

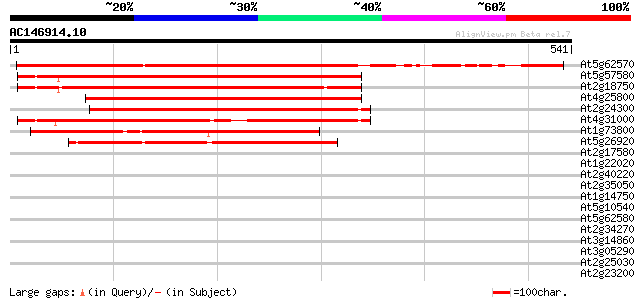
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146914.10 + phase: 0
(541 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g62570 unknown protein 451 e-127
At5g57580 calmodulin-binding protein 349 2e-96
At2g18750 putative calmodulin-binding protein 331 5e-91
At4g25800 putative calmodulin-binding protein 316 2e-86
At2g24300 putative calmodulin-binding protein 279 2e-75
At4g31000 putative calmodulin-binding protein 266 3e-71
At1g73800 putative calmodulin-binding protein 261 7e-70
At5g26920 calmodulin-binding - like protein 235 4e-62
At2g17580 poly(A) polymerase like protein 32 0.69
At1g22020 unknown protein 32 0.89
At2g40220 AP2 domain transcription factor (ABI4:abscisic acid-in... 31 1.5
At2g35050 putative protein kinase 30 3.4
At1g14750 cyclin, putative 30 3.4
At5g10540 oligopeptidase A - like protein 30 4.4
At5g62580 unknown protein 29 7.6
At2g34270 hypothetical protein 29 7.6
At3g14860 unknown protein 28 9.9
At3g05290 unknown protein 28 9.9
At2g25030 putative ATP-dependent CLPB protein 28 9.9
At2g23200 putative protein kinase 28 9.9
>At5g62570 unknown protein
Length = 494
Score = 451 bits (1160), Expect = e-127
Identities = 256/530 (48%), Positives = 345/530 (64%), Gaps = 56/530 (10%)
Query: 7 FSVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKKHLSSIKQTCGKEMNTSESRTL 66
FSVV+EVM+LQ+V++ +EP+LEPL+R+VV+EEVELAL KHL+ IK C KE + ESR L
Sbjct: 12 FSVVQEVMRLQTVKHFLEPVLEPLIRKVVKEEVELALGKHLAGIKWICEKETHPLESRNL 71
Query: 67 QLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEE 126
QL+F N++SLPVFT ARIEG++G +R+ L+D TG++ +GP SSAK+E+ V+EGDF
Sbjct: 72 QLKFLNNLSLPVFTSARIEGDEGQAIRVGLIDPSTGQIFSSGPASSAKLEVFVVEGDFNS 131
Query: 127 ESDIWMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFRLG 186
SD W ED +NNIVRER+GKKPLL G+V L DG+ ++ EIS+TDNSSWTRSR+FRLG
Sbjct: 132 VSD-WTDEDIRNNIVREREGKKPLLNGNVFAVLNDGIGVMDEISFTDNSSWTRSRKFRLG 190
Query: 187 VRVVDNFDGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSR 246
VR+VD FD ++IREA T+SF+VRDHRGELYKKHHPPSL DEVWRLEKIGKDGAFHRRL+
Sbjct: 191 VRIVDQFDYVKIREAITESFVVRDHRGELYKKHHPPSLFDEVWRLEKIGKDGAFHRRLNL 250
Query: 247 EKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFA-SHSQQ 305
I TVKDFLT +L+ +KLR +LGTGMS+KMWE+T++HAR+CVLD++ HV A ++
Sbjct: 251 SNINTVKDFLTHFHLNSSKLRQVLGTGMSSKMWEITLDHARSCVLDSSVHVYQAPGFQKK 310
