expect = 2e-12
Identities = 47/134 (35%), Positives = 68/134 (50%), Gaps = 10/134 (7%)
Query: 13 GDKVEVCSKEEGFVGSYFEATIV-SCLESG--KYVIRYKNLLKDDESELLMETLFPKDLR 69
G +VEV S EEGF ++F + + +SG K +RY LL DD L+E + P+ +R
Sbjct: 8 GSEVEVSSTEEGFADAWFRGILQENPTKSGRKKLRVRYLTLLNDDALSPLIENIEPRFIR 67
Query: 70 PLPP-RVHNPWRFELNQKVDVFDNDGWWVGEIASEKILMEKSYYYSVYFDYCHQTIYYPC 128
P+PP +N E VD DGWW G I + +E ++ VY+D I +
Sbjct: 68 PVPPENEYNGIVLEEGTVVDADHKDGWWTGVIIKK---LENGKFW-VYYDSPPDIIEFER 123
Query: 129 DQIRVHQELVWGDW 142
+Q+R H L W W
Sbjct: 124 NQLRPH--LRWSGW 135
>At1g26540 unknown protein
Length = 695
Score = 65.9 bits (159), Expect = 7e-12
Identities = 48/135 (35%), Positives = 66/135 (48%), Gaps = 13/135 (9%)
Query: 16 VEVCSKEEGFVGSYFEATIVSCLESG---KYVIRYKNLLKDDESELLMETLFPKDLRPLP 72
VEV S+EEGF G++F A + + K +RY LL D S L+E + + +RP+P
Sbjct: 14 VEVSSEEEGFEGAWFRAVLEENPGNSSRRKLRVRYSTLLDMDGSSPLIEHIEQRFIRPVP 73
Query: 73 PRVHNPWRFELNQ--KVDVFDNDGWWVGEIASEKILMEKSYYYSVYFDYCHQTIYYPCDQ 130
P + L + VD DGWW G + + ME Y VYFD I + Q
Sbjct: 74 PEENQQKDVVLEEGLLVDADHKDGWWTGVVVKK---MEDDNYL-VYFDLPPDIIQFERKQ 129
Query: 131 IRVHQELVW--GDWI 143
+R H L+W G WI
Sbjct: 130 LRTH--LIWTGGTWI 142
>At1g11420 hypothetical protein
Length = 604
Score = 56.2 bits (134), Expect = 6e-09
Identities = 43/134 (32%), Positives = 66/134 (49%), Gaps = 8/134 (5%)
Query: 14 DKVEVCSKEEGFVGSYFEATIV-SCLESG--KYVIRYKNLLKDDESELLMETLFPKDLRP 70
DKVEV S+EE GSY+ A + + +SG K +RY L + L E + + +RP
Sbjct: 11 DKVEVFSEEEELKGSYYRAILEDNPTKSGHNKLKVRYLTQLNEHRLAPLTEFVDQRFIRP 70
Query: 71 LPPR-VHNPWRFELNQKVDVFDNDGWWVGEIASEKILMEKSYYYSVYFDYCHQTIYYPCD 129
+P V++ F VD + DGWW G + K + ++ + VYFD I +
Sbjct: 71 VPSEDVNDGVVFVEGLMVDAYLKDGWWTGVVV--KTMEDEKFL--VYFDCPPDIIQFEKK 126
Query: 130 QIRVHQELVWGDWI 143
++RVH + WI
Sbjct: 127 KLRVHLDWTGFKWI 140
>At5g58610 putative protein
Length = 1065
Score = 52.0 bits (123), Expect = 1e-07
Identities = 43/152 (28%), Positives = 64/152 (41%), Gaps = 30/152 (19%)
Query: 20 SKEEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKD------------ 67
S EEG +GS++ T+ S + + IRY N+L DD S L+ET+ D
Sbjct: 27 SLEEGSLGSWYLGTVTSAKKRRRRCIRYDNILSDDGSGNLVETVDVSDIVEGLDDCTDVS 86
Query: 68 ------LRPLPPRVH-NPWRFELNQKVDVFDNDGWWVGEIASEKILMEKSYYYSVYFDYC 120
LRP+PP++ VDVF +D WW G + + EK V+F
Sbjct: 87 DTFRGRLRPVPPKLDVAKLNLAYGLCVDVFFSDAWWEGVLFDHENGSEKR---RVFFPDL 143
Query: 121 HQTIYYPCDQIRVHQEL-----VW---GDWIF 144
+ +R+ Q+ W G W+F
Sbjct: 144 GDELDADLQSLRITQDWNEATETWECRGSWLF 175
>At1g03300 unknown protein
Length = 670
Score = 40.8 bits (94), Expect = 3e-04
Identities = 34/135 (25%), Positives = 57/135 (42%), Gaps = 10/135 (7%)
Query: 15 KVEVCSKEEGFVGSYFEATIVS-----CLESGKYVIRYKNLLKDDESELLMETLFPKDLR 69
+VE+ S+E+GF +++ A + ES K Y + E T+ + +R
