

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146941.10 - phase: 0
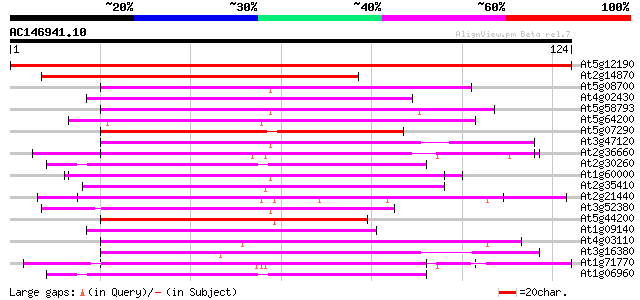
(124 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g12190 unknown protein 229 3e-61
At2g14870 hypothetical protein 110 1e-25
At5g08700 putative protein 52 6e-08
At4g02430 unknown protein 50 4e-07
At5g58793 putative protein (possibly pseudogene) 49 6e-07
At5g64200 unknown protein 48 1e-06
At5g07290 Mei2-like protein 48 1e-06
At3g47120 putative RNA-binding protein 48 1e-06
At2g36660 putative poly(A) binding protein 48 1e-06
At2g30260 putative small nuclear ribonucleoprotein U2B 48 1e-06
At1g60000 unknown protein 48 1e-06
At2g35410 putative chloroplast RNA binding protein precursor 47 2e-06
At2g21440 unknown protein (At2g21440) 47 2e-06
At3g52380 RNA-binding protein cp33 precursor 46 4e-06
At5g44200 nuclear cap-binding protein; CBP20 (gb|AAD29697.1) 45 7e-06
At1g09140 SF2/ASF splicing modulator, Srp30 like protein 45 9e-06
At4g03110 putative ribonucleoprotein 44 2e-05
At3g16380 putative poly(A) binding protein 44 2e-05
At1g71770 polyadenylate-binding protein 5 44 2e-05
At1g06960 putative spliceosomal protein U2B 44 2e-05
>At5g12190 unknown protein
Length = 124
Score = 229 bits (584), Expect = 3e-61
Identities = 113/124 (91%), Positives = 117/124 (94%)
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
MTTISLRK NTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIG +K T+GTA
Sbjct: 1 MTTISLRKSNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGCDKATKGTA 60
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS 120
FVVYEDIYDAK AVDHLSGFNVANRYLIVLYYQ AKMSKKFDQKK EDEI ++QEKYGVS
Sbjct: 61 FVVYEDIYDAKNAVDHLSGFNVANRYLIVLYYQHAKMSKKFDQKKSEDEITKLQEKYGVS 120
Query: 121 TKDK 124
TKDK
Sbjct: 121 TKDK 124
>At2g14870 hypothetical protein
Length = 101
Score = 110 bits (276), Expect = 1e-25
Identities = 51/70 (72%), Positives = 59/70 (83%)
Query: 8 KGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDI 67
K N RLPPEV R+LY+ NLPF+ITSE+ YD+FG+Y IRQ+RIG K T+GTAFVVYEDI
Sbjct: 9 KQNPRLPPEVTRLLYICNLPFSITSEDTYDLFGRYSTIRQVRIGCEKGTKGTAFVVYEDI 68
Query: 68 YDAKTAVDHL 77
YDAK AVDHL
Sbjct: 69 YDAKKAVDHL 78
>At5g08700 putative protein
Length = 768
Score = 52.4 bits (124), Expect = 6e-08
Identities = 32/85 (37%), Positives = 46/85 (53%), Gaps = 3/85 (3%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIYDAKTAVDHL 77
L+V LP++ T EE+ + F K+G I ++ + +KDT RG AFV+Y AK A+D L
Sbjct: 257 LFVHGLPYSTTEEELMEHFSKFGDISEVHLVLDKDTRSCRGMAFVLYLIPESAKMAMDKL 316
Query: 78 SGFNVANRYLIVLYYQQAKMSKKFD 102
R L +L + MS K D
Sbjct: 317 DKLPFQGRTLHILPAKPRAMSAKQD 341
Score = 35.0 bits (79), Expect = 0.009
Identities = 21/72 (29%), Positives = 42/72 (58%), Gaps = 5/72 (6%)
Query: 7 RKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYED 66
RKG+ + +L V++LPF T +E+ +F K+G++ +I + T+ A VV+ +