Query: 306 PHVVFNAVGEVTGLLAESEYVAVDKLSETEKAFISDHPLVSLADAQISVISALNQC-DFA 364
VVFN V +V GLL + +Y+ +KLSE EK A A++ VI AL+ +
Sbjct: 311 TAVVFNVVAQVLGLLVDFQYIPAEKLSEIEK-----------AQAEVMVIDALSHLNEVI 359
Query: 365 SFEDEVSLMDGYSHLTNVHYSPSSPRTEGSSANKLLALQKTGGFNYTQESASSTDIMPSI 424
S++DEVS+M NV +P+S +GS A G +Y+ S +S D +
Sbjct: 360 SYDDEVSMM------RNVLNAPAS---QGSVA----------GIDYSGLSLTSLDGYGFV 400
Query: 425 YSVGGTSSLDDYGLPNFESLGLRYDQHLGFPVQVSNSLICDMDSIVHAFGDEDHLQFFDA 484
S+ T+ + + + + H G N C H G E+
Sbjct: 401 SSLHNTAECSG---KHSDDVDMEVTPH-GLYEDYDNLWNCS-----HILGLEEP------ 445
Query: 485 DLQSQCHIEADLHSAVDSFMPVSSTSMTKGKAQRRWRKVVNVLKWFMVKK 534
+++L SA+D FM + S+ +RW K+ +V +W V K
Sbjct: 446 --------QSELQSALDDFMSQKNASVGGKAHSKRWTKLFSVSRWLSVFK 487
>At5g57580 calmodulin-binding protein
Length = 647
Score = 349 bits (896), Expect = 2e-96
Identities = 173/338 (51%), Positives = 251/338 (74%), Gaps = 7/338 (2%)
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKK----HLSSIKQTCGKEMNTSES 63
SV+ E +K+ S++ L LEP++RRVV EE+E AL K L+ + K + +
Sbjct: 33 SVIVEALKVDSLQKLCSS-LEPILRRVVSEELERALAKLGPARLTGSSGSSPKRIEGPDG 91
Query: 64 RTLQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGD 123
R LQL F++ +SLP+FTG ++EGE G+ + + L+DA TG+ V GPE+SAK+ IVVLEGD
Sbjct: 92 RKLQLHFKSRLSLPLFTGGKVEGEQGAVIHVVLIDANTGRAVVYGPEASAKLHIVVLEGD 151
Query: 124 FEEESDI-WMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRR 182
F E D W E+F++++V+ER GK+PLLTG+V + LK+G+ +GE+ +TDNSSW RSR+
Sbjct: 152 FNTEDDEDWTQEEFESHVVKERSGKRPLLTGEVYVTLKEGVGTLGELVFTDNSSWIRSRK 211
Query: 183 FRLGVRVVDNF-DGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFH 241
FRLG+RVV DG+RIREAKT++F+V+DHRGELYKKH+PP+L+D+VWRL+KIGKDGAFH
Sbjct: 212 FRLGLRVVSGCCDGMRIREAKTEAFVVKDHRGELYKKHYPPALNDDVWRLDKIGKDGAFH 271
Query: 242 RRLSREKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFAS 301
++L+ E I TV+DFL ++ D KLRTILG+GMS KMW+ VEHA+TCV + ++ +A
Sbjct: 272 KKLTAEGINTVEDFLRVMVKDSPKLRTILGSGMSNKMWDALVEHAKTCVQSSKLYIYYAE 331
Query: 302 HSQQPHVVFNAVGEVTGLLAESEYVAVDKLSETEKAFI 339
S+ VVFN + E++GL++ +Y + D L++++K ++
Sbjct: 332 DSRNVGVVFNNIYELSGLISGDQYFSADSLTDSQKVYV 369
>At2g18750 putative calmodulin-binding protein
Length = 622
Score = 331 bits (849), Expect = 5e-91
Identities = 171/338 (50%), Positives = 242/338 (71%), Gaps = 11/338 (3%)
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKK----HLSSIKQTCGKEMNTSES 63
