

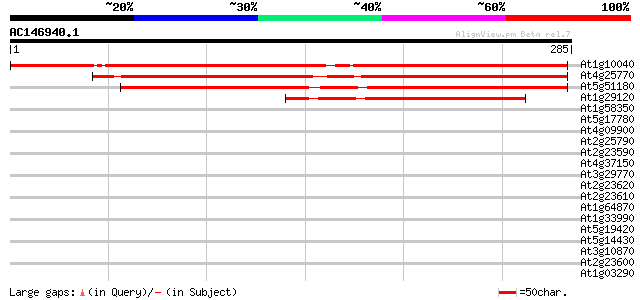
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146940.1 - phase: 0
(285 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g10040 unknown protein 307 4e-84
At4g25770 unknown protein 254 3e-68
At5g51180 unknown protein 232 2e-61
At1g29120 hypothetical protein 112 2e-25
At1g58350 putative protein 40 0.001
At5g17780 putative protein 30 1.1
At4g09900 putative host response protein 30 1.9
At2g25790 putative receptor-like protein kinase 29 3.3
At2g23590 putative acetone-cyanohydrin lyase 28 4.3
At4g37150 hydroxynitrile lyase like protein 28 7.4
At3g29770 alpha/beta hydrolase, putative 28 7.4
At2g23620 putative acetone-cyanohydrin lyase 28 7.4
At2g23610 putative acetone-cyanohydrin lyase 28 7.4
At1g64870 hypothetical protein 28 7.4
At1g33990 unknown protein 28 7.4
At5g19420 putative protein 27 9.6
At5g14430 unknown protein 27 9.6
At3g10870 putative alpha-hydroxynitrile lyase 27 9.6
At2g23600 putative acetone-cyanohydrin lyase 27 9.6
At1g03290 unknown protein 27 9.6
>At1g10040 unknown protein
Length = 412
Score = 307 bits (787), Expect = 4e-84
Identities = 160/284 (56%), Positives = 208/284 (72%), Gaps = 8/284 (2%)
Query: 1 MESQQIPKVQHSVVDVQINNKNIKKLKFPKF-GCFRIQHDATGDGFDIEVVDASGHRSNP 59
+E + PK + + ++K KK K + GC R + D +G+ D+ VD G R+ P
Sbjct: 22 LEGIRKPKKMKKMRSRKSDDKETKKKKKKYWMGCLRAESDESGN-VDL-TVDFPGERTEP 79
Query: 60 THLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRLAEEV 119
THL++MVNGLIGSA NW++AAKQ LK+YP D++VHCS+ N ST TFDGVDV G RLAEEV
Sbjct: 80 THLVVMVNGLIGSAQNWRFAAKQMLKKYPQDLLVHCSKRNHSTQTFDGVDVMGERLAEEV 139
Query: 120 ISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQISNQECHVR 179
SVIKRHPS++KISF+ HSLGGLIARYAI +LYE++ +EL H I + +C +
Sbjct: 140 RSVIKRHPSLQKISFVGHSLGGLIARYAIGRLYEQESREELP----HNSDDIGD-KCSIE 194
Query: 180 KYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTASRLSRFLGKTGKHLFLT 239
+ + +IAGLEP+ FITSATPHLG RGHKQVPL G ++LE+ A+R+S LGKTGKHLFL
Sbjct: 195 EPKARIAGLEPVYFITSATPHLGSRGHKQVPLFSGSYTLERLATRMSGCLGKTGKHLFLA 254
Query: 240 DGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDRIL 283
D KPPLLL+MV+DS D+KF+SAL+ FKRR+AYAN +D ++
Sbjct: 255 DSDGGKPPLLLRMVKDSRDLKFISALQCFKRRIAYANTSFDHLV 298
>At4g25770 unknown protein
Length = 418
Score = 254 bits (650), Expect = 3e-68
Identities = 130/242 (53%), Positives = 174/242 (71%), Gaps = 14/242 (5%)
Query: 43 DGFDIEVVDASGHRSNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSST 102
D FD +V++++ P HL++MVNG++GSA +WKYAA+QF+K++P V+VH SE NS+T
Sbjct: 78 DFFDADVMESA---EKPDHLVVMVNGIVGSAADWKYAAEQFVKKFPDKVLVHRSESNSAT 134