Sbjct: 8 EVEIFSEEDGFRNAWYRAILEETPTNPTSESKKLRFSYMTKSLNKEGSSSPPTVEQRFIR 67
Query: 70 PLPP-RVHNPWRFELNQKVDVFDNDGWWVGEIASEKILMEKSYYYSVYFDYCHQTIYYPC 128
P+PP ++N FE VD W G + ++ ME Y V FD I +
Sbjct: 68 PVPPENLYNGVVFEEGTMVDADYKHRWRTGVVINK---MENDSYL-VLFDCPPDIIQFET 123
Query: 129 DQIRVHQELVWGDWI 143
+R H + +W+
Sbjct: 124 KHLRAHLDWTGSEWV 138
>At4g32440 unknown protein
Length = 393
Score = 39.7 bits (91), Expect = 6e-04
Identities = 33/103 (32%), Positives = 51/103 (49%), Gaps = 7/103 (6%)
Query: 13 GDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKDLRPLP 72
G +VEV S +E G++ A IVS Y +R+ + + E E +ME + K +RP P
Sbjct: 6 GSRVEVFSNKEAPYGAWRCAEIVSG-NGHTYNVRFYSFQIEHE-EAVMEKVPRKIIRPCP 63
Query: 73 PRVHNPWRFELNQKVDVFDNDGWWVGEIASEKILMEKSYYYSV 115
P V + R++ + V+V DN W + E +YY V
Sbjct: 64 PLV-DVERWDTGELVEVLDNFSWKAATVREEL----SGHYYVV 101
>At1g68580 unknown protein
Length = 648
Score = 39.3 bits (90), Expect = 7e-04
Identities = 37/153 (24%), Positives = 70/153 (45%), Gaps = 25/153 (16%)
Query: 11 KVGDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLK-DDESELLMETLFP---- 65
K G +EV S++ G G +F+A ++ K ++Y+++ DDES+ L E +
Sbjct: 364 KKGSLIEVLSEDSGIRGCWFKALVLK-KHKDKVKVQYQDIQDADDESKKLEEWILTSRVA 422
Query: 66 -------------KDLRPL--PPRVHNPWRFELNQKVDVFDNDGWWVGEIASEKILMEKS 110
K +RP+ P + ++ + VDV+ DGWW G I +++ EK
Sbjct: 423 AGDHLGDLRIKGRKVVRPMLKPSKENDVCVIGVGMPVDVWWCDGWWEG-IVVQEVSEEK- 480
Query: 111 YYYSVYFDYCHQTIYYPCDQIRVHQELVWGDWI 143
+ VY + + + +R +E + +W+
Sbjct: 481 --FEVYLPGEKKMSAFHRNDLRQSREWLDDEWL 511
>At5g23800 putative protein
Length = 552
Score = 37.0 bits (84), Expect = 0.004
Identities = 37/139 (26%), Positives = 58/139 (41%), Gaps = 7/139 (5%)
Query: 4 PVKRVDYKVGDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETL 63
P + + G +VE+ K G +++A + + S LLKDD S L E
Sbjct: 6 PSESLSLSEGCEVEISYKNNGNESVWYKAILEAKPNSIFKEELSVRLLKDDFSTPLNELR 65
Query: 64 FPKDLRPLPP-RVHNPWRFELNQKVDVFDNDGWWVGEIASEKILMEKSYYYSVYFDYCHQ 122
+RP+PP V E+ VD D WW G + K++ + ++ V FD
Sbjct: 66 HKVLIRPIPPTNVQACIDIEIGTFVDADYKDAWWAGFVV--KVIDDDKFW--VCFDSPPD 121
Query: 123 TIYYPCDQIRVHQELVWGD 141
I + D+ + L W D
Sbjct: 122 IIQF--DRNHLRPTLEWVD 138
Score = 32.7 bits (73), Expect = 0.070
Identities = 22/99 (22%), Positives = 46/99 (46%), Gaps = 5/99 (5%)
Query: 10 YKVGDKVEVCSKEEGFVGSYFEATIVSCLESG---KYVIRYKNLLKDDESELLMETLFPK 66
+ G VE+CSK + + A + + +Y+++ K L+ +T+ +
Sbjct: 165 FSPGTIVELCSKRDEGEVVWVPALVYKEFKENDEYRYIVKDKPLIGRSYKSRPSKTVDLR 224
Query: 67 DLRPLPPRVHNPWRFELNQKVDVF-DNDGWWVGEIASEK 104
LRP+PP + + L++ ++V+ D GW G + +
Sbjct: 225 SLRPIPPPIRVK-EYRLDEYIEVYHDGIGWRQGRVVKSE 262
>At2g25590 hypothetical protein
Length = 381
Score = 36.2 bits (82), Expect = 0.006