SV+ E +K+ S++ L LEP++RRVV EEVE AL K LS +++ K +
Sbjct: 39 SVIVEALKMDSLQRLCSS-LEPILRRVVSEEVERALAKLGPARLS--ERSSPKRIEGIGG 95
Query: 64 RTLQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGD 123
R LQLQF + +S+P+FTG +IEGE G+ + + L+D TG V+ GPE+SAK+++VVL+GD
Sbjct: 96 RNLQLQFRSRLSVPLFTGGKIEGEQGAAIHVVLLDMTTGHVLTVGPEASAKLDVVVLDGD 155
Query: 124 FE-EESDIWMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRR 182
F E+ D W E+F+ ++V+ER GK+PLLTGDV + LK+G+ +GE+ +TDNSSW R R+
Sbjct: 156 FNTEDDDGWSGEEFEGHLVKERQGKRPLLTGDVQVTLKEGVGTLGELIFTDNSSWIRCRK 215
Query: 183 FRLGVRVVDNF-DGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFH 241
FRLG+RV + +G+R+REAKT++F V+DHRGELYKKH+PP+L DEVWRLEKIGKDGAFH
Sbjct: 216 FRLGLRVSSGYCEGMRVREAKTEAFTVKDHRGELYKKHYPPALDDEVWRLEKIGKDGAFH 275
Query: 242 RRLSREKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFAS 301
++L++ I VK+FL L+ D KLRTILG+GMS +MWE EH++TCVL +V +
Sbjct: 276 KKLNKAGIYNVKEFLRLMVKDSQKLRTILGSGMSNRMWETLAEHSKTCVLSEMLYVYYPE 335
Query: 302 HSQQPHVVFNAVGEVTGLLAESEYVAVDKLSETEKAFI 339
S VVFN + E +GL++ +Y D LS+ +K ++
Sbjct: 336 DS--VGVVFNNIYEFSGLISGKQYYPADSLSDNQKGYV 371
>At4g25800 putative calmodulin-binding protein
Length = 536
Score = 316 bits (809), Expect = 2e-86
Identities = 151/268 (56%), Positives = 207/268 (76%), Gaps = 2/268 (0%)
Query: 74 ISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEEESDI-WM 132
+SLP+FTG R+EGE G+ + + L+DA TG+ V GPE+S K+E+VVL GDF E D W
Sbjct: 10 LSLPLFTGGRVEGEQGATIHVVLIDANTGRPVTVGPEASLKLEVVVLGGDFNNEDDEDWT 69
Query: 133 PEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFRLGVRVVDN 192
E+F++++V+ER+GK+PLLTGD+ + LK+G+ +GEI +TDNSSW RSR+FRLG+RV
Sbjct: 70 QEEFESHVVKEREGKRPLLTGDLFVVLKEGVGTLGEIVFTDNSSWIRSRKFRLGLRVPSG 129
Query: 193 F-DGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRT 251
+ DGIRIREAKT++F V+DHRGELYKKH+PP+L+DEVWRLEKIGKDGAFH+RL+ I T
Sbjct: 130 YCDGIRIREAKTEAFSVKDHRGELYKKHYPPALNDEVWRLEKIGKDGAFHKRLTAAGIVT 189
Query: 252 VKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHSQQPHVVFN 311
V+ FL L D KLR ILG+GMS KMW++ VEHA+TCVL ++ + S+ VVFN
Sbjct: 190 VEGFLRQLVRDSTKLRAILGSGMSNKMWDLLVEHAKTCVLSGKLYIYYTEDSRSVGVVFN 249
Query: 312 AVGEVTGLLAESEYVAVDKLSETEKAFI 339
+ E++GL+ E +Y++ D LSE++K ++
Sbjct: 250 NIYELSGLITEDQYLSADSLSESQKVYV 277
>At2g24300 putative calmodulin-binding protein
Length = 503