Query: 103 LTFDGVDVTGNRLAEEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQ 162
LTFDGVD G RLA EV+ V+K ++KISF+AHSLGGL+ARYAI KLYE+
Sbjct: 135 LTFDGVDKMGERLANEVLGVVKHRSGLKKISFVAHSLGGLVARYAIGKLYEQ-------P 187
Query: 163 GNVHCEGQISNQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTA 222
G V S ++ G+IAGLEP+NFIT ATPHLG RGH+Q P+LCG LE+TA
Sbjct: 188 GEVDSLDSPSKEK---SARGGEIAGLEPMNFITFATPHLGSRGHRQFPILCGLPFLERTA 244
Query: 223 SRLSRF-LGKTGKHLFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDR 281
S+ + G+TGKHLFL D + PLL++M DS+D+KF+SAL +FKRRVAYAN+ +D
Sbjct: 245 SQTAHLAAGRTGKHLFLVDNDDGNAPLLIRMATDSDDLKFISALNAFKRRVAYANVNFDS 304
Query: 282 IL 283
++
Sbjct: 305 MV 306
>At5g51180 unknown protein
Length = 357
Score = 232 bits (591), Expect = 2e-61
Identities = 122/228 (53%), Positives = 161/228 (70%), Gaps = 10/228 (4%)
Query: 57 SNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRLA 116
S+ HL++MV+G++GS +WK+ A+QF+K+ P V VHCSE N S LT DGVDV G RLA
Sbjct: 31 SSADHLVVMVHGILGSTDDWKFGAEQFVKKMPDKVFVHCSEKNVSALTLDGVDVMGERLA 90
Query: 117 EEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQISNQEC 176
EV+ +I+R P++ KISF+AHSLGGL ARYAI KLY K +Q +V S+QE
Sbjct: 91 AEVLDIIQRKPNICKISFVAHSLGGLAARYAIGKLY-----KPANQEDVKDSVADSSQET 145
Query: 177 HVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKTASRLSRFLGK-TGKH 235
+G I GLE +NFIT ATPHLG G+KQVP L GF S+EK A + ++ K TG+H
Sbjct: 146 P----KGTICGLEAMNFITVATPHLGSMGNKQVPFLFGFSSIEKVAGLIIHWIFKRTGRH 201
Query: 236 LFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDRIL 283
LFL D + KPPLL +MV D++D F+SALR+FKRRVAY+N+ +D ++
Sbjct: 202 LFLKDEEEGKPPLLRRMVEDTDDCHFISALRAFKRRVAYSNVGHDHVV 249
>At1g29120 hypothetical protein
Length = 145
Score = 112 bits (280), Expect = 2e-25
Identities = 63/123 (51%), Positives = 81/123 (65%), Gaps = 9/123 (7%)
Query: 141 GLIARYAIAKLYERDISKELSQGNVHCEGQISNQECHVRKYEGKIAGLEPINFITSATPH 200
G++AR+A+A LY S ++Q + Q N G+IAGLEPINFIT ATPH
Sbjct: 31 GILARHAVAVLY----SAAMAQVSDVAVSQSGNSNL----LRGRIAGLEPINFITLATPH 82
Query: 201 LGCRGHKQVPLLCGFHSLEKTASRLSR-FLGKTGKHLFLTDGKNEKPPLLLQMVRDSEDI 259
LG RG KQ+P L G LEK A+ ++ F+G+TG LFLTDGK +KPPLLL+M D ED+
Sbjct: 83 LGVRGRKQLPFLLGVPILEKLAAPIAPFFVGRTGSQLFLTDGKADKPPLLLRMASDGEDL 142
Query: 260 KFM 262
KF+
Sbjct: 143 KFL 145
>At1g58350 putative protein
Length = 794
Score = 40.0 bits (92), Expect = 0.001
Identities = 31/97 (31%), Positives = 48/97 (48%), Gaps = 11/97 (11%)
Query: 62 LIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRLAEEVIS 121
+++ V+G G + + Q+L P + SE N D ++ G RLA+EV+S
Sbjct: 514 IVVFVHGFQGHHLDLRLVRNQWLLIDP-KIEFLMSEANEEKTHGDFREM-GQRLAQEVVS 571
