

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146910.5 - phase: 0 /pseudo
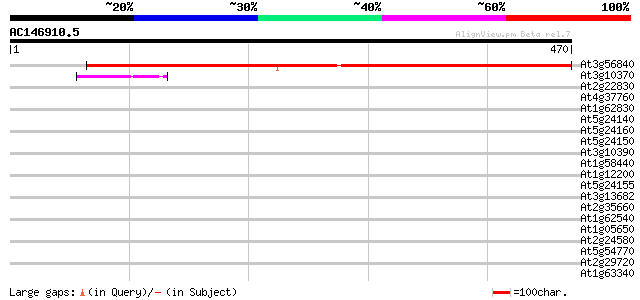
(470 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g56840 unknown protein 545 e-155
At3g10370 putative glycerol-3-phosphate dehydrogenase 42 7e-04
At2g22830 putative squalene epoxidase 37 0.031
At4g37760 unknown protein 35 0.068
At1g62830 flavin-containing amine oxidase 35 0.068
At5g24140 squalene monooxygenase 2 (squalene epoxidase 2) (SE 2)... 34 0.15
At5g24160 squalene monooxygenase 1,2 (squalene epoxidase 1,2) (s... 34 0.20
At5g24150 squalene monooxygenase 34 0.20
At3g10390 hypothetical protein 34 0.20
At1g58440 unknown protein 34 0.20
At1g12200 unknown protein 34 0.20
At5g24155 putative protein 33 0.34
At3g13682 hypothetical protein 33 0.44
At2g35660 CTF2A 33 0.44
At1g62540 unknown protein 33 0.44
At1g05650 putative polygalacturonase 33 0.44
At2g24580 putative sarcosine oxidase 32 0.58
At5g54770 thiazole biosynthetic enzyme precursor (ARA6) (sp|Q38814) 32 0.75
At2g29720 Hydroxylase/Oxygenase (CTF2B) 32 0.75
At1g63340 unknown protein 32 0.75
>At3g56840 unknown protein
Length = 483
Score = 545 bits (1403), Expect = e-155
Identities = 268/411 (65%), Positives = 339/411 (82%), Gaps = 8/411 (1%)
Query: 65 SVPRERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYP 124
++ +ERVD VVIGAGVVG+AVAR L+L+GREV+++++A SFGT TSSRNSEVVHAGIYYP
Sbjct: 74 TIAKERVDTVVIGAGVVGLAVARELSLRGREVLILDAASSFGTVTSSRNSEVVHAGIYYP 133
Query: 125 HHSLKAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGL 184
+SLKA FCV+GRE+LY+YC++++IPH++ GKLIVAT SSEIPKL ++++ G QN V GL
Sbjct: 134 PNSLKAKFCVRGRELLYKYCSEYEIPHKKIGKLIVATGSSEIPKLDLLMHLGTQNRVSGL 193
Query: 185 KMMDGVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLV-----LHIDRNINPMLVQGEA 239
+M++G +AM+MEP+L+CVKA+LSP SGI+D+HS MLSLV + R+ N + +QGEA
Sbjct: 194 RMLEGFEAMRMEPQLRCVKALLSPESGILDTHSFMLSLVEKSFDFMVYRDNNNLRLQGEA 253
Query: 240 ENHGATFTYNSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLS 299
+N+ ATF+YN+ V+ G +E ++ L+V++T+ + + + +L LIP LVVNSAGL
Sbjct: 254 QNNHATFSYNTVVLNGRVEEKKMHLYVADTRFSES---RCEAEAQLELIPNLVVNSAGLG 310
Query: 300 ALALAKRFTGLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNG 359
A ALAKR GL+++ +P ++YARGCYFTLS KA PF L+YPIPE+GGLGVHVT+DLNG
Sbjct: 311 AQALAKRLHGLDHRFVPSSHYARGCYFTLSGIKAPPFNKLVYPIPEEGGLGVHVTVDLNG 370
Query: 360 QVKFGPDVEWIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIR 419
VKFGPDVEWI+ DD SSF NKFDY V R+EKFYPEIRKYYP+LKDGSLEPGYSGIR
Sbjct: 371 LVKFGPDVEWIECTDDTSSFLNKFDYRVNPQRSEKFYPEIRKYYPDLKDGSLEPGYSGIR 430