Score = 279 bits (714), Expect = 2e-75
Identities = 140/273 (51%), Positives = 195/273 (71%), Gaps = 4/273 (1%)
Query: 78 VFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEEESDI-WMPEDF 136
+FTG ++EGE GS + + L+DA TG VV TG ES++K+ +VVLEGDF +E D W E F
Sbjct: 5 LFTGGKVEGERGSAIHVVLIDANTGNVVQTGEESASKLNVVVLEGDFNDEDDEDWTREHF 64
Query: 137 KNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFRLGVRVVDNF-DG 195
++ V+ER+GK+P+LTGD + LK+G+ +GE+++TDNSSW RSR+FRLGV+ + D
Sbjct: 65 ESFEVKEREGKRPILTGDTQIVLKEGVGTLGELTFTDNSSWIRSRKFRLGVKPASGYGDS 124
Query: 196 IRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRTVKDF 255
IREAKT+ F V+DHRGELYKKH+PP++ DEVWRL++I KDG H++L + I TV+DF
Sbjct: 125 FCIREAKTEPFAVKDHRGELYKKHYPPAVHDEVWRLDRIAKDGVLHKKLLKANIVTVEDF 184
Query: 256 LTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHSQQPHVVFNAVGE 315
L LL DP KLR +LG+GMS +MWE TVEHA+TCVL +V + + VVFN + E
Sbjct: 185 LRLLVKDPQKLRNLLGSGMSNRMWENTVEHAKTCVLGGKLYVFYTDQTHATGVVFNHIYE 244
Query: 316 VTGLLAESEYVAVDKLSETEKAFISDHPLVSLA 348
GL+ ++++++ L+ +K IS LV LA
Sbjct: 245 FRGLITNGQFLSLESLNHDQK--ISADILVKLA 275
>At4g31000 putative calmodulin-binding protein
Length = 548
Score = 266 bits (679), Expect = 3e-71
Identities = 151/345 (43%), Positives = 217/345 (62%), Gaps = 24/345 (6%)
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELA---LKKHLSSIKQTCGKEMNTSESR 64
SV+ E +K+ S++ L LEPL RR+V EEVE A L+ S+ + T ++ + R
Sbjct: 33 SVIVEAVKVDSLQRLCSS-LEPLFRRIVSEEVERAISRLENSKSTSRSTEPNKIQGLDGR 91
Query: 65 TLQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDF 124
LQL+F + +FTG ++EGE GS + + L+DA TG V+ TG ES K+ IVVL+GDF
Sbjct: 92 NLQLRFRTRMPPHLFTGGKVEGEQGSAIHVVLIDANTGNVIQTGEESMTKLNIVVLDGDF 151
Query: 125 EEESDI-WMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRF 183
+E D W E F++ V+ER+GK+P+LTGD + +K+G+ +G++++TDNSSW RSR+F
Sbjct: 152 NDEDDKDWTREHFESFEVKEREGKRPILTGDRHVIIKEGVGTLGKLTFTDNSSWIRSRKF 211
Query: 184 RLGVRVVDNFDGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRR 243
RLGV+ F IREAKT+ F V+DHRGE L+KI KDGA H++
Sbjct: 212 RLGVKPATGF---HIREAKTEPFAVKDHRGE--------------HELDKIAKDGALHKK 254
Query: 244 LSREKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHS 303
L + I TV+DFL +L DP KLR++LG+GMS +MW+ TVEHA+TCVL + + +
Sbjct: 255 LLKSNIVTVEDFLQILMKDPQKLRSLLGSGMSNRMWDNTVEHAKTCVLGGKLYAYYTDQT 314
Query: 304 QQPHVVFNAVGEVTGLLAESEYVAVDKLSETEKAFISDHPLVSLA 348