Query: 122 VIKR---------HPSVRKISFIAHSLGGLIARYAIA 149
+KR H K+SF+ HS+G +I R AIA
Sbjct: 572 FLKRKKDRYARQGHLKSIKLSFVGHSIGNVIIRTAIA 608
>At5g17780 putative protein
Length = 419
Score = 30.4 bits (67), Expect = 1.1
Identities = 23/93 (24%), Positives = 43/93 (45%), Gaps = 2/93 (2%)
Query: 59 PTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVI-VHCSECNSSTLTFDGVDVTGNRLAE 117
P+ +I ++G +GS+H W + +++ Y ++ + S D + + +
Sbjct: 148 PSENVIFIHGFMGSSHFWTETVFEHIQKDDYRLLAIDLLGFGESPKPRDSLYTLKDHVDT 207
Query: 118 EVISVIKRHPSVRKISFIAHSLGGLIARYAIAK 150
SVIK + + +AHS+G LIA AK
Sbjct: 208 IERSVIKPY-QLDSFHVVAHSMGCLIALALAAK 239
>At4g09900 putative host response protein
Length = 256
Score = 29.6 bits (65), Expect = 1.9
Identities = 22/83 (26%), Positives = 37/83 (44%), Gaps = 12/83 (14%)
Query: 89 YDVIVHCSECNSSTLTFD----GVDVTG-------NRLAEEVISVIKRHPSVRKISFIAH 137
Y I E S +T D G ++T ++ +I +I+ P+ K+ + H
Sbjct: 20 YKTIASLEESGLSPVTVDLAGSGFNMTDANSVSTLEEYSKPLIELIQNLPAEEKVILVGH 79
Query: 138 SLGGLIARYAIAKLYERDISKEL 160
S GG YA+ + E+ ISK +
Sbjct: 80 STGGACVSYALERFPEK-ISKAI 101
>At2g25790 putative receptor-like protein kinase
Length = 960
Score = 28.9 bits (63), Expect = 3.3
Identities = 24/86 (27%), Positives = 39/86 (44%), Gaps = 14/86 (16%)
Query: 127 PSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCE-GQISNQEC--------- 176
PS+R ++ ++ G I R + LY D+S + G ++ + G SN
Sbjct: 123 PSLRYLNLSNNNFSGSIPRGFLPNLYTLDLSNNMFTGEIYNDIGVFSNLRVLDLGGNVLT 182
Query: 177 -HVRKYEGKIAGLEPINFITSATPHL 201
HV Y G ++ LE F+T A+ L
Sbjct: 183 GHVPGYLGNLSRLE---FLTLASNQL 205
>At2g23590 putative acetone-cyanohydrin lyase
Length = 272
Score = 28.5 bits (62), Expect = 4.3
Identities = 22/92 (23%), Positives = 42/92 (44%), Gaps = 3/92 (3%)
Query: 64 IMVNGLIGSAHNWKYAAKQFLKRYPYDVI-VHCSECNSSTLTFDGVDVTGNRLAEEVISV 122
++V+G A W Y K L+ + V + + C T + + T + +E +I +
Sbjct: 29 VLVHGSCLGAWCW-YKVKPLLEASGHRVTALDLAACGIDTRSITDIS-TCEQYSEPLIQL 86
Query: 123 IKRHPSVRKISFIAHSLGGLIARYAIAKLYER 154
+ P+ K+ + HS GGL A+ K ++
Sbjct: 87 MTSLPNDEKVVLVGHSYGGLTLAIAMDKFPDK 118
>At4g37150 hydroxynitrile lyase like protein
Length = 256
Score = 27.7 bits (60), Expect = 7.4
Identities = 29/101 (28%), Positives = 46/101 (44%), Gaps = 16/101 (15%)
Query: 64 IMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDV-------TGNRLA 116
++V+G A W Y K L+ + V V LT GV++ T A
Sbjct: 5 VLVHGGCHGAWCW-YKVKPMLEHSGHRVTVF-------DLTAHGVNMSRVEDIQTLEDFA 56
Query: 117 EEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDIS 157
+ ++ V++ S K+ +AHSLGG+ A A A ++ IS
Sbjct: 57 KPLLEVLESFGSDDKVVLVAHSLGGIPAALA-ADMFPSKIS 96
>At3g29770 alpha/beta hydrolase, putative