Identities = 27/87 (31%), Positives = 48/87 (55%), Gaps = 3/87 (3%)
Query: 10 YKVGDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKDLR 69
++ G +VEV S +E G + A I+S Y +RY + + +E++ + + K +R
Sbjct: 3 FRRGSRVEVFSIKEASYGVWRSAEIISG-NGHTYNVRYYS-FEIANNEVVEDRVPRKIIR 60
Query: 70 PLPPRVHNPWRFELNQKVDVFDNDGWW 96
P PP+V + R+E + V+V DN+ W
Sbjct: 61 PCPPQV-DVDRWEAGELVEVLDNNISW 86
>At3g62300 putative protein
Length = 708
Score = 34.7 bits (78), Expect = 0.018
Identities = 26/103 (25%), Positives = 48/103 (46%), Gaps = 4/103 (3%)
Query: 9 DYKVGDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKDL 68
D+ G VEV +K + + A ++ E G +++ K LK++E ++ ++
Sbjct: 160 DFSAGKSVEVRTKVDKLGDVWAPAMVIKEDEDGTMLVKLKT-LKEEEVNCTKISVSYSEI 218
Query: 69 RPLPPRVHNPWRFELNQKVDVFDNDGWWVGEIASEKILMEKSY 111
RP P + ++L + VD GW G ++ K+L K Y
Sbjct: 219 RPSPLPI-GLRDYKLMENVDALVESGWCPGVVS--KVLAGKRY 258
Score = 30.8 bits (68), Expect = 0.27
Identities = 37/143 (25%), Positives = 64/143 (43%), Gaps = 13/143 (9%)
Query: 6 KRVDYKVGDKVEVCSKEEGF-VGSYFEATIV----SCLESGKYVIRYKN-LLKDDESELL 59
+++ G ++E+ S+E + G+ + I+ + + K +R+ + LLK D S L
Sbjct: 9 EKLSVSKGSEIEISSQEYEYGSGNVWYCVILEENLAKSKRKKLSVRHLDPLLKYDYSPPL 68
Query: 60 METLFPKDLRPLPPRVHNP-WRFELNQKVDVFDNDGWWVGEIASEKILMEKSYYYSVYFD 118
++T + +RP+PP P FE VD GW G + K+L + + VY
Sbjct: 69 IKTTVHRFMRPVPPPDPFPEVDFEEGDVVDAAYKGGWCSGSVV--KVLGNRRFL--VYLR 124
Query: 119 YCHQTIYYPCDQIRVHQELVWGD 141
+ I +R H VW D
Sbjct: 125 FQPDVIELLRKDLRPH--FVWKD 145
>At4g17330 G2484-1 protein
Length = 2037
Score = 33.5 bits (75), Expect = 0.041
Identities = 38/161 (23%), Positives = 66/161 (40%), Gaps = 28/161 (17%)
Query: 6 KRVDYKVGDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFP 65
K D K G VEV +E G +++ A ++S LE K + + +L + ++ L E +
Sbjct: 1569 KNEDIKEGSNVEVFKEEPGLRTAWYSANVLS-LEDDKAYVLFSDLSVEQGTDKLKEWVAL 1627
Query: 66 K-------DLRPLPPRVHNPWR--------------FELNQKVDVFDNDGWWVGEIASEK 104
K +RP P+ +++ +VD + +D W G I +
Sbjct: 1628 KGEGDQAPKIRPARSVTALPYEGTRKRRRAALGDHIWKIGDRVDSWVHDSWLEGVITEKN 1687
Query: 105 ILMEKSYYYSVYFDYCHQTIYYPCDQIRVHQELVW--GDWI 143
E + +V+F +T+ +R LVW G WI
Sbjct: 1688 KKDENT--VTVHFPAEEETLTIKAWNLR--PSLVWKDGKWI 1724
>At5g55600 unknown protein
Length = 663
Score = 32.0 bits (71), Expect = 0.12
Identities = 37/153 (24%), Positives = 61/153 (39%), Gaps = 26/153 (16%)
Query: 11 KVGDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPK---- 66
K K+E ++ G G +F T++ S K V + ++D++ +E P
Sbjct: 382 KTDAKIEFLCQDSGIRGCWFRCTVLDV--SRKQVKLQYDDIEDEDGYGNLEEWVPAFKSA 439
Query: 67 -------------DLRPLPPRVHNPWRFELN--QKVDVFDNDGWWVG-EIASEKILMEKS 110
+RP P V + F+L + VD + NDGWW G IA+ K E
Sbjct: 440 MPDKLGIRLSNRPTIRPAPRDVKTAY-FDLTIGEAVDAWWNDGWWEGVVIATGKPDTEDL 498
Query: 111 YYYSVYFDYCHQTIYYPCDQIRVHQELVWGDWI 143