Q VVFN + E GL+A +++ + L+ +K IS LV A
Sbjct: 315 HQTAVVFNHIYEFQGLIANGHFLSSESLNHDQK--ISADTLVKTA 357
>At1g73800 putative calmodulin-binding protein
Length = 451
Score = 261 bits (667), Expect = 7e-70
Identities = 133/281 (47%), Positives = 189/281 (66%), Gaps = 7/281 (2%)
Query: 21 NLMEPILEPLVRRVVREEVELALKKHLSSIKQTCGK-EMNTSESRTLQLQFENSISLPVF 79
N + +LEP++R+VVR+EVE + K + + + E + + TL+L F ++ P+F
Sbjct: 44 NTLRSVLEPVIRKVVRQEVEYGISKRFRLSRSSSFRIEAPEATTPTLKLIFRKNLMTPIF 103
Query: 80 TGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEEESDIWMPEDFKNN 139
TG++I D + L I LVD V P K++IV L GDF D W ++F++N
Sbjct: 104 TGSKISDVDNNPLEIILVDDSNKPVNLNRP---IKLDIVALHGDFPS-GDKWTSDEFESN 159
Query: 140 IVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFRLGVRVV--DNFDGIR 197
I++ERDGK+PLL G+V + +++G+ +GEI +TDNSSW RSR+FR+G +V + G+
Sbjct: 160 IIKERDGKRPLLAGEVSVTVRNGVATIGEIVFTDNSSWIRSRKFRIGAKVAKGSSGQGVV 219
Query: 198 IREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRTVKDFLT 257
+ EA T++ +VRDHRGELYKKHHPP L DEVWRLEKIGKDGAFH++LS I TV+DFL
Sbjct: 220 VCEAMTEAIVVRDHRGELYKKHHPPMLEDEVWRLEKIGKDGAFHKKLSSRHINTVQDFLK 279
Query: 258 LLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVS 298
L +D +LR ILG GMS + WEVT++HAR C+L ++S
Sbjct: 280 LSVVDVDELRQILGPGMSDRKWEVTLKHARECILGNKLYIS 320
>At5g26920 calmodulin-binding - like protein
Length = 492
Score = 235 bits (600), Expect = 4e-62
Identities = 130/261 (49%), Positives = 174/261 (65%), Gaps = 8/261 (3%)
Query: 57 EMNTSESRTLQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVE 116
E +S SR L+L F NS +FTG++IE EDGS L I LVDA T +V TGP SS++VE
Sbjct: 7 ETPSSRSR-LKLCFINSPPSSIFTGSKIEAEDGSPLVIELVDATTNTLVSTGPFSSSRVE 65
Query: 117 IVVLEGDFEEESDIWMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMV-GEISYTDNS 175
+V L DF EES W E F NI+ +R+GK+PLLTGD+ + LK+G+ ++ G+I+++DNS
Sbjct: 66 LVPLNADFTEES--WTVEGFNRNILTQREGKRPLLTGDLTVMLKNGVGVITGDIAFSDNS 123
Query: 176 SWTRSRRFRLGVRVVDNFDGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIG 235
SWTRSR+FRLG ++ G EA++++F RD RGE YKKHHPP SDEVWRLEKI
Sbjct: 124 SWTRSRKFRLGAKLT----GDGAVEARSEAFGCRDQRGESYKKHHPPCPSDEVWRLEKIA 179
Query: 236 KDGAFHRRLSREKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTR 295
KDG RL+ KI TVKDF L ++ +L I+G G+S K W V HA CVLD T
Sbjct: 180 KDGVSATRLAERKILTVKDFRRLYTVNRNELHNIIGAGVSKKTWNTIVSHAMDCVLDETE 239