Query: 420 PKLSGPCQSPVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIADFISTKF 470
PKLSGP QSP DFVIQGE+ HGVPGL+NLFGIESPGLTSSLAIA+ I+ KF
Sbjct: 431 PKLSGPKQSPADFVIQGEETHGVPGLVNLFGIESPGLTSSLAIAEHIANKF 481
>At3g10370 putative glycerol-3-phosphate dehydrogenase
Length = 629
Score = 42.0 bits (97), Expect = 7e-04
Identities = 24/76 (31%), Positives = 42/76 (54%), Gaps = 4/76 (5%)
Query: 57 SRETTTSYSVPRERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEV 116
++E+ + + +D +VIG G G VA +G V ++E F +GTSSR++++
Sbjct: 60 AQESALIAATASDPLDVLVIGGGATGSGVALDAVTRGLRVGLVERE-DFSSGTSSRSTKL 118
Query: 117 VHAGIYYPHHSLKAIF 132
+H G+ Y KA+F
Sbjct: 119 IHGGVRYLE---KAVF 131
>At2g22830 putative squalene epoxidase
Length = 585
Score = 36.6 bits (83), Expect = 0.031
Identities = 24/63 (38%), Positives = 34/63 (53%), Gaps = 5/63 (7%)
Query: 41 RSTTQNDTNTTTSYPVSRETTTSYSVPRE---RVDCVVIGAGVVGIAVARALALKGREVI 97
RS+ +N N VS+ T S ++ E D +++GAGV G A+A L +GR V
Sbjct: 92 RSSNKNKKNR--GLVVSQNDTVSKNLETEVDSGTDVIIVGAGVAGSALAHTLGKEGRRVH 149
Query: 98 VIE 100
VIE
Sbjct: 150 VIE 152
>At4g37760 unknown protein
Length = 525
Score = 35.4 bits (80), Expect = 0.068
Identities = 16/30 (53%), Positives = 21/30 (69%)
Query: 71 VDCVVIGAGVVGIAVARALALKGREVIVIE 100
VD +++GAGV G A+A L +GR V VIE
Sbjct: 55 VDIIIVGAGVAGAALAHTLGKEGRRVHVIE 84
>At1g62830 flavin-containing amine oxidase
Length = 844
Score = 35.4 bits (80), Expect = 0.068
Identities = 28/118 (23%), Positives = 49/118 (40%), Gaps = 18/118 (15%)
Query: 13 KSSSSSTRTRDHVHMKWNNPFNNWV------RNIRSTTQNDTNTTTSY------------ 54
K ++ R+H+ W + +NW+ +IR+ + +T ++
Sbjct: 190 KDQANYIVVRNHIIALWRSNVSNWLTRDHALESIRAEHKTLVDTAYNFLLEHGYINFGLA 249
Query: 55 PVSRETTTSYSVPRERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSR 112
PV +E E + VV+GAG+ G+ AR L G V+V+E G +R
Sbjct: 250 PVIKEAKLRSFDGVEPPNVVVVGAGLAGLVAARQLLSMGFRVLVLEGRDRPGGRVKTR 307
>At5g24140 squalene monooxygenase 2 (squalene epoxidase 2) (SE 2)
(sp|O65403)
Length = 516
Score = 34.3 bits (77), Expect = 0.15
Identities = 16/29 (55%), Positives = 21/29 (72%)
Query: 72 DCVVIGAGVVGIAVARALALKGREVIVIE 100
D +++GAGV G ++A ALA GR V VIE
Sbjct: 45 DVIIVGAGVAGASLAYALAKDGRRVHVIE 73
>At5g24160 squalene monooxygenase 1,2 (squalene epoxidase 1,2) (se
1,2) (sp|O65402)
Length = 517
Score = 33.9 bits (76), Expect = 0.20
Identities = 17/29 (58%), Positives = 21/29 (71%)
Query: 72 DCVVIGAGVVGIAVARALALKGREVIVIE 100
D +++GAGV G A+A ALA GR V VIE
Sbjct: 47 DVIIVGAGVGGSALAYALAKDGRRVHVIE 75
>At5g24150 squalene monooxygenase
Length = 516
Score = 33.9 bits (76), Expect = 0.20