Sbjct: 451 RKGDVK--NRSKHILLVKHLPFASTEKELAQMFRKFGSLDKIVL---PPTKTMALVVFLE 505
Query: 67 IYDAKTAVDHLS 78
+A+ A++ L+
Sbjct: 506 AAEARAAMNGLA 517
>At4g02430 unknown protein
Length = 294
Score = 49.7 bits (117), Expect = 4e-07
Identities = 27/72 (37%), Positives = 39/72 (53%)
Query: 18 NRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAKTAVDHL 77
+R +YV NLP +I E+ D+F KYG + QI + G AFV +ED DA A+
Sbjct: 6 SRTIYVGNLPGDIREREVEDLFSKYGPVVQIDLKIPPRPPGYAFVEFEDARDADDAIYGR 65
Query: 78 SGFNVANRYLIV 89
G++ +L V
Sbjct: 66 DGYDFDGHHLRV 77
>At5g58793 putative protein (possibly pseudogene)
Length = 172
Score = 48.9 bits (115), Expect = 6e-07
Identities = 28/94 (29%), Positives = 51/94 (53%), Gaps = 7/94 (7%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIYDAKTAVDHL 77
+YV LP+++T ++ +F +YG + + + +K T + AFV YED AVD++
Sbjct: 37 VYVGELPYDLTEGDLLAVFSQYGEVVDLNLVRDKGTGRSKRFAFVAYEDQRSTNLAVDNI 96
Query: 78 SGFNVANRYLIV----LYYQQAKMSKKFDQKKKE 107
+G V R + V Y ++ + ++ QKK+E
Sbjct: 97 NGAKVLGRIIKVEHCGKYLKREEEDEETKQKKRE 130
>At5g64200 unknown protein
Length = 303
Score = 48.1 bits (113), Expect = 1e-06
Identities = 29/95 (30%), Positives = 52/95 (54%), Gaps = 5/95 (5%)
Query: 14 PPEVNRV--LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNK---DTRGTAFVVYEDIY 68
PP+++ L V N+ F T++++Y +F KYG + + I ++ D+RG AFV Y+
Sbjct: 9 PPDISDTYSLLVLNITFRTTADDLYPLFAKYGKVVDVFIPRDRRTGDSRGFAFVRYKYKD 68
Query: 69 DAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQ 103
+A AV+ L G V R + V + + ++K +
Sbjct: 69 EAHKAVERLDGRVVDGREITVQFAKYGPNAEKISK 103
>At5g07290 Mei2-like protein
Length = 907
Score = 48.1 bits (113), Expect = 1e-06
Identities = 24/67 (35%), Positives = 43/67 (63%), Gaps = 2/67 (2%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAKTAVDHLSGF 80
L+V NL +I++EE++ IF YG IR++R +++++ ++ + D+ AK A+ L+G
Sbjct: 297 LWVNNLDSSISNEELHGIFSSYGEIREVRRTMHENSQ--VYIEFFDVRKAKVALQGLNGL 354
Query: 81 NVANRYL 87
VA R L
Sbjct: 355 EVAGRQL 361
Score = 35.8 bits (81), Expect = 0.006
Identities = 24/75 (32%), Positives = 39/75 (52%), Gaps = 2/75 (2%)
Query: 17 VNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAKTAVDH 76
++R+L+VRN+ +I E+ +F ++G +R + T RG V Y DI A+ A
Sbjct: 209 LSRILFVRNVDSSIEDCELGVLFKQFGDVRALH--TAGKNRGFIMVSYYDIRAAQKAARA 266
Query: 77 LSGFNVANRYLIVLY 91
L G + R L + Y
Sbjct: 267 LHGRLLRGRKLDIRY 281
>At3g47120 putative RNA-binding protein
Length = 352
Score = 47.8 bits (112), Expect = 1e-06
Identities = 30/99 (30%), Positives = 52/99 (52%), Gaps = 9/99 (9%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIYDAKTAVDHL 77
+YV +PF++T ++ +F +YG I + + +K T +G AF+ YED AVD+L
Sbjct: 38 VYVGGIPFDLTEGDLLAVFSQYGEIVDVNLIRDKGTGKSKGFAFLAYEDQRSTILAVDNL 97
Query: 78 SGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEK 116
+G V R + V + + + ++EDE R Q +
Sbjct: 98 NGALVLGRTIKVDH------CGAYKKHEEEDEETRRQNR 130
>At2g36660 putative poly(A) binding protein