Query: 296 HVSFASHSQQPHVVFNAVGEV 316
+ +++ ++FN+V E+
Sbjct: 240 CYIYNANTPGVTLLFNSVYEL 260
>At2g17580 poly(A) polymerase like protein
Length = 757
Score = 32.3 bits (72), Expect = 0.69
Identities = 21/64 (32%), Positives = 31/64 (47%), Gaps = 3/64 (4%)
Query: 279 WEVTVEHAR---TCVLDTTRHVSFASHSQQPHVVFNAVGEVTGLLAESEYVAVDKLSETE 335
W VE AR T V+ VS +S + + AV E T LL +++YV DK + E
Sbjct: 395 WSKAVEFAREHETSVIGYAPEVSKSSRKRSDEDLAEAVSEFTCLLKDTQYVLTDKEALRE 454
Query: 336 KAFI 339
++
Sbjct: 455 ALYL 458
>At1g22020 unknown protein
Length = 599
Score = 32.0 bits (71), Expect = 0.89
Identities = 21/95 (22%), Positives = 41/95 (43%), Gaps = 4/95 (4%)
Query: 378 HLTNVHYSPSSPRTEGSS----ANKLLALQKTGGFNYTQESASSTDIMPSIYSVGGTSSL 433
H+++ +Y+P + G+S + +TG +Y + + D P I GG+S
Sbjct: 264 HMSHGYYTPGGKKVSGASIFFESFPYKVDPRTGYIDYDKLEEKALDYRPKILICGGSSYP 323
Query: 434 DDYGLPNFESLGLRYDQHLGFPVQVSNSLICDMDS 468
D+ P F + + L F + + L+ +S
Sbjct: 324 RDWEFPRFRHIADKCGAVLMFDMAQISGLVAAKES 358
>At2g40220 AP2 domain transcription factor (ABI4:abscisic
acid-insensitive 4 )
Length = 328
Score = 31.2 bits (69), Expect = 1.5
Identities = 20/82 (24%), Positives = 37/82 (44%), Gaps = 5/82 (6%)
Query: 363 FASFEDEVSLMDG-----YSHLTNVHYSPSSPRTEGSSANKLLALQKTGGFNYTQESASS 417
FA+ ED D Y ++ +PSSP + SS++ + A + + ++
Sbjct: 84 FATAEDAARAYDRAAVYLYGSRAQLNLTPSSPSSVSSSSSSVSAASSPSTSSSSTQTLRP 143
Query: 418 TDIMPSIYSVGGTSSLDDYGLP 439
P+ +VGG ++ YG+P
Sbjct: 144 LLPRPAAATVGGGANFGPYGIP 165
>At2g35050 putative protein kinase
Length = 1257
Score = 30.0 bits (66), Expect = 3.4
Identities = 43/208 (20%), Positives = 81/208 (38%), Gaps = 23/208 (11%)
Query: 320 LAESEYVAVDKLSETEKAFISDHPLVSLADAQISVISALNQCDFASFEDEVSLMDGYSHL 379
L +SE + LS E +H + + +S +S+ + + ++E + S+
Sbjct: 552 LRKSEDHVENNLSAKEPKMRKEHSTTRVNEYSVSSVSSDSMVPDQALKEEAPISMKISNS 611
Query: 380 T----NVHYSPSSPRTEGSSANKLLALQKTGGFNYTQESASSTDIMPSIYSVGGTSSLDD 435
T ++ Y S RT + +KTG F+ T E + + + G S+
Sbjct: 612 TPDPKSLVYPEKSLRT---------SQEKTGAFDTTNEGMKKNQ--DNQFCLLGGFSVSG 660
Query: 436 YGLPNFESLGLRYDQHLGFPVQVSNSLICDMDSIVHAFGDEDHLQFFDADLQSQCHIEAD 495
+G N S + F V+ + + V + + L D L SQ +
Sbjct: 661 HGTSNNSSSNVS-----NFDQPVTQQRVFHSERTVRDPTETNRLSKSDDSLASQFVMA-- 713
Query: 496 LHSAVDSFMPVSSTSMTKGKAQRRWRKV 523
+ D+F+P+S +S T +A + V
Sbjct: 714 -QTTSDAFLPISESSETSHEANMESQNV 740
>At1g14750 cyclin, putative
Length = 614
Score = 30.0 bits (66), Expect = 3.4