Identities = 17/29 (58%), Positives = 21/29 (71%)
Query: 72 DCVVIGAGVVGIAVARALALKGREVIVIE 100
D +++GAGV G A+A ALA GR V VIE
Sbjct: 47 DVIIVGAGVGGSALAYALAKDGRRVHVIE 75
>At3g10390 hypothetical protein
Length = 789
Score = 33.9 bits (76), Expect = 0.20
Identities = 23/114 (20%), Positives = 47/114 (41%), Gaps = 15/114 (13%)
Query: 22 RDHVHMKWNNPFNNWVRN---IRSTTQNDTNTTTS------------YPVSRETTTSYSV 66
R+H+ KW ++WV + S ++ ++ S + +++ +
Sbjct: 121 RNHIISKWRENISSWVTKEMFLNSIPKHCSSLLDSAYNYLVTHGYINFGIAQAIKDKFPA 180
Query: 67 PRERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAG 120
+ +++GAG+ G+A AR L G +V V+E G ++ E G
Sbjct: 181 QSSKSSVIIVGAGLSGLAAARQLMRFGFKVTVLEGRKRPGGRVYTKKMEANRVG 234
>At1g58440 unknown protein
Length = 531
Score = 33.9 bits (76), Expect = 0.20
Identities = 23/69 (33%), Positives = 32/69 (46%), Gaps = 9/69 (13%)
Query: 32 PFNNWVRNIRSTTQNDTNTTTSYPVSRETTTSYSVPRERVDCVVIGAGVVGIAVARALAL 91
P N +R+ R T T+ S ++ +T D +V+GAGV G A+A L
Sbjct: 31 PKRNGLRHDRKTVSTVTSDVGSVNITGDTVA---------DVIVVGAGVAGSALAYTLGK 81
Query: 92 KGREVIVIE 100
R V VIE
Sbjct: 82 DKRRVHVIE 90
>At1g12200 unknown protein
Length = 465
Score = 33.9 bits (76), Expect = 0.20
Identities = 17/45 (37%), Positives = 22/45 (48%)
Query: 62 TSYSVPRERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFG 106
TS+ P VIGAG G+ AR L +G V+V+E G
Sbjct: 3 TSHPDPTTSRHVAVIGAGAAGLVAARELRREGHSVVVLERGSQIG 47
>At5g24155 putative protein
Length = 121
Score = 33.1 bits (74), Expect = 0.34
Identities = 15/29 (51%), Positives = 21/29 (71%)
Query: 72 DCVVIGAGVVGIAVARALALKGREVIVIE 100
D +++GAGV G A+A +LA GR V+ IE
Sbjct: 49 DVIIVGAGVGGSALAYSLAKDGRRVLAIE 77
>At3g13682 hypothetical protein
Length = 746
Score = 32.7 bits (73), Expect = 0.44
Identities = 25/94 (26%), Positives = 39/94 (40%), Gaps = 15/94 (15%)
Query: 22 RDHVHMKWNNPFNNWVRN--IRSTTQNDTNTTTSYPVSRETTTSYS-----------VPR 68
R+H+ +W W+ IR T +D S Y +P
Sbjct: 96 RNHIVARWRGNVGIWLLKDQIRETVSSDFEHLISAAYDFLLFNGYINFGVSPLFAPYIPE 155
Query: 69 ERVD--CVVIGAGVVGIAVARALALKGREVIVIE 100
E + +V+GAG+ G+A AR L G +V+V+E
Sbjct: 156 EGTEGSVIVVGAGLAGLAAARQLLSFGFKVLVLE 189
>At2g35660 CTF2A
Length = 439
Score = 32.7 bits (73), Expect = 0.44
Identities = 23/62 (37%), Positives = 34/62 (54%), Gaps = 1/62 (1%)
Query: 51 TTSYPVSRETTTSYSVPRERVDCVVI-GAGVVGIAVARALALKGREVIVIESAPSFGTGT 109
T S PV T + + ++ + VVI GAG+ G+A A +L G +V+E A S TG
Sbjct: 24 TRSKPVCLALTNAQTNGGDQEEKVVIVGAGIGGLATAVSLHRLGIRSVVLEQAESLRTGG 83
Query: 110 SS 111
+S
Sbjct: 84 TS 85
>At1g62540 unknown protein
Length = 457
Score = 32.7 bits (73), Expect = 0.44