Length = 609
Score = 47.8 bits (112), Expect = 1e-06
Identities = 30/98 (30%), Positives = 53/98 (53%), Gaps = 7/98 (7%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD--TRGTAFVVYEDIYDAKTAVDHLS 78
++V+NLP ++T+ + D+F K+G I ++ T +D +RG FV +E A A+ L+
Sbjct: 114 VFVKNLPESVTNAVLQDMFKKFGNIVSCKVATLEDGKSRGYGFVQFEQEDAAHAAIQTLN 173
Query: 79 GFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEK 116
VA++ + V K KK D+ K E++ + K
Sbjct: 174 STIVADKEIYV-----GKFMKKTDRVKPEEKYTNLYMK 206
Score = 40.0 bits (92), Expect = 3e-04
Identities = 30/120 (25%), Positives = 63/120 (52%), Gaps = 8/120 (6%)
Query: 6 LRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGT--NKDTRGTAFVV 63
++K + P E LY++NL +++ + + + F ++G I + I N+ RG AFV
Sbjct: 188 MKKTDRVKPEEKYTNLYMKNLDADVSEDLLREKFAEFGKIVSLAIAKDENRLCRGYAFVN 247
Query: 64 YEDIYDAKTAVDHLSGFNVANRYLIVLYYQ-----QAKMSKKFDQKKKEDE-IARMQEKY 117
+++ DA+ A + ++G ++ L V Q + + ++F +K +E + IA++ Y
Sbjct: 248 FDNPEDARRAAETVNGTKFGSKCLYVGRAQKKAEREQLLREQFKEKHEEQKMIAKVSNIY 307
Score = 30.4 bits (67), Expect = 0.23
Identities = 25/100 (25%), Positives = 44/100 (44%), Gaps = 13/100 (13%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNK--DTRGTAFVVYEDIYDAKTAVDHLS 78
+YV+N+ +T EE+ F + G I ++ ++ ++G FV + +A AV
Sbjct: 306 IYVKNVNVAVTEEELRKHFSQCGTITSTKLMCDEKGKSKGFGFVCFSTPEEAIDAVKTFH 365
Query: 79 GFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYG 118
G + L V Q KKED ++Q ++G
Sbjct: 366 GQMFHGKPLYVAIAQ-----------KKEDRKMQLQVQFG 394
>At2g30260 putative small nuclear ribonucleoprotein U2B
Length = 232
Score = 47.8 bits (112), Expect = 1e-06
Identities = 28/84 (33%), Positives = 45/84 (53%), Gaps = 4/84 (4%)
Query: 9 GNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIY 68
G +PP N +L+++NLP TS + +F +Y ++IR+ K G AFV YED
Sbjct: 150 GQETMPP--NNILFIQNLPHETTSMMLQLLFEQYPGFKEIRMIDAKP--GIAFVEYEDDV 205
Query: 69 DAKTAVDHLSGFNVANRYLIVLYY 92
A A+ L GF + + +V+ +
Sbjct: 206 QASIAMQPLQGFKITPQNPMVISF 229
>At1g60000 unknown protein
Length = 258
Score = 47.8 bits (112), Expect = 1e-06
Identities = 28/90 (31%), Positives = 43/90 (47%), Gaps = 3/90 (3%)
Query: 14 PPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIYDA 70
P VN LY NLP+N+ S + I + + + N+DT RG AFV ++ D
Sbjct: 80 PAAVNTKLYFGNLPYNVDSATLAQIIQDFANPELVEVLYNRDTGQSRGFAFVTMSNVEDC 139
Query: 71 KTAVDHLSGFNVANRYLIVLYYQQAKMSKK 100
+D+L G R L V + + K +K+
Sbjct: 140 NIIIDNLDGTEYLGRALKVNFADKPKPNKE 169
Score = 44.7 bits (104), Expect = 1e-05
Identities = 29/87 (33%), Positives = 43/87 (49%), Gaps = 3/87 (3%)
Query: 13 LPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIYD 69
L PE L+V NL + +TSE + F + G + R+ + DT RG FV Y +
Sbjct: 171 LYPETEHKLFVGNLSWTVTSESLAGAFRECGDVVGARVVFDGDTGRSRGYGFVCYSSKAE 230
Query: 70 AKTAVDHLSGFNVANRYLIVLYYQQAK 96
+TA++ L GF + R + V Q K
Sbjct: 231 METALESLDGFELEGRAIRVNLAQGKK 257