Length = 390
Score = 27.7 bits (60), Expect = 7.4
Identities = 21/100 (21%), Positives = 48/100 (48%), Gaps = 4/100 (4%)
Query: 60 THLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVI-VHCSECNSSTLTFDGVDVTGNRLAEE 118
T+ ++V+G A W Y L+ + V + + C +++ +G+ + ++ +
Sbjct: 136 TNHFVLVHGGSFGAWCW-YKTIALLEEDGFKVTAIDLAGCGINSININGI-ASLSQYVKP 193
Query: 119 VISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISK 158
+ ++++ P K+ + H GG YA+ +L+ ISK
Sbjct: 194 LTDILEKLPIGEKVILVGHDFGGACISYAM-ELFPSKISK 232
>At2g23620 putative acetone-cyanohydrin lyase
Length = 263
Score = 27.7 bits (60), Expect = 7.4
Identities = 22/92 (23%), Positives = 39/92 (41%), Gaps = 2/92 (2%)
Query: 64 IMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDV-TGNRLAEEVISV 122
++V+G A W Y K L+ + V + T D+ T + +E + +
Sbjct: 11 VLVHGSCHGAWCW-YKVKPLLEAVGHRVTAVDLAASGIDTTRSITDIPTCEQYSEPLTKL 69
Query: 123 IKRHPSVRKISFIAHSLGGLIARYAIAKLYER 154
+ P+ K+ + HS GGL A+ K E+
Sbjct: 70 LTSLPNDEKVVLVGHSFGGLNLAIAMEKFPEK 101
>At2g23610 putative acetone-cyanohydrin lyase
Length = 263
Score = 27.7 bits (60), Expect = 7.4
Identities = 20/81 (24%), Positives = 37/81 (44%), Gaps = 2/81 (2%)
Query: 63 IIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDV-TGNRLAEEVIS 121
+++V+G A W Y K L+ + V + +T D+ T + +E ++
Sbjct: 10 VVLVHGACHGAWCW-YKVKPQLEASGHRVTAVDLAASGIDMTRSITDISTCEQYSEPLMQ 68
Query: 122 VIKRHPSVRKISFIAHSLGGL 142
++ P K+ + HSLGGL
Sbjct: 69 LMTSLPDDEKVVLVGHSLGGL 89
>At1g64870 hypothetical protein
Length = 304
Score = 27.7 bits (60), Expect = 7.4
Identities = 38/159 (23%), Positives = 64/159 (39%), Gaps = 28/159 (17%)
Query: 104 TFDGVDVTGNRLAEEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQG 163
TF ++V N +A + + P+ F H L G A + +A ER + S+G
Sbjct: 161 TFGSINVLPNIVARDNL------PNANSFLF-PHGLWGWNANH-VATDKERTVFLTFSRG 212
Query: 164 N-------VHCEGQISNQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPL----- 211
+H +I ++C Y + G S+ + C GH+Q PL
Sbjct: 213 FPVSHAEVIHLFTEIYGEDCVESVYMPEDGG-------NSSNDNTNCNGHQQQPLFAKMV 265
Query: 212 LCGFHSLEKTASRLSRFLGK-TGKHLFLTDGKNEKPPLL 249
L ++++ S + K GKH++ KN K L+
Sbjct: 266 LDSVVTVDRILSGQEKQKYKINGKHIWARKFKNNKDGLI 304
>At1g33990 unknown protein
Length = 348
Score = 27.7 bits (60), Expect = 7.4
Identities = 20/83 (24%), Positives = 37/83 (44%), Gaps = 12/83 (14%)
Query: 89 YDVIVHCSECNSSTLTFD----GVDVTGNRL-------AEEVISVIKRHPSVRKISFIAH 137
Y ++ E S +T D G ++T ++ +I +++ P K+ + H
Sbjct: 112 YKMVASLEESGLSPVTVDLTGCGFNMTDTNTVSTLEEYSKPLIDLLENLPEEEKVILVGH 171
Query: 138 SLGGLIARYAIAKLYERDISKEL 160
S GG YA+ + E+ ISK +
Sbjct: 172 STGGASISYALERFPEK-ISKAI 193