Identities = 15/33 (45%), Positives = 20/33 (60%)
Query: 74 VVIGAGVVGIAVARALALKGREVIVIESAPSFG 106
VVIGAG G+ AR L+ +G V+V+E G
Sbjct: 14 VVIGAGAAGLVAARELSREGHTVVVLEREKEVG 46
>At1g05650 putative polygalacturonase
Length = 394
Score = 32.7 bits (73), Expect = 0.44
Identities = 23/75 (30%), Positives = 31/75 (40%), Gaps = 2/75 (2%)
Query: 26 HMKWNNPFNNWVRNIRSTTQNDTNTTTSYPVSRET--TTSYSVPRERVDCVVIGAGVVGI 83
HM N N VRNIR D+ T + V T T + S + DCV IG G
Sbjct: 170 HMTLNGCTNVAVRNIRLVAPGDSPNTDGFTVQFSTGVTLTGSTVQTGDDCVAIGQGTRNF 229
Query: 84 AVARALALKGREVIV 98
+++ G V +
Sbjct: 230 LISKLACGPGHGVSI 244
>At2g24580 putative sarcosine oxidase
Length = 416
Score = 32.3 bits (72), Expect = 0.58
Identities = 13/31 (41%), Positives = 23/31 (73%)
Query: 70 RVDCVVIGAGVVGIAVARALALKGREVIVIE 100
R D +V+GAGV+G + A LA +G++ +++E
Sbjct: 8 RFDVIVVGAGVMGSSAAYQLAKRGQKTLLLE 38
>At5g54770 thiazole biosynthetic enzyme precursor (ARA6) (sp|Q38814)
Length = 349
Score = 32.0 bits (71), Expect = 0.75
Identities = 27/80 (33%), Positives = 39/80 (48%), Gaps = 10/80 (12%)
Query: 39 NIRSTTQN-DTNTTTSYP-----VSRETTTSYS---VPRERVDCVVIGAGVVGIAVARAL 89
++R+TT D N T P VSRE T Y + D VV+GAG G++ A +
Sbjct: 43 SVRATTAGYDLNAFTFDPIKESIVSREMTRRYMTDMITYAETDVVVVGAGSAGLSAAYEI 102
Query: 90 ALKGR-EVIVIESAPSFGTG 108
+ +V +IE + S G G
Sbjct: 103 SKNPNVQVAIIEQSVSPGGG 122
>At2g29720 Hydroxylase/Oxygenase (CTF2B)
Length = 427
Score = 32.0 bits (71), Expect = 0.75
Identities = 16/38 (42%), Positives = 23/38 (60%)
Query: 74 VVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSS 111
V++G G+ G+A A AL G +V+E A S TG +S
Sbjct: 46 VIVGGGIGGLATAVALHRLGIRSVVLEQAESLRTGGAS 83
>At1g63340 unknown protein
Length = 398
Score = 32.0 bits (71), Expect = 0.75
Identities = 19/52 (36%), Positives = 23/52 (43%), Gaps = 7/52 (13%)
Query: 55 PVSRETTTSYSVPRERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFG 106
P TTS+ V VIGAG G+ AR L +G V+V E G
Sbjct: 3 PAVNPPTTSHHV-------AVIGAGAAGLVAARELRREGHSVVVFERGNQIG 47
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,694,822
Number of Sequences: 26719
Number of extensions: 469239
Number of successful extensions: 1298
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 41
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 1247
Number of HSP's gapped (non-prelim): 63
length of query: 470
length of database: 11,318,596
effective HSP length: 103
effective length of query: 367
effective length of database: 8,566,539
effective search space: 3143919813
effective search space used: 3143919813
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146910.5