>At2g35410 putative chloroplast RNA binding protein precursor
Length = 308
Score = 47.4 bits (111), Expect = 2e-06
Identities = 25/82 (30%), Positives = 46/82 (55%), Gaps = 2/82 (2%)
Query: 17 VNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD--TRGTAFVVYEDIYDAKTAV 74
+ R L+V NLP++++ ++ ++FG+ G + + I KD RG AFV +A+ A+
Sbjct: 93 LKRKLFVFNLPWSMSVNDISELFGQCGTVNNVEIIRQKDGKNRGFAFVTMASGEEAQAAI 152
Query: 75 DHLSGFNVANRYLIVLYYQQAK 96
D F V+ R + V + ++ K
Sbjct: 153 DKFDTFQVSGRIISVSFARRFK 174
>At2g21440 unknown protein (At2g21440)
Length = 1003
Score = 47.4 bits (111), Expect = 2e-06
Identities = 33/102 (32%), Positives = 53/102 (51%), Gaps = 8/102 (7%)
Query: 16 EVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTR---GTAFVVYEDI---YD 69
+ R L++RNLPF++T EE+ F +G + + + +K T+ GTAFV ++
Sbjct: 558 DFERTLFIRNLPFDVTKEEVKQRFTVFGEVESLSLVLHKVTKRPEGTAFVKFKTADASVA 617
Query: 70 AKTAVDHLSGFNV--ANRYLIVLYYQQAKMSKKFDQKKKEDE 109
A +A D SG V R L V+ K +K + KK E++
Sbjct: 618 AISAADTASGVGVLLKGRQLNVMRAVGKKAAKDIELKKTEEK 659
Score = 42.0 bits (97), Expect = 8e-05
Identities = 33/122 (27%), Positives = 55/122 (45%), Gaps = 8/122 (6%)
Query: 7 RKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNK---DTRGTAFVV 63
RK P + V LP++IT+ ++ + F + G +R+ + TNK + RG AFV
Sbjct: 8 RKDGEEKSPHAAATVCVSGLPYSITNAQLEEAFSEVGPVRRCFLVTNKGSDEHRGFAFVK 67
Query: 64 YEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQK--KKEDEIARMQEKYGVST 121
+ D A++ +G V R + V +QA ++ K + I+ G S
Sbjct: 68 FALQEDVNRAIELKNGSTVGGRRITV---KQAAHRPSLQERRTKAAEGISVPDNSQGQSD 124
Query: 122 KD 123
KD
Sbjct: 125 KD 126
Score = 32.7 bits (73), Expect = 0.047
Identities = 21/72 (29%), Positives = 33/72 (45%), Gaps = 3/72 (4%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIYDAKTAVDHL 77
L +RNLPF ++ +F G + + I N +T +G AFV + DA A+
Sbjct: 333 LIIRNLPFQAKPSDIKVVFSAVGFVWDVFIPKNFETGLPKGFAFVKFTCKKDAANAIKKF 392
Query: 78 SGFNVANRYLIV 89
+G R + V
Sbjct: 393 NGHMFGKRPIAV 404
>At3g52380 RNA-binding protein cp33 precursor
Length = 329
Score = 46.2 bits (108), Expect = 4e-06
Identities = 30/81 (37%), Positives = 42/81 (51%), Gaps = 4/81 (4%)
Query: 8 KGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVY 64
K T+ E R LYV NLP+ ITS E+ IFG+ G + ++I +K T RG FV
Sbjct: 106 KQTTQASGEEGR-LYVGNLPYTITSSELSQIFGEAGTVVDVQIVYDKVTDRSRGFGFVTM 164
Query: 65 EDIYDAKTAVDHLSGFNVANR 85
I +AK A+ + + R
Sbjct: 165 GSIEEAKEAMQMFNSSQIGGR 185
Score = 35.4 bits (80), Expect = 0.007
Identities = 21/83 (25%), Positives = 41/83 (49%), Gaps = 3/83 (3%)
Query: 8 KGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVY 64
+ N R + +Y NL +N+TS+ + D FG + ++ ++T RG F+ +
Sbjct: 208 RDNNRSYVDSPHKVYAGNLGWNLTSQGLKDAFGDQPGVLGAKVIYERNTGRSRGFGFISF 267
Query: 65 EDIYDAKTAVDHLSGFNVANRYL 87
E + ++A+ ++G V R L
Sbjct: 268 ESAENVQSALATMNGVEVEGRAL 290
>At5g44200 nuclear cap-binding protein; CBP20 (gb|AAD29697.1)
Length = 257