Y + C + IR+ ++ V W+
Sbjct: 499 KIYIPGENLCLTVLR---KDIRISRDWVGDSWV 528
>At5g20420 putative protein
Length = 1261
Score = 28.1 bits (61), Expect = 1.7
Identities = 13/46 (28%), Positives = 25/46 (54%)
Query: 9 DYKVGDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLKDD 54
DY++ +++ +C + G VGS + E K+ I K++ +DD
Sbjct: 590 DYRLEEEIGMCCRLCGHVGSEIKDVSAPFAEHKKWTIETKHIEEDD 635
>At5g20030 unknown protein
Length = 326
Score = 26.6 bits (57), Expect = 5.0
Identities = 24/86 (27%), Positives = 39/86 (44%), Gaps = 8/86 (9%)
Query: 10 YKVGDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKDLR 69
+ G KVEV SK G++ A I+S Y + Y + +D +E + K +R
Sbjct: 3 FNKGTKVEVLSKSSVPSGAWRSAEIISG-NGHYYTVMYDH---NDGTERVPR----KSMR 54
Query: 70 PLPPRVHNPWRFELNQKVDVFDNDGW 95
P PPR+ + ++VF + W
Sbjct: 55 PEPPRLQVLDAWCPGDILEVFQSCSW 80
Score = 25.8 bits (55), Expect = 8.6
Identities = 15/53 (28%), Positives = 23/53 (43%)
Query: 80 RFELNQKVDVFDNDGWWVGEIASEKILMEKSYYYSVYFDYCHQTIYYPCDQIR 132
RF KV+V G S +I+ +YY+V +D+ T P +R
Sbjct: 2 RFNKGTKVEVLSKSSVPSGAWRSAEIISGNGHYYTVMYDHNDGTERVPRKSMR 54
>At2g03250 unknown protein
Length = 719
Score = 26.6 bits (57), Expect = 5.0
Identities = 9/22 (40%), Positives = 16/22 (71%)
Query: 44 VIRYKNLLKDDESELLMETLFP 65
++R +N+L+DD + M T+FP
Sbjct: 359 IVRTRNILQDDGQKQYMNTMFP 380
>At5g45150 putative protein
Length = 957
Score = 26.2 bits (56), Expect = 6.6
Identities = 23/98 (23%), Positives = 39/98 (39%), Gaps = 2/98 (2%)
Query: 13 GDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYK--NLLKDDESELLMETLFPKDLRP 70
G E+CS E + + E ++ + + VI N+ +D L E + + L+
Sbjct: 217 GKSFEICSTELFSLSTVSENSLTDEMSQEEVVIDEDSPNVEPEDVKGKLFEICYTRKLQI 276
Query: 71 LPPRVHNPWRFELNQKVDVFDNDGWWVGEIASEKILME 108
NP +E+ K V D D V + L+E
Sbjct: 277 QTGSSGNPLTYEMTTKQMVVDKDSLHVEPVNGRGELIE 314
>At5g37790 protein kinase - like protein
Length = 552
Score = 26.2 bits (56), Expect = 6.6
Identities = 12/38 (31%), Positives = 26/38 (67%), Gaps = 1/38 (2%)
Query: 52 KDDESELLMETLFPKDLRPLPPRVHNPWRFELNQKVDV 89
+++++ LL ET P+++ P P VH+ +RF L ++++
Sbjct: 93 ENNKAWLLAETA-PENMNPDPHSVHSSFRFSLCSQIEL 129
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.141 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,703,109
Number of Sequences: 26719
Number of extensions: 150569
Number of successful extensions: 399
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 355
Number of HSP's gapped (non-prelim): 32
length of query: 146
length of database: 11,318,596
effective HSP length: 90
effective length of query: 56
effective length of database: 8,913,886
effective search space: 499177616
effective search space used: 499177616
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146941.12