Identities = 21/96 (21%), Positives = 41/96 (41%), Gaps = 1/96 (1%)
Query: 339 ISDHPLVSLADAQISVISALNQCDFASFEDEVSLMDGYSHLTNVHYSPSSPRTEGSSANK 398
I+D+ +S + + ++ ++ +FE E D S ++ V Y G + N+
Sbjct: 152 INDNDEISFSRSDVTFAGHVSNSRSLNFESENKESDVVSVISGVEYCSKFGSVTGGADNE 211
Query: 399 LLALQKTGGFNYTQES-ASSTDIMPSIYSVGGTSSL 433
+ + K F S S+ ++ P + VG S L
Sbjct: 212 EIEISKPSSFVEADSSLGSAKELKPELEIVGCVSDL 247
>At5g10540 oligopeptidase A - like protein
Length = 701
Score = 29.6 bits (65), Expect = 4.4
Identities = 31/124 (25%), Positives = 57/124 (45%), Gaps = 20/124 (16%)
Query: 15 KLQSVRNLMEP----ILEPLVRRVVREEVELALKKHLSSIKQTCGKEMNTSESRTLQLQF 70
+L+ + +EP ++EPL + + R V + HL ++K T E + +++F
Sbjct: 43 ELEQLEKAVEPSWPKLVEPLEKIIDRLSVVWGMINHLKAVKDTPELRAAIEEVQPEKVKF 102
Query: 71 ENSI--SLPVFTGARI--EGEDGSNL---RIRLVDALTGKVVCTGPESSAKVEIVVLEGD 123
+ + S P++ + E D ++L R RLV+A + V +G + LE D
Sbjct: 103 QLRLGQSKPIYNAFKAIRESPDWNSLSEARQRLVEAQIKEAVLSG---------IALEDD 153
Query: 124 FEEE 127
EE
Sbjct: 154 KREE 157
>At5g62580 unknown protein
Length = 615
Score = 28.9 bits (63), Expect = 7.6
Identities = 21/84 (25%), Positives = 38/84 (45%), Gaps = 9/84 (10%)
Query: 13 VMKLQSVRNLMEPILEPLVRRVVREEVELALKKHLSSIKQTCGKEMNTSESRTLQLQFEN 72
V+ + N + P L ++ R+ R L+ SSI+ TC ++ SRT + F +
Sbjct: 74 VLSVSLPLNSLSPFLSKILTRITRR-----LRDPDSSIRSTCVAAVSAISSRTTKPPFYS 128
Query: 73 SISLP----VFTGARIEGEDGSNL 92
+ P +FT + + G+ L
Sbjct: 129 AFMKPLADTLFTEQEVNAQIGAAL 152
>At2g34270 hypothetical protein
Length = 374
Score = 28.9 bits (63), Expect = 7.6
Identities = 33/158 (20%), Positives = 66/158 (40%), Gaps = 24/158 (15%)
Query: 10 VREVMKLQSVRNLMEPI------LEPLVRRVVREEVELALKKHLSSIKQ---TCGKEMNT 60
V ++MKL + NL+EP+ L LVR + + S +++ T
Sbjct: 75 VIKLMKLMKLMNLLEPLGNRVSRLMGLVRNAEEHRERIVKRNQESDVREIFKIAASRTKT 134
Query: 61 SESRTLQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVL 120
+ LQ + +S FT A+ ++ +L IRL AL + ++ + ++++
Sbjct: 135 RFHKALQKESSSSSMKWGFTEAK---DEVLSLGIRLTHALIKLTLLRVKKNDREKQLLLR 191
Query: 121 EGDFEEESD------------IWMPEDFKNNIVRERDG 146
+ F+E + ++ E+F ++V E G
Sbjct: 192 DWIFDESYEKQSALMYLLDYLLYKAEEFAASVVVEESG 229
>At3g14860 unknown protein
Length = 493
Score = 28.5 bits (62), Expect = 9.9
Identities = 19/48 (39%), Positives = 25/48 (51%), Gaps = 5/48 (10%)
Query: 42 ALKKHLSSI-KQTCGKEMNTSESRTLQLQFENSISLPVFTGARIEGED 88