Score = 45.4 bits (106), Expect = 7e-06
Identities = 21/62 (33%), Positives = 40/62 (63%), Gaps = 3/62 (4%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTR---GTAFVVYEDIYDAKTAVDHL 77
+Y+ N+ F T E++Y++F + G I++I +G +K+T+ G FV++ D + AV ++
Sbjct: 36 VYIGNVSFYTTEEQLYELFSRAGEIKKIIMGLDKNTKTPCGFCFVLFYSREDTEDAVKYI 95
Query: 78 SG 79
SG
Sbjct: 96 SG 97
>At1g09140 SF2/ASF splicing modulator, Srp30 like protein
Length = 253
Score = 45.1 bits (105), Expect = 9e-06
Identities = 26/64 (40%), Positives = 35/64 (54%)
Query: 18 NRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAKTAVDHL 77
NR +YV NLP +I E+ D+F KYG I I + G AFV +ED DA A+
Sbjct: 6 NRTIYVGNLPGDIRKCEVEDLFYKYGPIVDIDLKIPPRPPGYAFVEFEDPRDADDAIYGR 65
Query: 78 SGFN 81
G++
Sbjct: 66 DGYD 69
>At4g03110 putative ribonucleoprotein
Length = 441
Score = 44.3 bits (103), Expect = 2e-05
Identities = 27/98 (27%), Positives = 52/98 (52%), Gaps = 5/98 (5%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRI--GTNKDTRGTAFVVYEDIYDAKTAVDHLS 78
L+V LP N++ E+ +F KYG I+ ++I G + ++G AF+ YE A +A++ ++
Sbjct: 108 LFVGMLPKNVSEAEVQSLFSKYGTIKDLQILRGAQQTSKGCAFLKYETKEQAVSAMESIN 167
Query: 79 GFNVANRYLIVLYYQQAKMSKKFDQK---KKEDEIARM 113
G + + L + A ++ + K + IAR+
Sbjct: 168 GKHKMEGSTVPLVVKWADTERERHTRRLQKAQSHIARL 205
>At3g16380 putative poly(A) binding protein
Length = 537
Score = 44.3 bits (103), Expect = 2e-05
Identities = 28/99 (28%), Positives = 51/99 (51%), Gaps = 13/99 (13%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAI--RQIRIGTNKDTRGTAFVVYEDIYDAKTAVDHLS 78
LYV+NL ++ + +IFG YG I ++ N ++G FV + + ++K A +L+
Sbjct: 306 LYVKNLSESMNETRLREIFGCYGQIVSAKVMCHENGRSKGFGFVCFSNCEESKQAKRYLN 365
Query: 79 GFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKY 117
GF V + ++V ++KED I R+Q+ +
Sbjct: 366 GFLVDGKPIVVRV-----------AERKEDRIKRLQQYF 393
Score = 36.6 bits (83), Expect = 0.003
Identities = 21/72 (29%), Positives = 41/72 (56%), Gaps = 4/72 (5%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQI---RIGTNKDTRGTAFVVYEDIYDAKTAVDHL 77
+YV+NL +T + ++ +F +YG + + R G + +RG FV + + +AK A++ L
Sbjct: 204 VYVKNLIETVTDDCLHTLFSQYGTVSSVVVMRDGMGR-SRGFGFVNFCNPENAKKAMESL 262
Query: 78 SGFNVANRYLIV 89
G + ++ L V
Sbjct: 263 CGLQLGSKKLFV 274
Score = 33.5 bits (75), Expect = 0.027
Identities = 29/104 (27%), Positives = 47/104 (44%), Gaps = 10/104 (9%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIR-IGTNKDTRGTAFVVYEDIYDAKTAVDHLSG 79
LYV+NL +ITS + +F +G+I + + N ++G FV ++ A +A L G
Sbjct: 114 LYVKNLDSSITSSCLERMFCPFGSILSCKVVEENGQSKGFGFVQFDTEQSAVSARSALHG 173
Query: 80 FNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVSTKD 123
V + L V KF K + +A Q+ V K+
Sbjct: 174 SMVYGKKLFV---------AKFINKDERAAMAGNQDSTNVYVKN 208
>At1g71770 polyadenylate-binding protein 5
Length = 668
Score = 44.3 bits (103), Expect = 2e-05