>At5g19420 putative protein
Length = 1121
Score = 27.3 bits (59), Expect = 9.6
Identities = 21/73 (28%), Positives = 31/73 (41%), Gaps = 1/73 (1%)
Query: 101 STLTFDGVDVTGNRLAEEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKEL 160
S+ F V G+R+A S I R PS + + +L GL + +R + L
Sbjct: 832 SSKKFFSASVPGSRIASRATSPISRRPSPPRSTTPTPTLSGLTTPKIVVDDTKRS-NDNL 890
Query: 161 SQGNVHCEGQISN 173
SQ V Q+ N
Sbjct: 891 SQEVVMLRSQVEN 903
>At5g14430 unknown protein
Length = 612
Score = 27.3 bits (59), Expect = 9.6
Identities = 12/31 (38%), Positives = 19/31 (60%)
Query: 46 DIEVVDASGHRSNPTHLIIMVNGLIGSAHNW 76
D+ V++ +S+P II GLIG+ H+W
Sbjct: 477 DVWVMNVMPVQSSPRMKIIYDRGLIGATHDW 507
>At3g10870 putative alpha-hydroxynitrile lyase
Length = 276
Score = 27.3 bits (59), Expect = 9.6
Identities = 22/94 (23%), Positives = 42/94 (44%), Gaps = 6/94 (6%)
Query: 59 PTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVD--VTGNRLA 116
P H ++ ++G+ + W Y K ++ + V C + SS + VD T ++
Sbjct: 18 PPHFVL-IHGMSLGSWCW-YKIKCLMEVSGFTVT--CIDLKSSGIDSSSVDSLTTFDQYN 73
Query: 117 EEVISVIKRHPSVRKISFIAHSLGGLIARYAIAK 150
+ +I + P ++ + HS GGL AI +
Sbjct: 74 QPLIDFLSSFPEQEQVILVGHSAGGLSLTSAIQR 107
>At2g23600 putative acetone-cyanohydrin lyase
Length = 263
Score = 27.3 bits (59), Expect = 9.6
Identities = 21/92 (22%), Positives = 40/92 (42%), Gaps = 2/92 (2%)
Query: 64 IMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDV-TGNRLAEEVISV 122
++V+G A W Y K L+ + V + T D+ T + +E ++ +
Sbjct: 11 VLVHGACHGAWCW-YKVKPLLEALGHRVTALDLAASGIDTTRSITDISTCEQYSEPLMQL 69
Query: 123 IKRHPSVRKISFIAHSLGGLIARYAIAKLYER 154
+ P+ K+ + HS GGL A+ K ++
Sbjct: 70 MTSLPNDEKVVLVGHSFGGLSLALAMDKFPDK 101
>At1g03290 unknown protein
Length = 571
Score = 27.3 bits (59), Expect = 9.6
Identities = 19/64 (29%), Positives = 26/64 (39%), Gaps = 5/64 (7%)
Query: 192 NFITSATPHLGCRGHKQVPLLCGFHSLEKTASRLSRFLGKTGKHLFLTDGKNEKPPLLLQ 251
NF S+ G ++V H LE ASR FLG +G + + L L
Sbjct: 55 NFTQSSYKSSGSISEREVE-----HGLEDVASRCRPFLGASGSKASTSSSSSSSETLPLV 109
Query: 252 MVRD 255
+ RD
Sbjct: 110 VTRD 113
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,380,278
Number of Sequences: 26719
Number of extensions: 266371
Number of successful extensions: 644
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 627
Number of HSP's gapped (non-prelim): 22
length of query: 285
length of database: 11,318,596
effective HSP length: 98
effective length of query: 187
effective length of database: 8,700,134
effective search space: 1626925058
effective search space used: 1626925058
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146940.1