+L KH+SS+ K T G S+S T LQFEN + +EG D
Sbjct: 44 SLIKHMSSVLKWTTGSSSKLSQSDTNVLQFENGYLVETV----VEGND 87
>At3g05290 unknown protein
Length = 322
Score = 28.5 bits (62), Expect = 9.9
Identities = 17/65 (26%), Positives = 32/65 (49%), Gaps = 1/65 (1%)
Query: 338 FISDHPLVSLADAQI-SVISALNQCDFASFEDEVSLMDGYSHLTNVHYSPSSPRTEGSSA 396
++SD +++ Q+ S+ L +F SF + YS+ VH + ++ G+ A
Sbjct: 49 YLSDVMWEAISKGQVFSLYQGLGTKNFQSFISQFIYFYSYSYFKRVHSERTGSKSIGTKA 108
Query: 397 NKLLA 401
N L+A
Sbjct: 109 NLLIA 113
>At2g25030 putative ATP-dependent CLPB protein
Length = 265
Score = 28.5 bits (62), Expect = 9.9
Identities = 40/143 (27%), Positives = 69/143 (47%), Gaps = 28/143 (19%)
Query: 16 LQSVRNLMEPILEPLVRRVVREEVELALK----KHLSSIKQ-TCGKEMNTSE-SRTLQLQ 69
L+++RN E I E + ++ VELA K K ++ I + + +N+SE S+ ++LQ
Sbjct: 116 LETIRN-NEDIKEAFYEMMKQQVVELARKTFRPKFMNRIDEYIVSQPLNSSEISKIVELQ 174
Query: 70 F--------ENSISLPVFTGARIE-----GEDGSNLRIRLVDALTGKVVCTGPESSAKVE 116
+N I+L +T ++ G D +N R V + K+V ++
Sbjct: 175 MRQVKKRLEQNKINLE-YTKEAVDLLAQLGFDPNN-GARPVKQMIEKLV------KKEIT 226
Query: 117 IVVLEGDFEEESDIWMPEDFKNN 139
+ VL+GDF E+ I + D NN
Sbjct: 227 LKVLKGDFAEDGTILIDADQPNN 249
>At2g23200 putative protein kinase
Length = 834
Score = 28.5 bits (62), Expect = 9.9
Identities = 37/154 (24%), Positives = 61/154 (39%), Gaps = 17/154 (11%)
Query: 293 TTRHVSFASHSQQPHVVFNAVGEVTGLLAESEYVAVDKLSETEKAFISDHPLVSLADAQI 352
T R A+ H+ + +T E++ + E E F+ DH ++L +A I
Sbjct: 127 TARFTVSATSGSNHHLKSFSPQNLTNTPRVEEFLLMMNSLEFEIRFVPDHSSLALINA-I 185
Query: 353 SVISALNQCDFASFEDEVSLMDGYSHLTNVH--YSPSSPRTEGSSANKLLA---LQKTGG 407
V SA + + S D+ N+H Y + + + N L L
Sbjct: 186 EVFSAPDDLEIPSASDK-----------NLHTIYRLNVGGEKITPDNDTLGRTWLPDDDD 234
Query: 408 FNYTQESASSTDIMPSIYSVGGTSSLDDYGLPNF 441
F Y ++SA + + + VGG SS D P+F
Sbjct: 235 FLYRKDSARNINSTQTPNYVGGLSSATDSTAPDF 268
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,021,245
Number of Sequences: 26719
Number of extensions: 521974
Number of successful extensions: 1325
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 1290
Number of HSP's gapped (non-prelim): 24
length of query: 541
length of database: 11,318,596
effective HSP length: 104
effective length of query: 437
effective length of database: 8,539,820
effective search space: 3731901340
effective search space used: 3731901340
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146914.10