Identities = 32/106 (30%), Positives = 54/106 (50%), Gaps = 10/106 (9%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNK--DTRGTAFVVYEDIYDAKTAVDHLS 78
+YV+NLP IT +E+ FGKYG I + ++ ++R FV + A AV+ ++
Sbjct: 227 VYVKNLPKEITDDELKKTFGKYGDISSAVVMKDQSGNSRSFGFVNFVSPEAAAVAVEKMN 286
Query: 79 GFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVSTKDK 124
G ++ L V + KK D +E+E+ R E+ +S +K
Sbjct: 287 GISLGEDVLYV-----GRAQKKSD---REEELRRKFEQERISRFEK 324
Score = 40.0 bits (92), Expect = 3e-04
Identities = 24/90 (26%), Positives = 45/90 (49%), Gaps = 7/90 (7%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD--TRGTAFVVYEDIYDAKTAVDHLS 78
LY++NL ++ E++ ++F +YG + ++ N +RG FV Y + +A A+ ++
Sbjct: 330 LYLKNLDDSVNDEKLKEMFSEYGNVTSCKVMMNSQGLSRGFGFVAYSNPEEALLAMKEMN 389
Query: 79 GFNVANRYLIVLYYQ-----QAKMSKKFDQ 103
G + + L V Q QA + F Q
Sbjct: 390 GKMIGRKPLYVALAQRKEERQAHLQSLFTQ 419
Score = 39.7 bits (91), Expect = 4e-04
Identities = 23/91 (25%), Positives = 51/91 (55%), Gaps = 4/91 (4%)
Query: 4 ISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTN--KDTRGTAF 61
+S R +TRL + N ++++NL +I ++ +Y+ F +G I ++ + ++G F
Sbjct: 119 LSNRDPSTRLSGKGN--VFIKNLDASIDNKALYETFSSFGTILSCKVAMDVVGRSKGYGF 176
Query: 62 VVYEDIYDAKTAVDHLSGFNVANRYLIVLYY 92
V +E A+ A+D L+G + ++ + V ++
Sbjct: 177 VQFEKEETAQAAIDKLNGMLLNDKQVFVGHF 207
Score = 28.9 bits (63), Expect = 0.68
Identities = 17/75 (22%), Positives = 38/75 (50%), Gaps = 2/75 (2%)
Query: 18 NRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTR--GTAFVVYEDIYDAKTAVD 75
N LYV +L ++ + D+F + + +R+ + R G A+V + + DA A++
Sbjct: 44 NSSLYVGDLDPSVNESHLLDLFNQVAPVHNLRVCRDLTHRSLGYAYVNFANPEDASRAME 103
Query: 76 HLSGFNVANRYLIVL 90
L+ + +R + ++
Sbjct: 104 SLNYAPIRDRPIRIM 118
>At1g06960 putative spliceosomal protein U2B
Length = 228
Score = 44.3 bits (103), Expect = 2e-05
Identities = 26/84 (30%), Positives = 43/84 (50%), Gaps = 4/84 (4%)
Query: 9 GNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIY 68
G +PP N +L++ NLP S + +F +Y ++IR+ K G AFV YED
Sbjct: 146 GQDTMPP--NNILFIHNLPIETNSMMLQLLFEQYPGFKEIRMIEAKP--GIAFVEYEDDV 201
Query: 69 DAKTAVDHLSGFNVANRYLIVLYY 92
+ A+ L GF + + +V+ +
Sbjct: 202 QSSMAMQALQGFKITPQNPMVVSF 225
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,613,815
Number of Sequences: 26719
Number of extensions: 94471
Number of successful extensions: 535
Number of sequences better than 10.0: 181
Number of HSP's better than 10.0 without gapping: 116
Number of HSP's successfully gapped in prelim test: 65
Number of HSP's that attempted gapping in prelim test: 326
Number of HSP's gapped (non-prelim): 247
length of query: 124
length of database: 11,318,596
effective HSP length: 87
effective length of query: 37
effective length of database: 8,994,043
effective search space: 332779591
effective search space used: 332779591
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 53 (25.0 bits)
Medicago: description of AC146941.10


